Renal Metabolome in Obese Mice Treated with Empagliflozin Suggests a Reduction in Cellular Respiration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Tissue Collection
2.3. Western Blotting
2.4. Glucose Measurement
2.5. Metabolite Analysis
2.6. Lipid Analysis
2.7. Targeted Metabolomics
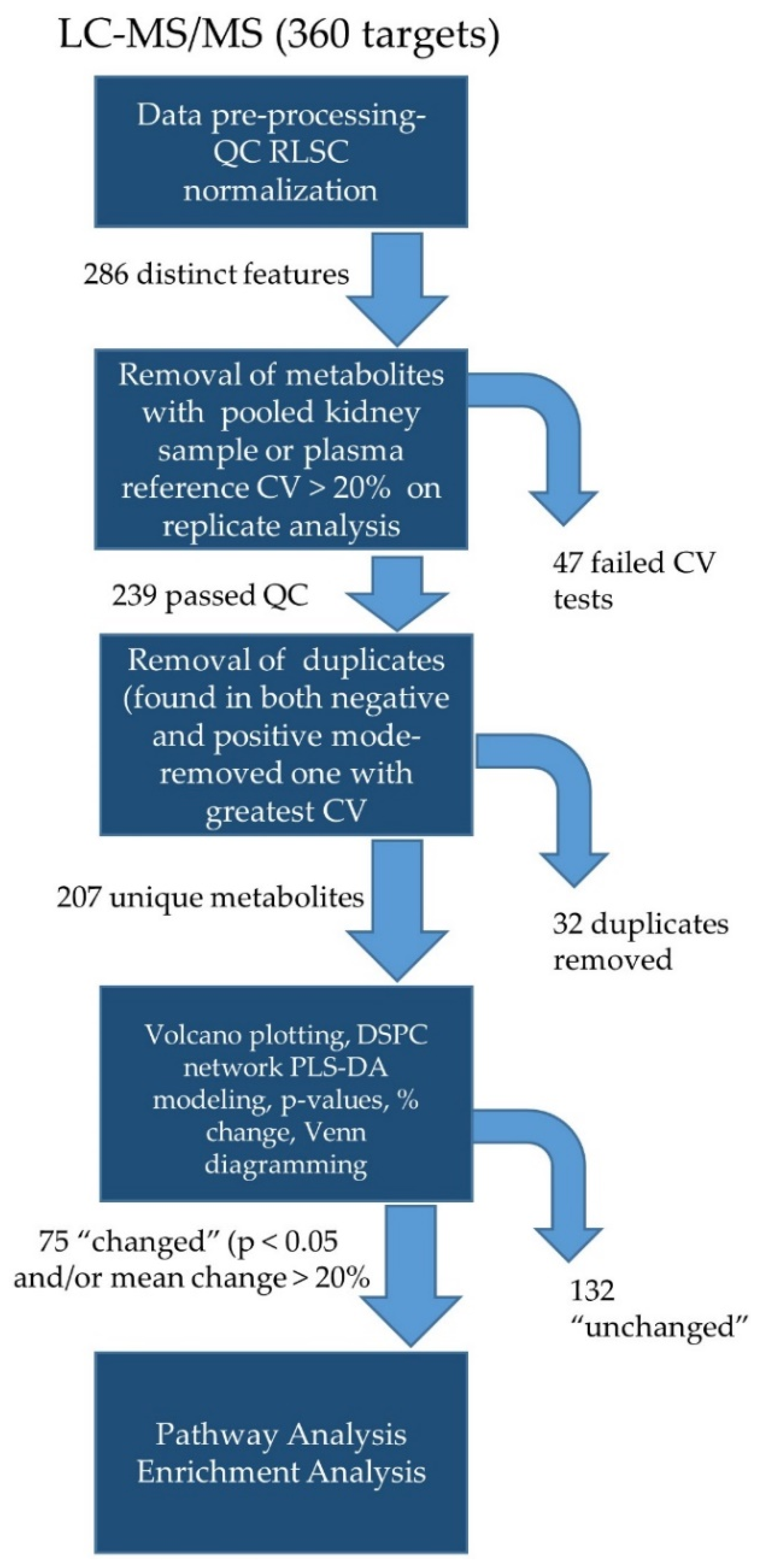
2.8. Targeted Lipidomics
2.9. Untargeted Analyses
3. Results
3.1. EMPA Did Not Affect Weights but Lowered Plasma Glucose
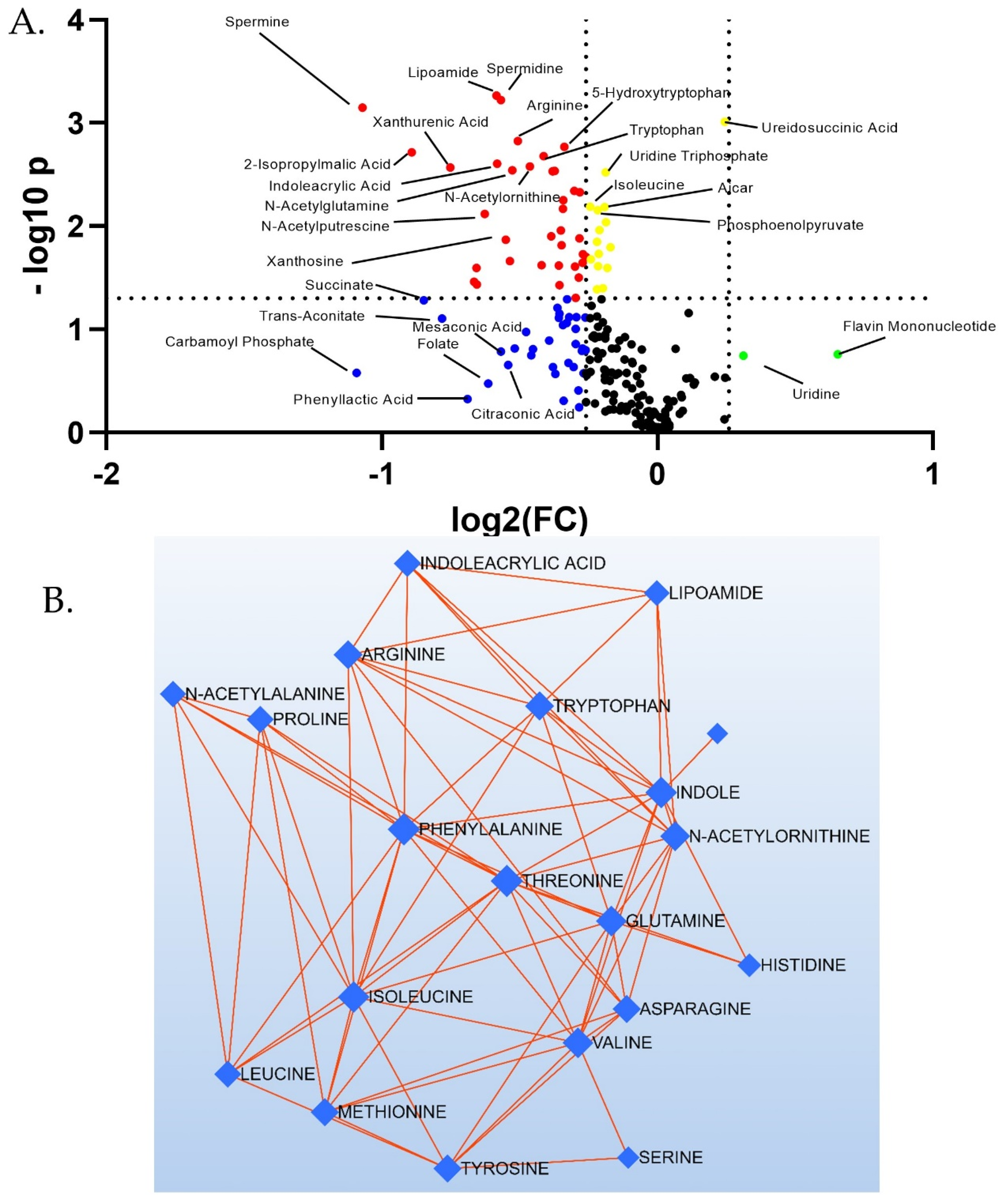
3.2. Targeted Metabolites Down-Regulated by EMPA in Kidney Cortex
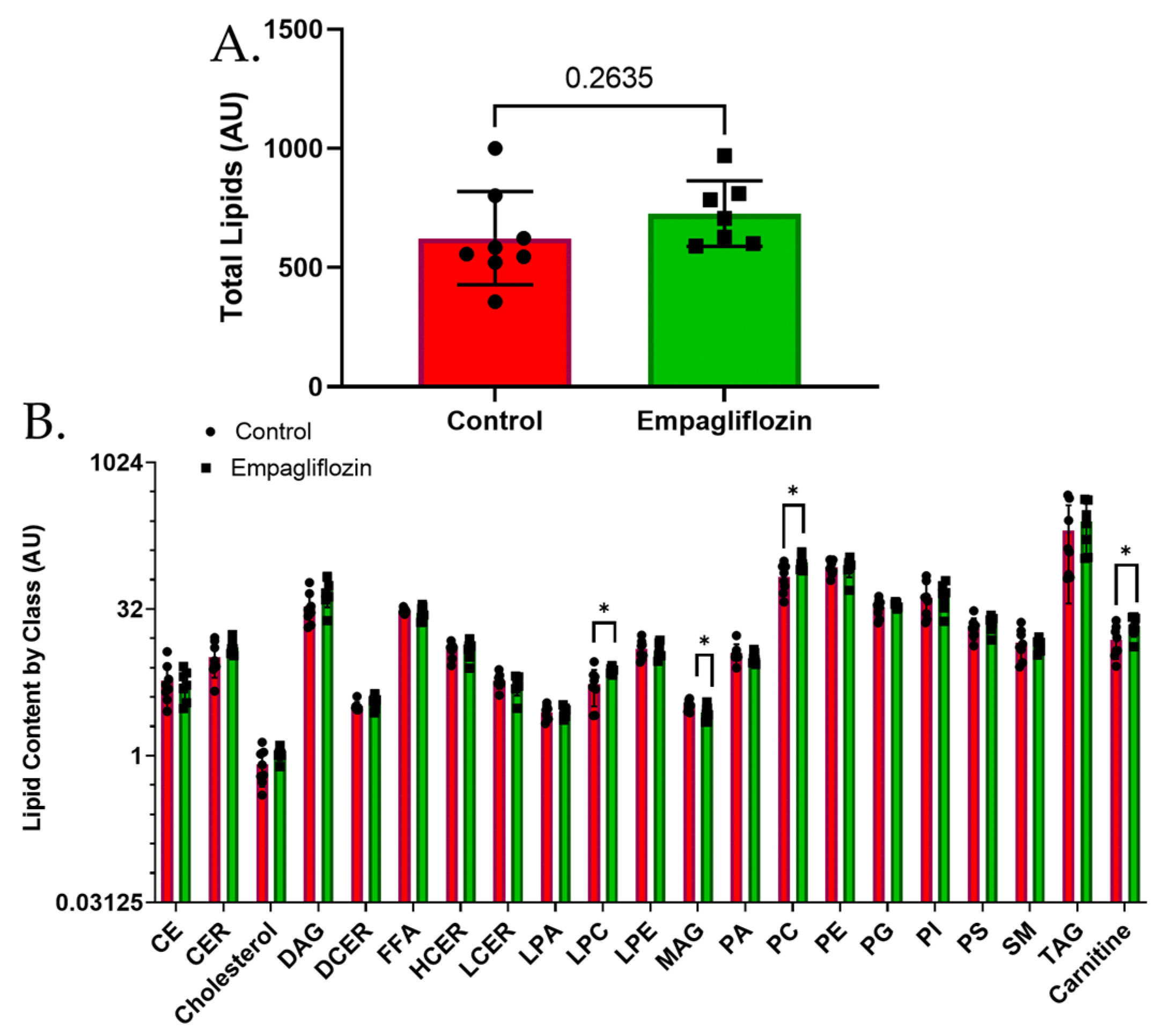
3.3. Targeted Lipidomics Reveals a Number of Membrane Lipid Species Up-Regulated by Empagliflozin
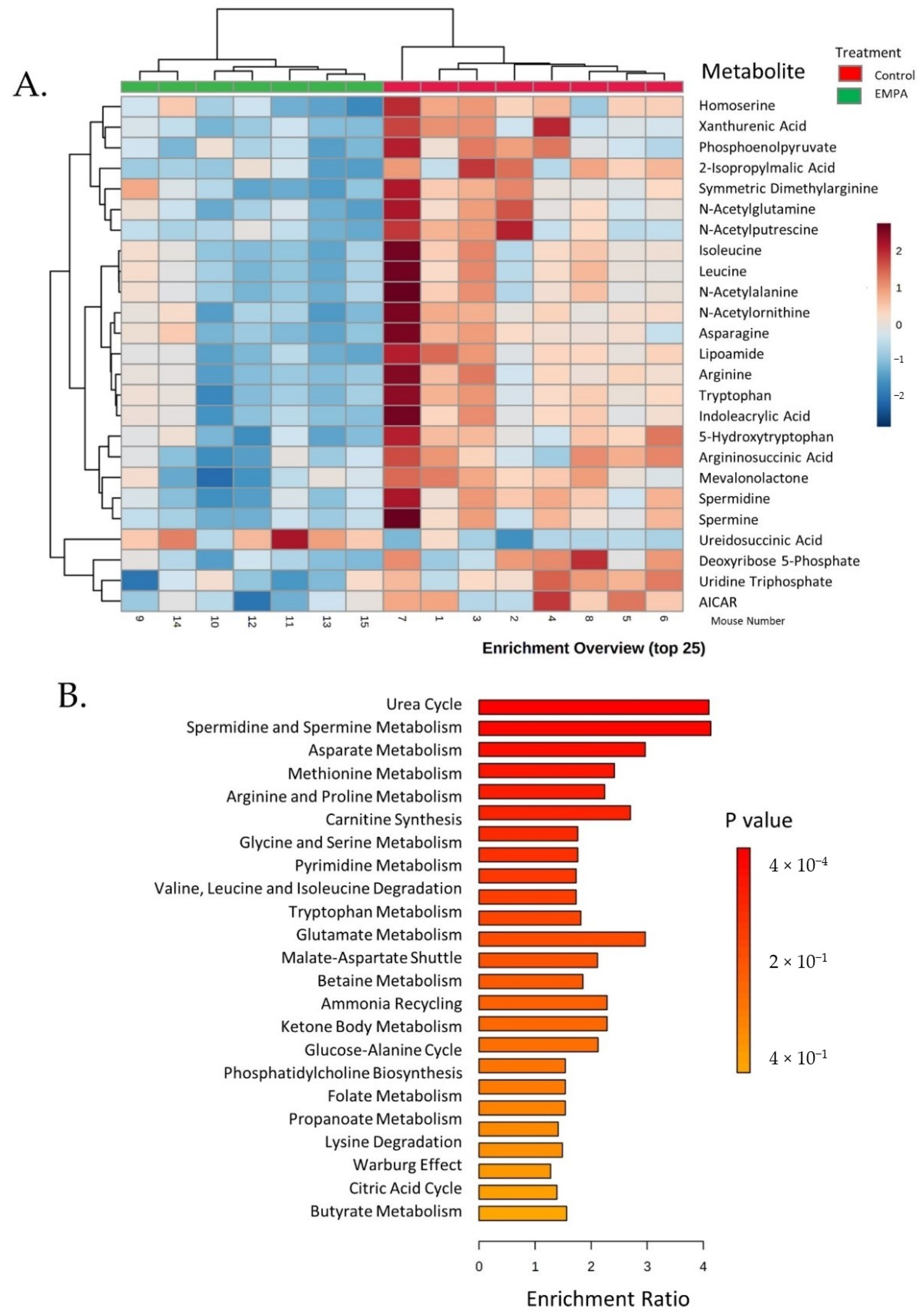
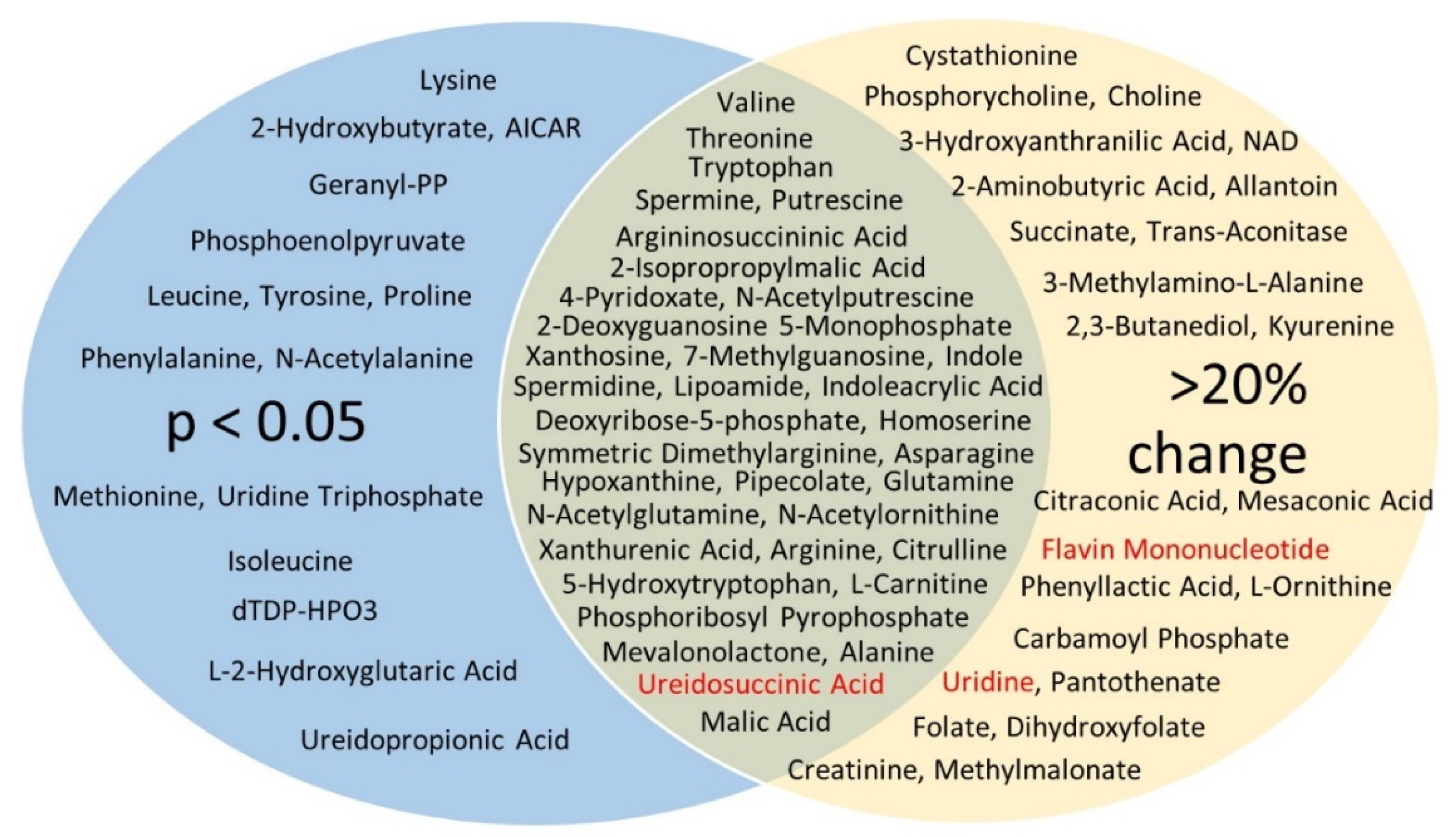
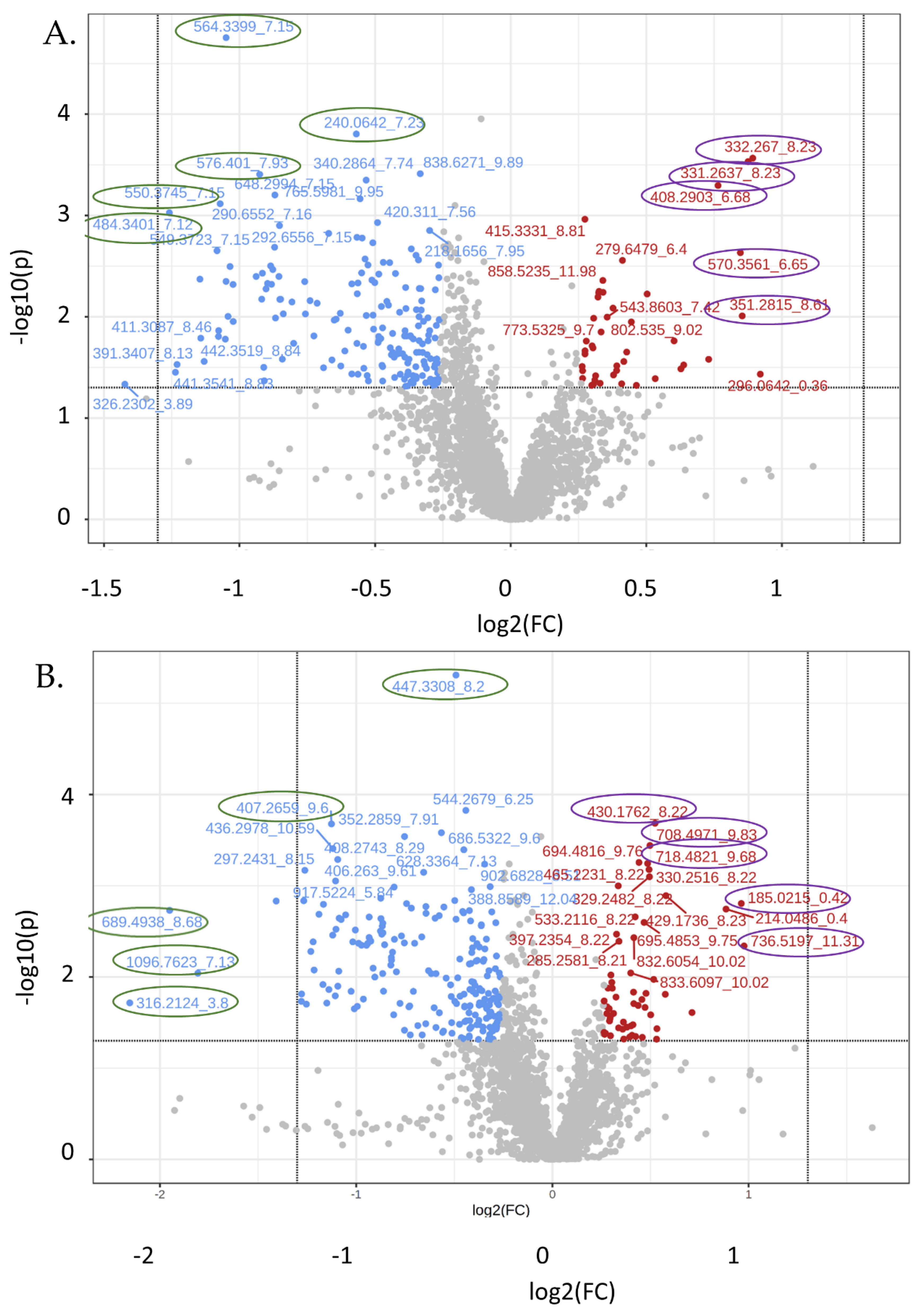
3.4. Untargeted Metabolite Analyses
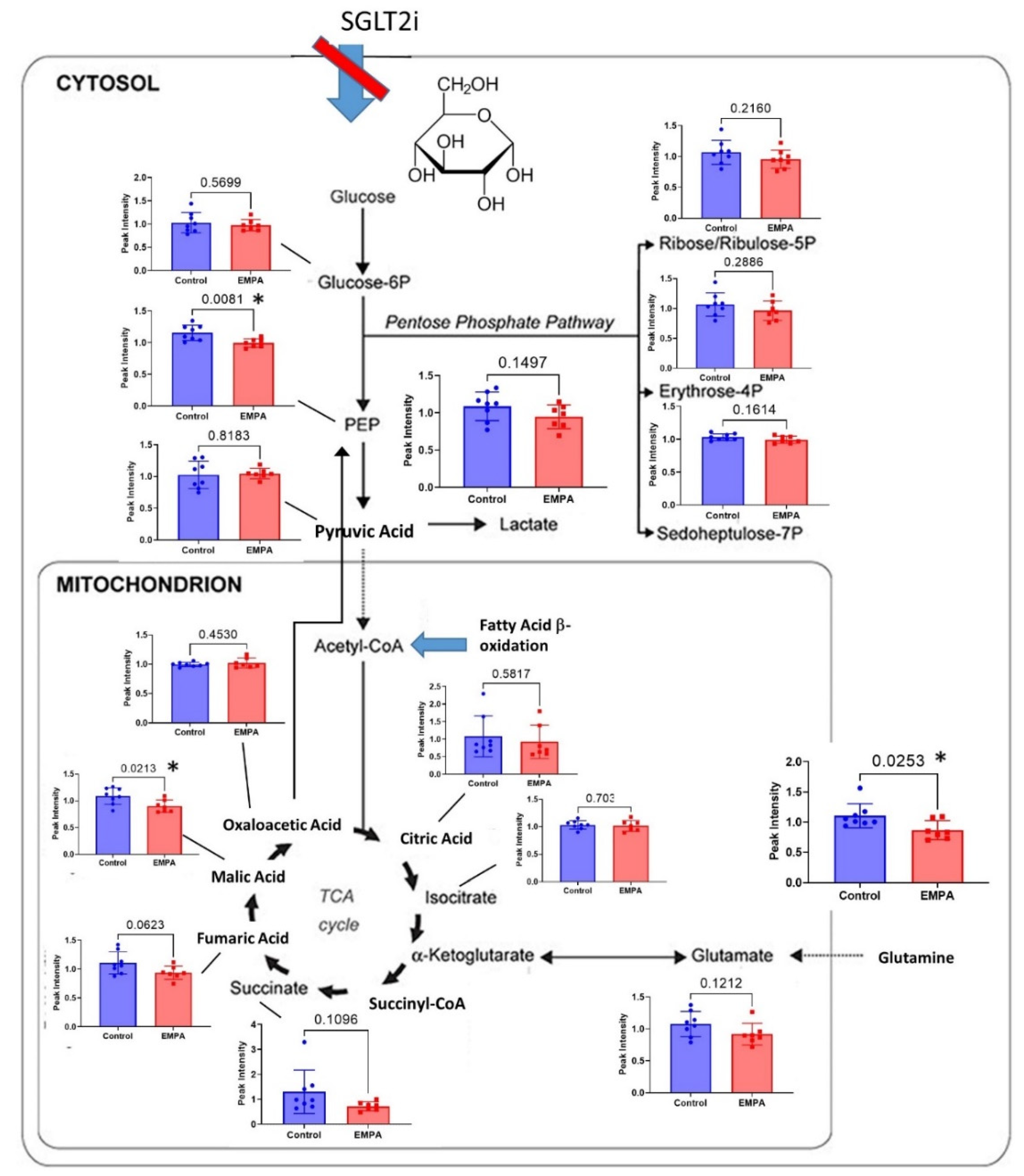
4. Discussion

5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rieg, T.; Vallon, V. Development of SGLT1 and SGLT2 inhibitors. Diabetologia 2018, 61, 2079–2086. [Google Scholar] [CrossRef]
- Mahaffey, K.W.; Jardine, M.J.; Bompoint, S.; Cannon, C.P.; Neal, B.; Heerspink, H.J.; Charytan, D.M.; Edwards, R.; Agarwal, R.; Bakris, G.; et al. Canagliflozin and Cardiovascular and Renal Outcomes in Type 2 Diabetes Mellitus and Chronic Kidney Disease in Primary and Secondary Cardiovascular Prevention Groups. Circulation 2019, 140, 739–750. [Google Scholar] [CrossRef]
- Perkovic, V.; Jardine, M.J.; Neal, B.; Bompoint, S.; Heerspink, H.J.L.; Charytan, D.M.; Edwards, R.; Agarwal, R.; Bakris, G.; Bull, S.; et al. Canagliflozin and Renal Outcomes in Type 2 Diabetes and Nephropathy. N. Engl. J. Med. 2019, 380, 2295–2306. [Google Scholar] [CrossRef]
- Heerspink, H.J.L.; Stefánsson, B.V.; Correa-Rotter, R.; Chertow, G.M.; Greene, T.; Hou, F.-F.; Mann, J.F.E.; McMurray, J.J.V.; Lindberg, M.; Rossing, P.; et al. Dapagliflozin in Patients with Chronic Kidney Disease. N. Engl. J. Med. 2020, 383, 1436–1446. [Google Scholar] [CrossRef]
- Muskiet, M.H.A.; van Raalte, D.H.; van Bommel, E.J.; Smits, M.M.; Tonneijck, L. Understanding EMPA-REG OUTCOME. Lancet Diabetes Endocrinol. 2015, 3, 928–929. [Google Scholar] [CrossRef]
