CAPTURE of the Human U2 snRNA Genes Expands the Repertoire of Associated Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. HEK293 Cell Culture
2.2. dCas9 and Guide Cloning and Guide Design
2.3. Stable Cell Line Production
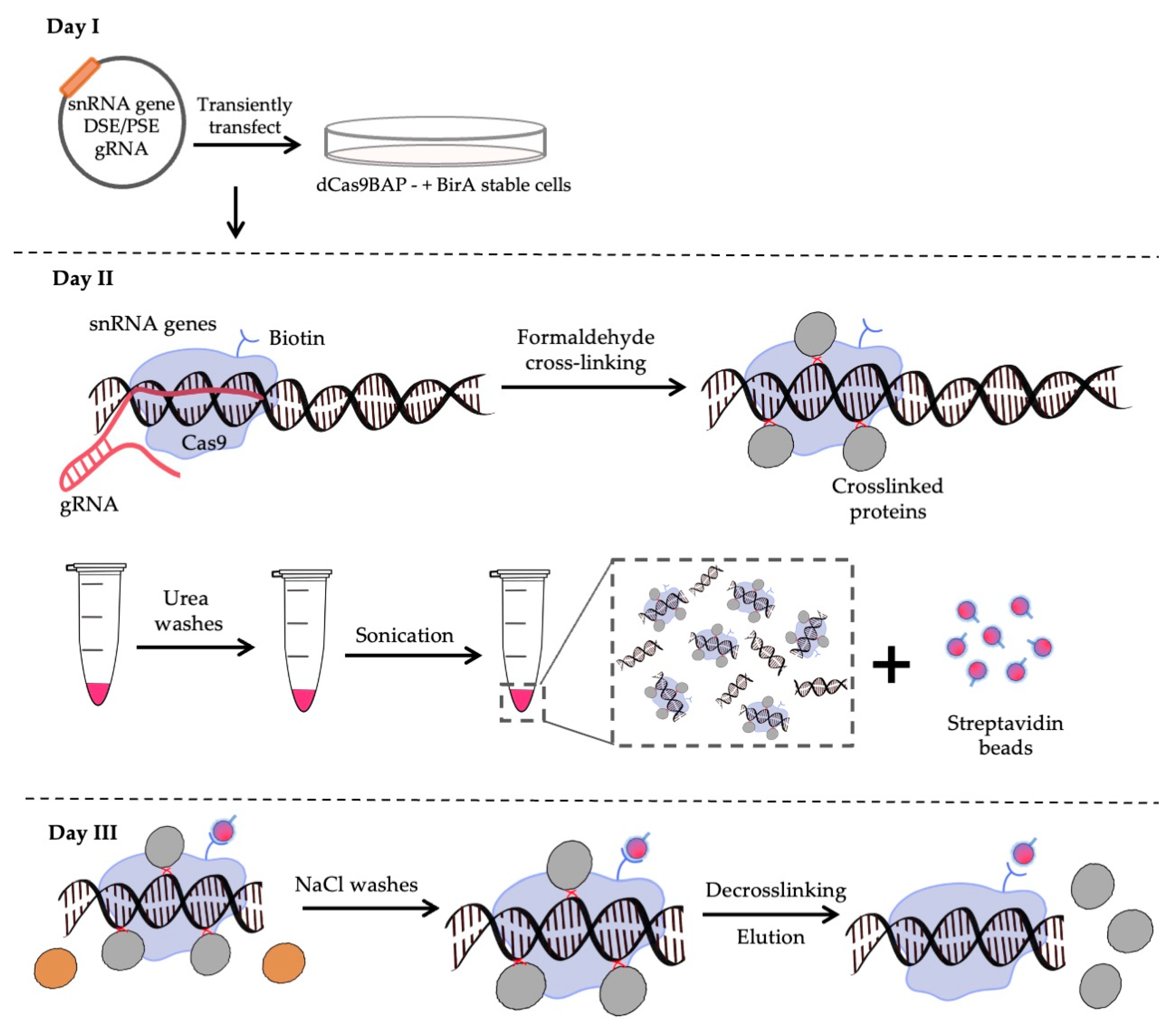
2.4. CAPTURE and Proteomics
2.5. Western Blotting
2.6. Chromatin Immunoprecipitation (ChIP)
2.7. ChIP-Seq
2.8. Bioinformatics Analysis
2.9. ChIP-Seq Data Processing
2.10. Chromatin RNA-Seq Data Processing
3. Results