- Zinman, B.; Wanner, C.; Lachin, J.M.; Fitchett, D.; Bluhmki, E.; Hantel, S.; Mattheus, M.; Devins, T.; Johansen, O.E.; Woerle, H.J.; et al. Empagliflozin, Cardiovascular Outcomes, and Mortality in Type 2 Diabetes. N. Engl. J. Med. 2015, 373, 2117–2128. [Google Scholar] [CrossRef]
- Oguz, F.; Demoulin, N.; Thissen, J.P.; Jadoul, M.; Morelle, J. Inhibition of sodium-glucose cotransporter 2 to slow the progression of chronic kidney disease. Acta Clin. Belg. 2021, 77, 805–814. [Google Scholar] [CrossRef]
- Ravindran, S.; Munusamy, S. Renoprotective mechanisms of sodium-glucose co-transporter 2 (SGLT2) inhibitors against the progression of diabetic kidney disease. J. Cell. Physiol. 2021, 237, 1182–1205. [Google Scholar] [CrossRef]
- Kim, J.H.; Stewart, T.P.; Soltani-Bejnood, M.; Wang, L.; Fortuna, J.M.; Mostafa, O.A.; Moustaid-Moussa, N.; Shoieb, A.M.; McEntee, M.F.; Wang, Y.; et al. Phenotypic characterization of polygenic type 2 diabetes in TALLYHO/JngJ mice. J. Endocrinol. 2006, 191, 437–446. [Google Scholar] [CrossRef]
- Geng, T.; Hu, W.; Broadwater, M.H.; Snider, J.M.; Bielawski, J.; Russo, S.B.; Schwacke, J.H.; Ross, J.; Cowart, L.A. Fatty acids differentially regulate insulin resistance through endoplasm reticulum stress-mediated induction of tribbles homologue 3: A potential link between dietary fat composition and the pathophysiological outcomes of obesity. Diabetologia 2013, 56, 2078–2087. [Google Scholar] [CrossRef]
- Geng, T.; Sutter, A.; Harland, M.D.; Law, B.A.; Ross, J.S.; Lewin, D.; Palanisamy, A.; Russo, S.B.; Chavin, K.D.; Cowart, L.A. SphK1 mediates hepatic inflammation in a mouse model of NASH induced by high saturated fat feeding and initiates proinflammatory signaling in hepatocytes. J. Lipid Res. 2015, 56, 2359–2371. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Xia, L.; Russo, S.; Kamara, D.; Cowart, L.A. Prosteatotic genes are associated with unsaturated fat suppression of saturated fat-induced hepatic steatosis in C57BL/6 mice. Nutr. Res. 2015, 35, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Petito-Da-Silva, T.I.; Souza-Mello, V.; Barbosa-Da-Silva, S. Empaglifozin mitigates NAFLD in high-fat-fed mice by alleviating insulin resistance, lipogenesis and ER stress. Mol. Cell. Endocrinol. 2019, 498, 110539. [Google Scholar] [CrossRef]
- Aljaylani, A.; Fluitt, M.; Piselli, A.; Shepard, B.D.; Tiwari, S.; Ecelbarger, C.M. Acid Loading Unmasks Glucose Homeostatic Instability in Proximal-Tubule-Targeted Insulin/Insulin-Like-Growth-Factor-1 Receptor Dual Knockout Mice. Cell. Physiol. Biochem. 2020, 54, 682–695. [Google Scholar] [CrossRef]
- Balen, D.; Ljubojević, M.; Breljak, D.; Brzica, H.; Zlender, V.; Koepsell, H.; Sabolić, I. Revised immunolocalization of the Na+-d-glucose cotransporter SGLT1 in rat organs with an improved antibody. Am. J. Physiol. Physiol. 2008, 295, C475–C489. [Google Scholar] [CrossRef]
- Vallon, V.; Platt, K.A.; Cunard, R.; Schroth, J.; Whaley, J.; Thomson, S.C.; Koepsell, H.; Rieg, T. SGLT2 Mediates Glucose Reabsorption in the Early Proximal Tubule. J. Am. Soc. Nephrol. 2010, 22, 104–112. [Google Scholar] [CrossRef]
- Li, Y.; Girgis, M.; Wise, S.Y.; Fatanmi, O.O.; Seed, T.M.; Maniar, M.; Cheema, A.K.; Singh, V.K. Analysis of the metabolomic profile in serum of irradiated nonhuman primates treated with Ex-Rad, a radiation countermeasure. Sci. Rep. 2021, 11, 11449. [Google Scholar] [CrossRef]
- Frolkis, A.; Knox, C.; Lim, E.; Jewison, T.; Law, V.; Hau, D.D.; Liu, P.; Gautam, B.; Ly, S.; Guo, A.C.; et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2009, 38, D480–D487. [Google Scholar] [CrossRef]
- Lin, W.-J.; Shen, P.-C.; Liu, H.-C.; Cho, Y.-C.; Hsu, M.-K.; Lin, I.-C.; Chen, F.-H.; Yang, J.-C.; Ma, W.-L.; Cheng, W.-C. LipidSig: A web-based tool for lipidomic data analysis. Nucleic Acids Res. 2021, 49, W336–W345. [Google Scholar] [CrossRef]
- Gil-De-La-Fuente, A.; Godzien, J.; Saugar, S.; Garcia-Carmona, R.; Badran, H.; Wishart, D.S.; Barbas, C.; Otero, A. CEU Mass Mediator 3.0: A Metabolite Annotation Tool. J. Proteome Res. 2018, 18, 797–802. [Google Scholar] [CrossRef]
- de la Fuente, A.G.; Godzien, J.; Fernández-López, M.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Norton, L.; Abdul-Ghani, M. Renal, metabolic and cardiovascular considerations of SGLT2 inhibition. Nat. Rev. Nephrol. 2016, 13, 11–26. [Google Scholar] [CrossRef]