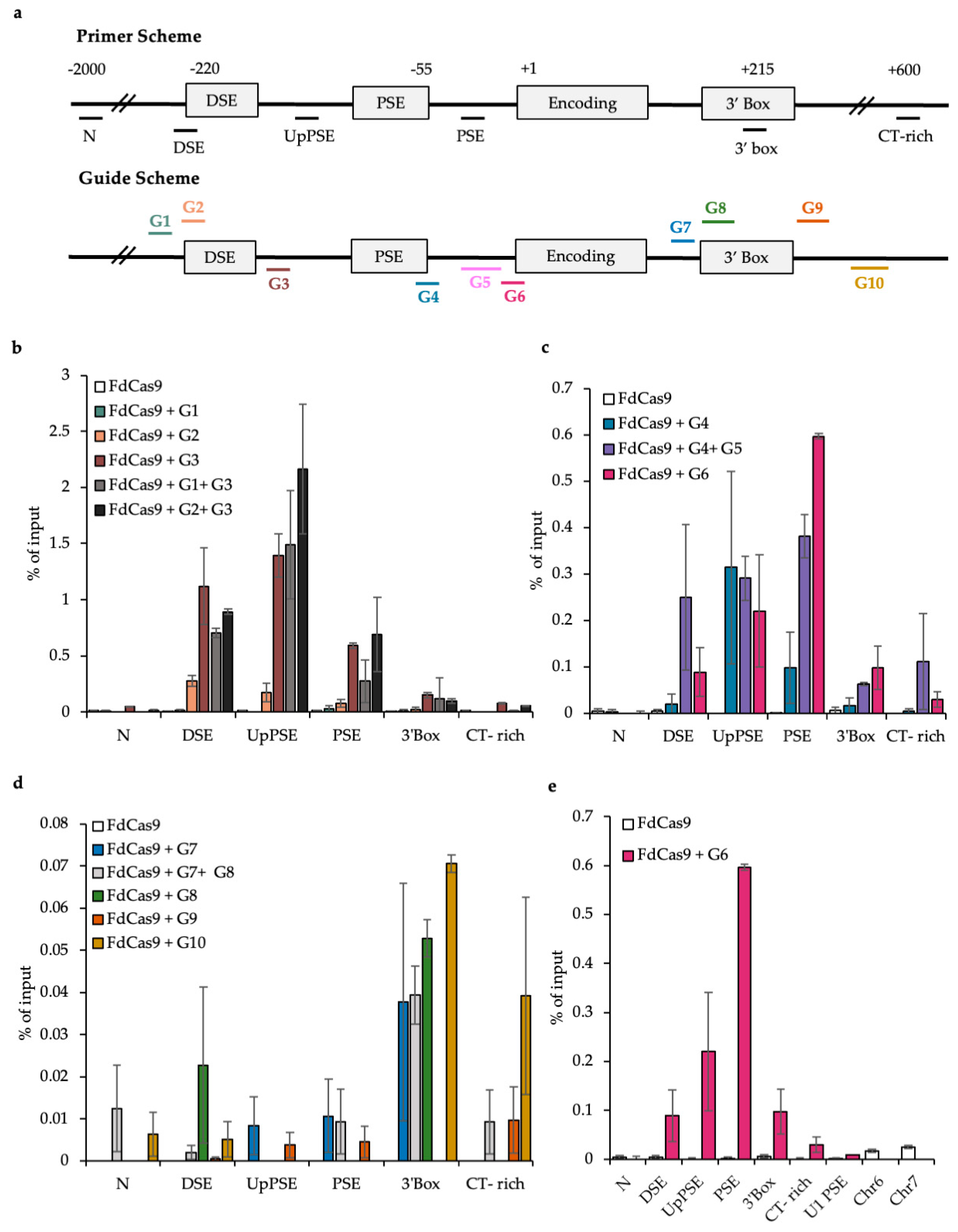
3.1. Designing Guides for DCas9 Targeting
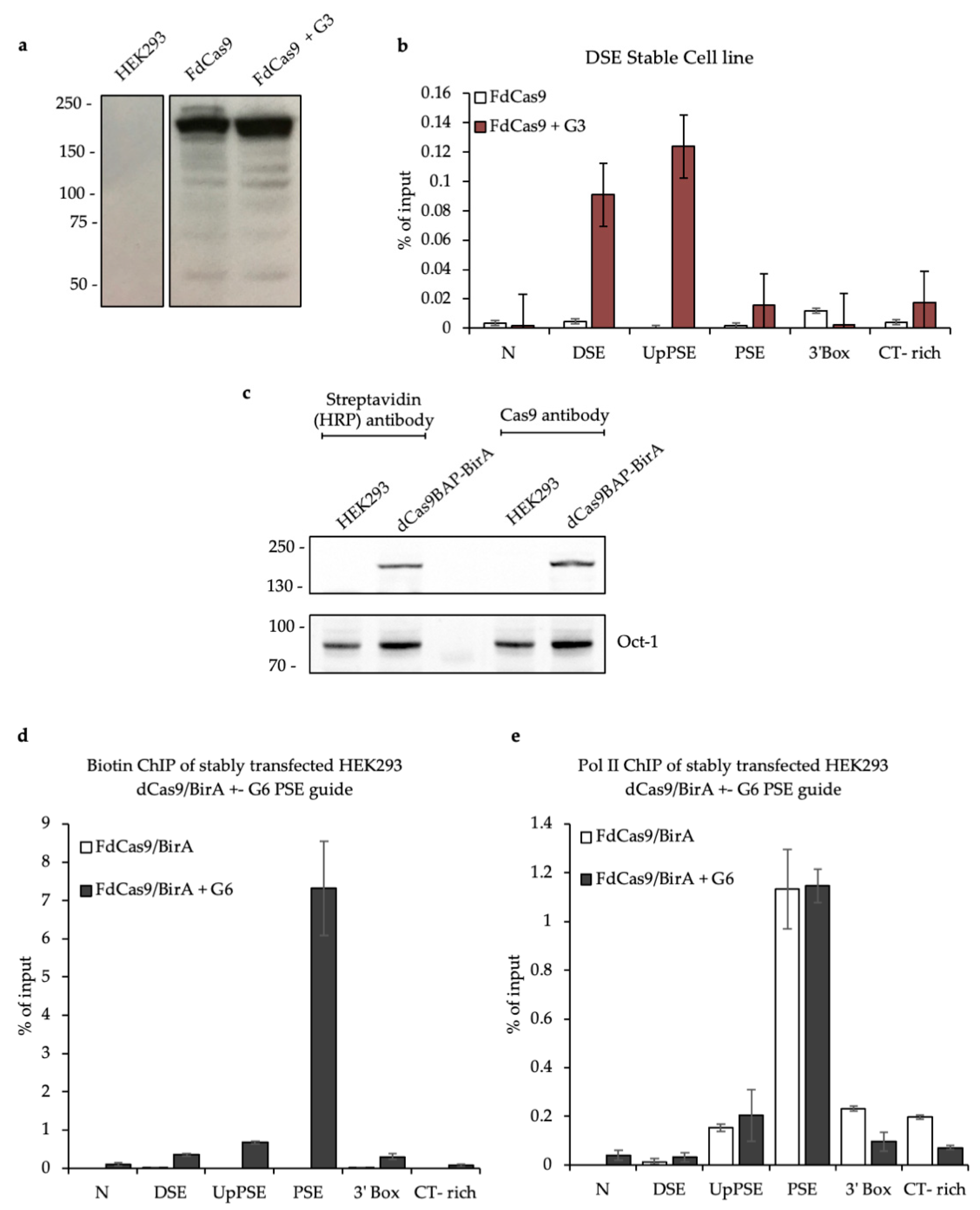
3.2. Testing the CAPTURE System
3.3. CAPTURE on the U2 snRNA Gene PSE and DSE
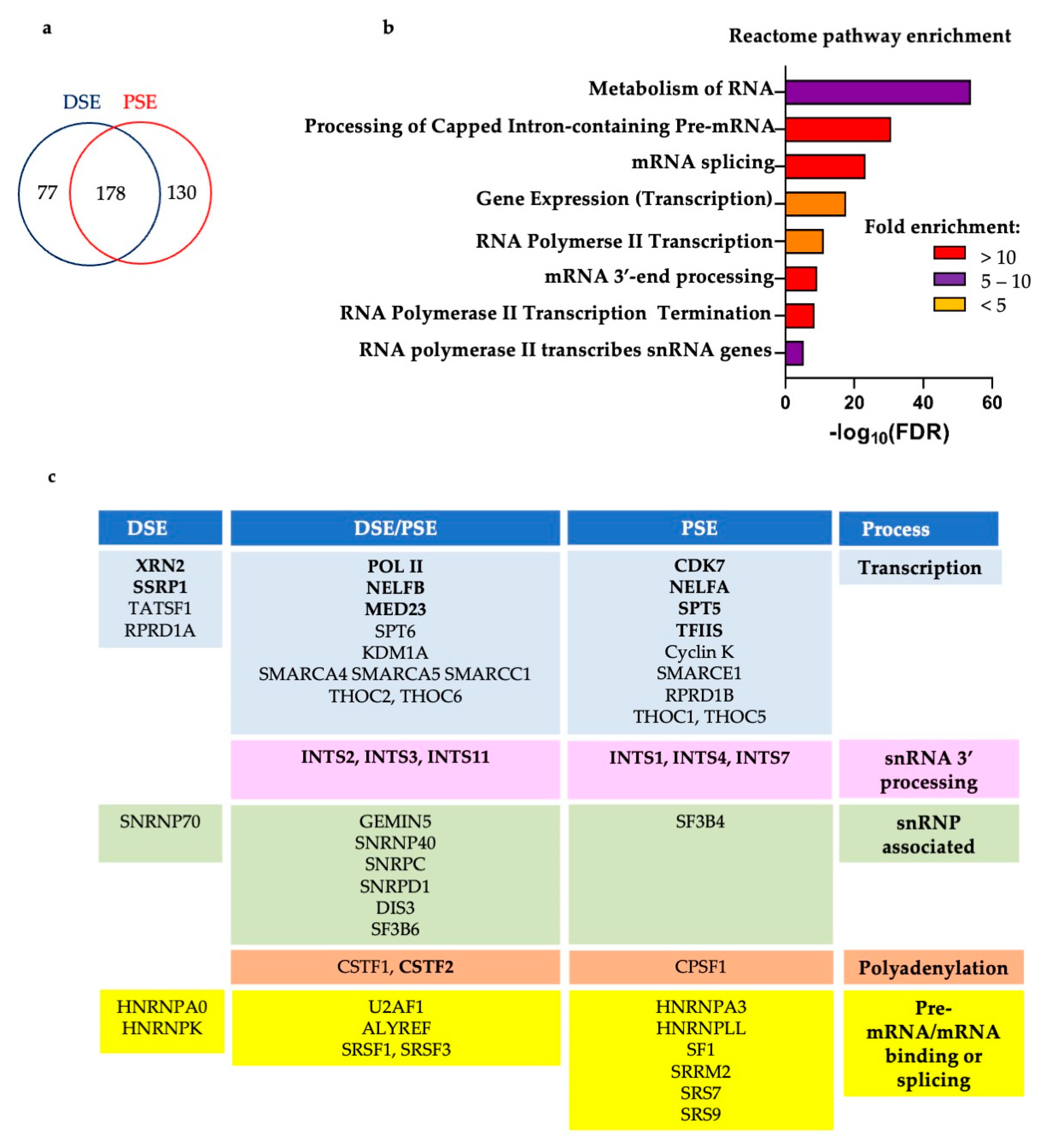
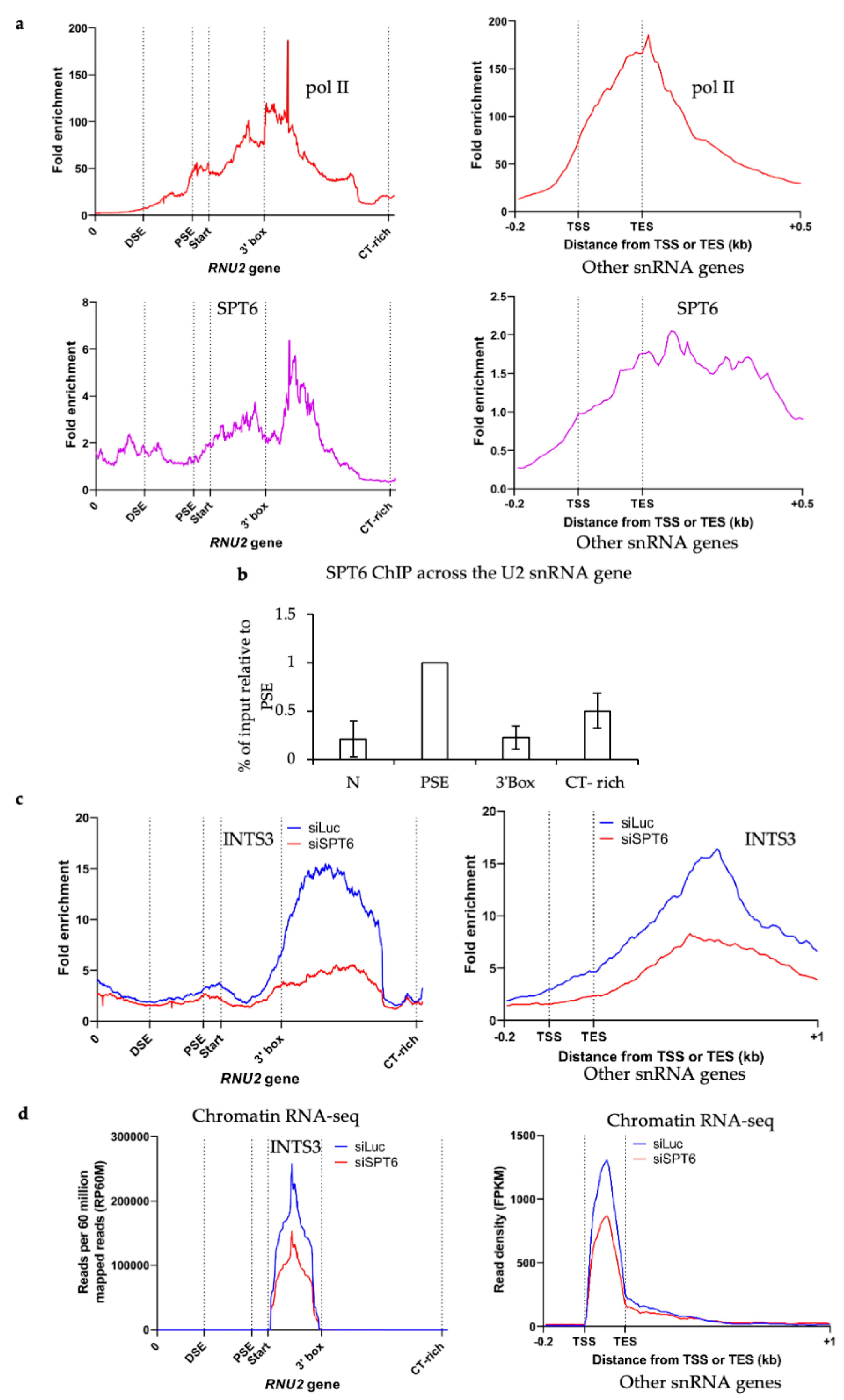
3.4. SPT6 Helps to Recruit Integrator to the Human snRNA Genes
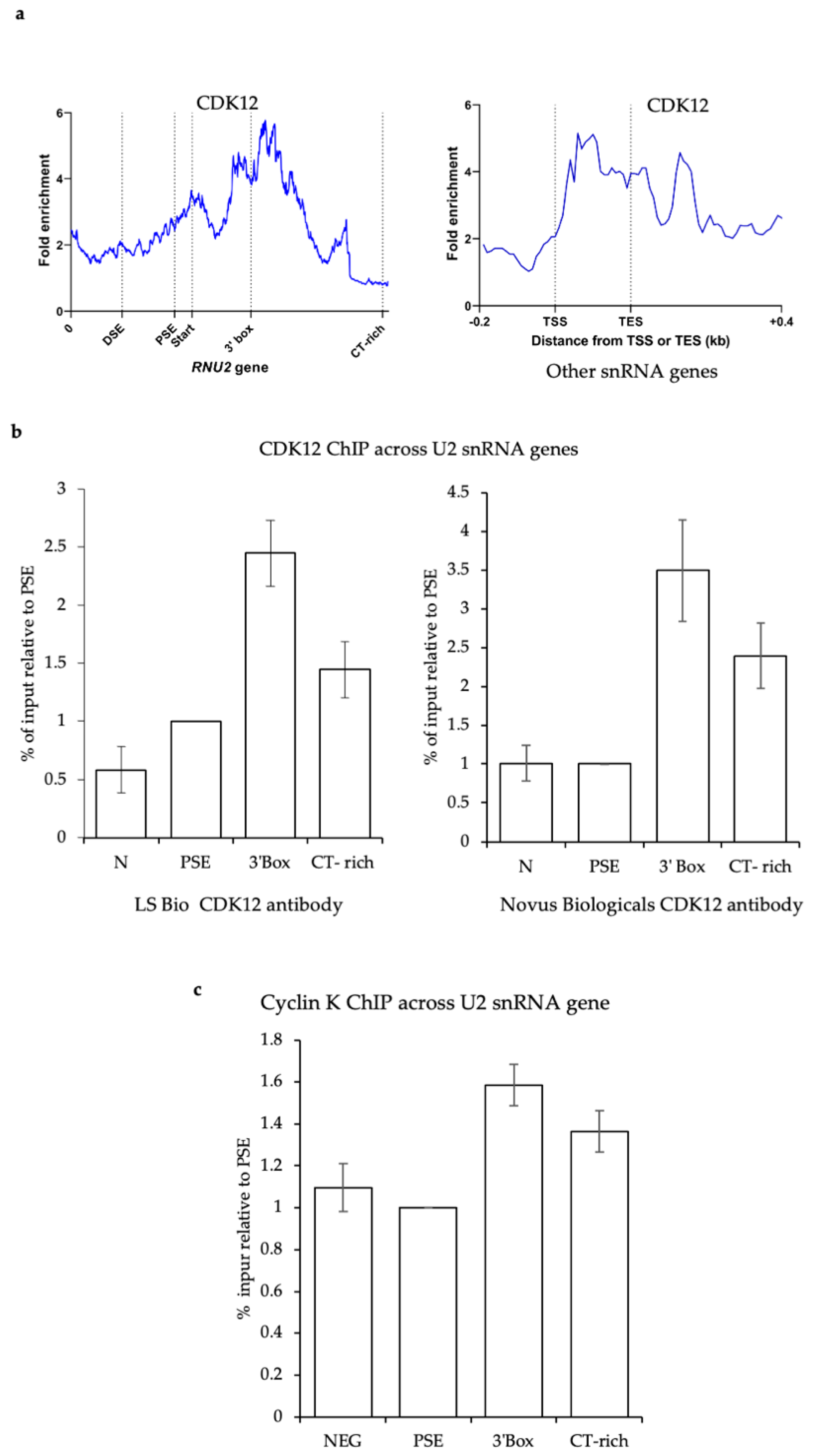
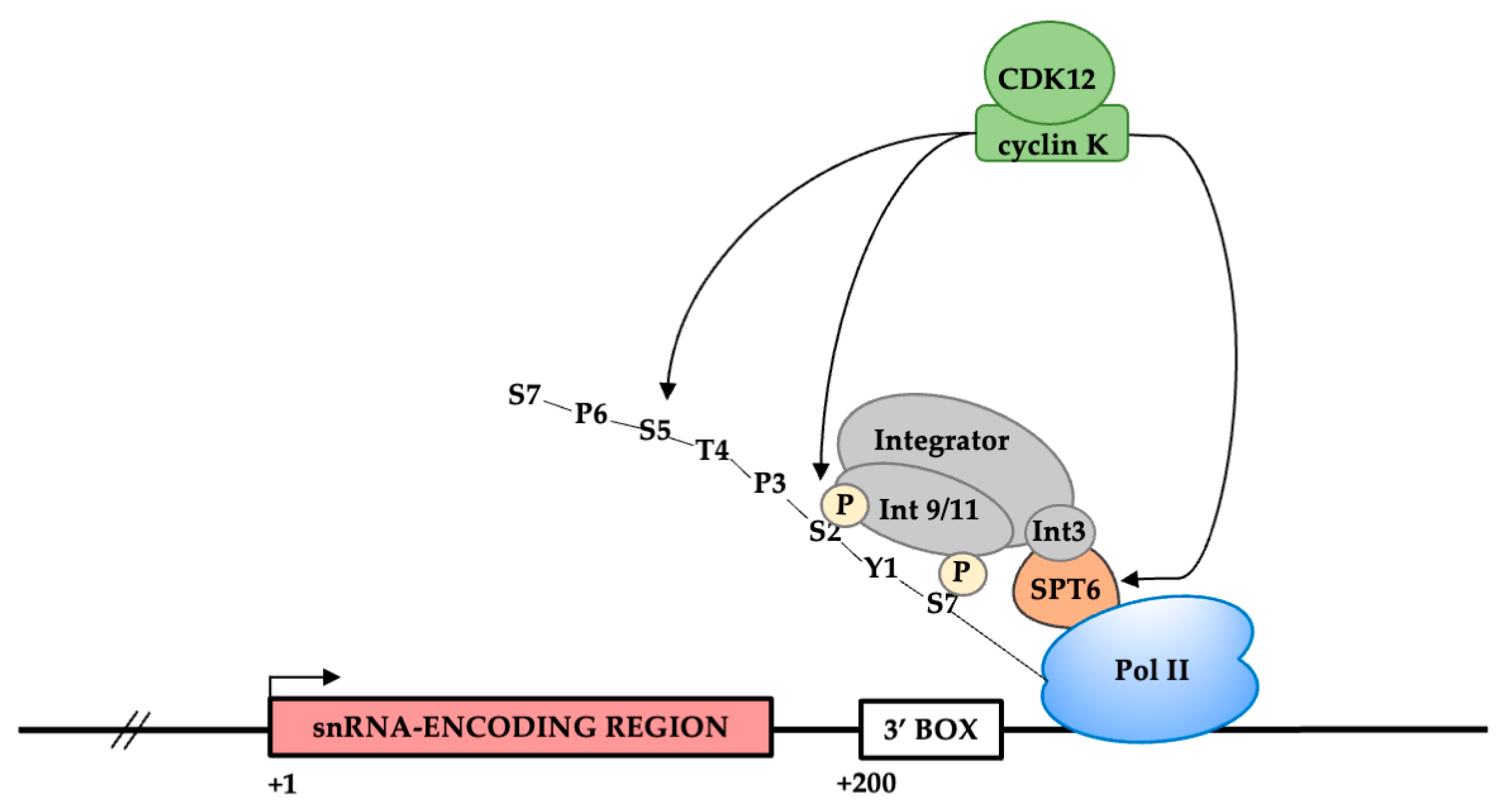