- Ferrannini, G.; Hach, T.; Crowe, S.; Sanghvi, A.; Hall, K.D.; Ferrannini, E. Energy Balance After Sodium–Glucose Cotransporter 2 Inhibition. Diabetes Care 2015, 38, 1730–1735. [Google Scholar] [CrossRef]
- Hallow, K.M.; Boulton, D.W.; Penland, R.C.; Helmlinger, G.; Nieves, E.H.; Van Raalte, D.H.; Heerspink, H.L.; Greasley, P.J. Renal Effects of Dapagliflozin in People with and without Diabetes with Moderate or Severe Renal Dysfunction: Prospective Modeling of an Ongoing Clinical Trial. J. Pharmacol. Exp. Ther. 2020, 375, 76–91. [Google Scholar] [CrossRef]
- Tuttle, K.R.; Brosius, F.C.; Cavender, M.A.; Fioretto, P.; Fowler, K.J.; Heerspink, H.J.; Manley, T.; McGuire, D.K.; Molitch, M.E.; Mottl, A.K.; et al. SGLT2 Inhibition for CKD and Cardiovascular Disease in Type 2 Diabetes: Report of a Scientific Workshop Sponsored by the National Kidney Foundation. Am. J. Kidney Dis. 2021, 77, 94–109. [Google Scholar] [CrossRef]
- Vallon, V. Molecular determinants of renal glucose reabsorption. Focus on “Glucose transport by human renal Na+/d-glucose cotransporters SGLT1 and SGLT2”. Am. J. Physiol. Physiol. 2011, 300, C6–C8. [Google Scholar] [CrossRef]
- Vallon, V. The proximal tubule in the pathophysiology of the diabetic kidney. Am. J. Physiol. Integr. Comp. Physiol. 2011, 300, R1009–R1022. [Google Scholar] [CrossRef]
- Vallon, V.; Komers, R. Pathophysiology of the Diabetic Kidney. Compr. Physiol. 2011, 1, 1175–1232. [Google Scholar] [CrossRef]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef]
- Gowans, G.J.; Hardie, D.G. AMPK: A cellular energy sensor primarily regulated by AMP. Biochem. Soc. Trans. 2014, 42, 71–75. [Google Scholar] [CrossRef] [Green Version]
- Rieg, J.A.D.; Xue, J.; Rieg, T. Tubular effects of sodium–glucose cotransporter 2 inhibitors: Intended and unintended consequences. Curr. Opin. Nephrol. Hypertens. 2020, 29, 523–530. [Google Scholar] [CrossRef]
- Castellino, P.; DeFronzo, R.A. Glucose metabolism and the kidney. Semin. Nephrol. 1990, 10, 458–463. [Google Scholar] [PubMed]
- Harris, A.N.; Lee, H.-W.; Osis, G.; Fang, L.; Webster, K.; Verlander, J.W.; Weiner, I.D. Differences in renal ammonia metabolism in male and female kidney. Am. J. Physiol. Physiol. 2018, 315, F211–F222. [Google Scholar] [CrossRef]
- Guder, W.G.; Wagner, S.; Wirthensohn, G. Metabolic fuels along the nephron: Pathways and intracellular mechanisms of interaction. Kidney Int. 1986, 29, 41–45. [Google Scholar] [CrossRef]
- Lin, L.; Yee, S.W.; Kim, R.B.; Giacomini, K.M. SLC transporters as therapeutic targets: Emerging opportunities. Nat. Rev. Drug Discov. 2015, 14, 543–560. [Google Scholar] [CrossRef]
- Verrey, F.; Singer, D.; Ramadan, T.; Vuille-Dit-Bille, R.N.; Mariotta, L.; Camargo, S.M.R. Kidney amino acid transport. Pflugers Arch. 2009, 458, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Aliaga, I.; Wagner, C.A. Regulation and function of the SLC38A3/SNAT3 glutamine transporter. Channels 2016, 10, 440–452. [Google Scholar] [CrossRef]
- Ni, Y.-Q.; Liu, Y.-S. New Insights into the Roles and Mechanisms of Spermidine in Aging and Age-Related Diseases. Aging Dis. 2021, 12, 1948. [Google Scholar] [CrossRef]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef]
- Kramer, D.L.; Chang, B.D.; Chen, Y.; Diegelman, P.; Alm, K.; Black, A.R.; Roninson, I.B.; Porter, C.W. Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res. 2001, 61, 7754–7762. [Google Scholar]
- Pegg, A.E. Toxicity of Polyamines and Their Metabolic Products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, S.; Xie, C.; Fang, J. Uridine Metabolism and Its Role in Glucose, Lipid, and Amino Acid Homeostasis. BioMed Res. Int. 2020, 2020, 7091718. [Google Scholar] [CrossRef]
- Le, T.T.; Urasaki, Y.; Pizzorno, G. Uridine Prevents Fenofibrate-Induced Fatty Liver. PLoS ONE 2014, 9, e87179. [Google Scholar] [CrossRef]
- Le, T.T.; Urasaki, Y.; Pizzorno, G. Uridine prevents tamoxifen-induced liver lipid droplet accumulation. BMC Pharmacol. Toxicol. 2014, 15, 27. [Google Scholar] [CrossRef]
- Alam, M.; Iqbal, S.; Naseem, I. Ameliorative effect of riboflavin on hyperglycemia, oxidative stress and DNA damage in type-2 diabetic mice: Mechanistic and therapeutic strategies. Arch. Biochem. Biophys. 2015, 584, 10–19. [Google Scholar] [CrossRef]