3.5. CDK12 Is Associated with the U2 snRNA Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiss, T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004, 117, 5949–5951. [Google Scholar] [CrossRef] [PubMed]
- Guiro, J.; Murphy, S. Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol. 2017, 7, 170073–170082. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, N. Small nuclear RNA genes: A model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001, 276, 26733–26736. [Google Scholar] [CrossRef] [PubMed]
- Jawdekar, G.W.; Henry, R.W. Transcriptional regulation of human small nuclear RNA genes. Biochim. Biophys. Acta 2008, 1779, 295–305. [Google Scholar] [CrossRef]
- Egloff, S.; O’Reilly, D.; Murphy, S. Expression of human snRNA genes from beginning to end. Biochem. Soc. Trans. 2008, 36, 590–594. [Google Scholar] [CrossRef]
- Baillat, D.; Hakimi, M.A.; Naar, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef]
- Baillat, D.; Wagner, E.J. Integrator: Surprisingly diverse functions in gene expression. Trends Biochem. Sci. 2015, 40, 257–264. [Google Scholar] [CrossRef]
- Mendoza-Figueroa, M.S.; Tatomer, D.C.; Wilusz, J.E. The Integrator Complex in Transcription and Development. Trends Biochem. Sci. 2020, 45, 923–934. [Google Scholar] [CrossRef]
- Murphy, S.; Yoon, J.B.; Gerster, T.; Roeder, R.G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 1992, 12, 3247–3261. [Google Scholar]
- Mittal, V.; Cleary, M.A.; Herr, W.; Hernandez, N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol. Cell. Biol. 