- Suwannasom, N.; Kao, I.; Pruß, A.; Georgieva, R.; Bäumler, H. Riboflavin: The Health Benefits of a Forgotten Natural Vitamin. Int. J. Mol. Sci. 2020, 21, 950. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef]
- Mulder, S.; Hammarstedt, A.; Nagaraj, S.B.; Nair, V.; Ju, W.; Hedberg, J.; Greasley, P.J.; Eriksson, J.W.; Oscarsson, J.; Heerspink, H.J.L. A metabolomics-based molecular pathway analysis of how the sodium-glucose co-transporter-2 inhibitor dapagliflozin may slow kidney function decline in patients with diabetes. Diabetes Obes. Metab. 2020, 22, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
| Compound Name | Control * | EMPA | p-Value ‡ | EMPA-to-Control Ratio |
|---|---|---|---|---|
| Carbamoyl phosphate | 1.38 ± 0.53 | 0.65 ± 0.14 | 0.23 | 0.47 |
| Spermine | 1.38 ± 0.18 | 0.66 ± 0.07 | 0.0036 | 0.48 |
| 2-Isopropylmalic acid | 1.27 ± 0.12 | 0.68 ± 0.08 | 0.0021 | 0.54 |
| Succinate | 1.30 ± 0.31 | 0.72 ± 0.07 | 0.11 | 0.56 |
| Trans-Aconitate | 1.23 ± 0.27 | 0.71 ± 0.12 | 0.12 | 0.58 |
| Xanthurenic acid | 1.29 ± 0.14 | 0.77 ± 0.06 | 0.0057 | 0.59 |
| Phenyllactic acid | 1.10 ± 0.43 | 0.68 ± 0.08 | 0.39 | 0.62 |
| 2-Deoxyguanosine 5-monophosphate | 1.33 ± 0.17 | 0.84 ± 0.11 | 0.036 | 0.63 |
| 4-Pyridoxate | 1.27 ± 0.17 | 0.76 ± 0.10 | 0.022 | 0.63 |
| Putrescine | 1.22 ± 0.14 | 0.77 ± 0.08 | 0.018 | 0.64 |
| Spermidine | 1.24 ± 0.07 | 0.84 ± 0.06 | 0.00078 | 0.67 |
| Metabolite Set | Total | Hits | Expect † | p-Value ‡ | Holm’s p ¥ | FDR € |
|---|---|---|---|---|---|---|
| Urea cycle | 29 | 8 | 1.95 | 0.0004 | 0.038 | 0.038 |
| Spermidine and spermine biosynthesis | 18 | 5 | 1.21 | 0.0052 | 0.502 | 0.223 |
| Aspartate metabolism | 35 | 7 | 2.36 | 0.0068 | 0.66 | 0.22 |
| Methionine metabolism | 43 | 7 | 2.90 | 0.021 | 1.0 | 0.42 |
| Arginine and proline metabolism | 53 | 8 | 3.57 | 0.022 | 1.0 | 0.42 |
| Carnitine synthesis | 22 | 4 | 1.48 | 0.055 | 1.0 | 0.90 |
| Glycine and serine metabolism | 59 | 7 | 3.98 | 0.094 | 1.0 | 0.94 |
| Pyrimidine metabolism | 59 | 7 | 3.98 | 0.094 | 1.0 | 0.94 |
| Valine, leucine, and isoleucine degradation | 60 | 7 | 4.04 | 0.10 | 1.0 | 0.94 |
| Trypotophan metabolism | 60 | 7 | 4.04 | 0.10 | 1.0 | 0.94 |
| Compound Name | Control | EMPA | p-Value | EMPA-to Control Ratio | Function |
|---|---|---|---|---|---|
| Symmetric Dimethylarginine | 1.16 ± 0.052 | 0.91 ± 0.059 | 0.0082 | 0.79 | Inhibitor of arginine transport; marker of GFR |
| Lipoic Acid | 0.87 ± 0.085 | 0.78 ± 0.11 | 0.52 | 0.90 | Anti-oxidant |
| AICAR | 1.08 ± 0.031 | 0.95 ± 0.027 | 0.0062 | 0.88 | AMPK activator, anti-oxidant |
| Nicotinamide Adenine Dinucleotide | 1.14 ± 0.12 | 0.83 ± 0.16 | 0.15 | 0.73 | Co-Factor in Redox Reactions |
| Pyruvic Acid | 1.02 ± 0.077 | 0.94 ± 0.071 | 0.40 | 0.89 | Component of Glycolysis; anti-oxidant |
| Sarcosine | 1.06 ± 0.079 | 0.90 ± 0.055 | 0.13 | 0.85 | Derivative of glycine; causes oxidative stress |
| Uridine | 0.93 ± 0.11 | 1.16 ± 0.098 | 0.16 | 1.24 | Glycosylated pyrimidine; anti-oxidant |
| Flavin Mononucleotide | 69 ± 0.046 | 1.08 ± 0.25 | 0.11 | 1.57 | Produced from riboflavin; prosthetic group of oxidoreductases |
| Carboxytridecanoylcarnitine ‡ | 0.61 ± 0.11 | 1.21 ± 0.10 | 0.0021 | 1.99 | Fat transport into mitochondria |
| Hexadecanoylcarnitine ‡ | 0.62 ± 0.12 | 1.17 ± 0.12 | 0.0068 | 1.88 | Fat transport into mitochondria |
| Pentadecanoylcarnitine ‡ | 0.83 ± 0.05 | 1.16 ± 0.089 | 0.024 | 1.40 | Fat transport into mitochondria |
| Hydroxyoctadecenoylcarnitine ‡ | 0.65 ± 0.12 | 1.04 ± 0.12 | 0.037 | 1.59 | Fat transport into mitochondria |
| Mz_rt * | FC | p-Value | Direction | Annotation † |
|---|---|---|---|---|
| Positive | ||||
| 332.3_8.23 | 1.86 | 0.000272 | Increased | 3; modified PA species |
| 331.3_8.23 | 1.83 | 0.000294 | Increased | >10; modified eicosadienoic acid species |
| 351.3_8.61 | 1.81 | 0.009813 | Increased | 6; modified SM or PE-ceramide |
| 570.3_8.61 | 1.80 | 0.002333 | Increased | >10; modified, LPC, CL, PC |
| 408.3_6.68 | 1.70 | 0.000506 | Increased | >10; Radarin & PG modified species |
| 484.3_7.12 | 0.42 | 0.000941 | Decreased | 5; modified PA, taurine, sulfonic acid |
| 550.3_7.15 | 0.48 | 0.000766 | Decreased | 2, modified vitamin D3 or lactone |
| 564.3_7.15 | 0.48 | 1.76 × 10−5 | Decreased | 1; 1-(2-methoxy-eicosanyl)-sn-glycero-3-phosphoethanolamine; M + K |
| 576.4_7.93 | 0.53 | 0.000393 | Decreased | >10; PE or PC modifications |
| 240.1_7.23 | 0.67 | 0.000157 | Decreased | >10; carboxylic acid, gluco, & galactoside |
| Negative | ||||
| 736.5_11.3 | 1.97 | 0.004575 | Increased | >10; PE or PC modifications |
| 185.0_0.42 | 1.95 | 0.001566 | Increased | Modified carbohydrate, e.g., ribulose |
| 430.2_8.22 | 1.44 | 0.000207 | Increased | >10; tripeptide |
| 708.5_9.83 | 1.41 | 0.000362 | Increased | >10; PE or PC modifications |
| 718.5_9.68 | 1.40 | 0.00057 | Increased | >10; PE modifications |
| 316.2_3.8 | 0.22 | 0.019235 | Decreased | 2; 3-hydroxynonanoyl carnitine and Tridecanoylglycine |
| 689.5_8.68 | 0.26 | 0.001863 | Decreased | >10; DG, PE, or PA modified species |
| 1096.8_7.13 | 0.29 | 0.009105 | Decreased | Nothing found |
| 407.3_9.6 | 0.46 | 0.00021 | Decreased | 1; Sinapoylspermine |
| 447.3_8.2 | 0.71 | 4.91 × 10−6 | Decreased | 9; vitamin K, quinone; cholesterol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bangarbale, S.; Shepard, B.D.; Bansal, S.; Jayatilake, M.M.; Kurtz, R.; Levi, M.; Ecelbarger, C.M. Renal Metabolome in Obese Mice Treated with Empagliflozin Suggests a Reduction in Cellular Respiration. Biomolecules 2022, 12, 1176. https://doi.org/10.3390/biom12091176
Bangarbale S, Shepard BD, Bansal S, Jayatilake MM, Kurtz R, Levi M, Ecelbarger CM. Renal Metabolome in Obese Mice Treated with Empagliflozin Suggests a Reduction in Cellular Respiration. Biomolecules. 2022; 12(9):1176. https://doi.org/10.3390/biom12091176
Chicago/Turabian StyleBangarbale, Surabhi, Blythe D. Shepard, Shivani Bansal, Meth M. Jayatilake, Ryan Kurtz, Moshe Levi, and Carolyn M. Ecelbarger. 2022. "Renal Metabolome in Obese Mice Treated with Empagliflozin Suggests a Reduction in Cellular Respiration" Biomolecules 12, no. 9: 1176. https://doi.org/10.3390/biom12091176
APA StyleBangarbale, S., Shepard, B. D., Bansal, S., Jayatilake, M. M., Kurtz, R., Levi, M., & Ecelbarger, C. M. (2022). Renal Metabolome in Obese Mice Treated with Empagliflozin Suggests a Reduction in Cellular Respiration. Biomolecules, 12(9), 1176. https://doi.org/10.3390/biom12091176