1996, 16, 1955–1965. [Google Scholar] [CrossRef]
- Murphy, S. Differential in vivo activation of the class II and class III snRNA genes by the POU-specific domain of Oct-1. Nucleic Acids Res. 1997, 25, 2068–2076. [Google Scholar] [CrossRef] [PubMed]
- Waldschmidt, R.; Wanandi, I.; Seifart, K.H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991, 10, 2595–2603. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, C.L.; Henry, R.W.; Lobo, S.M.; Hernandez, N. Targeting TBP to a non-TATA box cis- regulatory element: A TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993, 7, 1535–1548. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.B.; Murphy, S.; Bai, L.; Wang, Z.; Roeder, R.G. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 1995, 15, 2019–2027. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.W.; Sadowski, C.L.; Kobayashi, R.; Hernandez, N. A TBP–TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature 1995, 374, 653–656. [Google Scholar] [CrossRef]
- Kuhlman, T.C.; Cho, H.; Reinberg, D.; Hernandez, N. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol. Cell. Biol. 1999, 19, 2130–2141. [Google Scholar] [CrossRef][Green Version]
- Zaborowska, J.; Taylor, A.; Roeder, R.G.; Murphy, S. A novel TBP–TAF complex on RNA polymerase II-transcribed snRNA genes. Transcription 2012, 3, 92–104. [Google Scholar] [CrossRef]
- Takahashi, H.; Takigawa, I.; Watanabe, M.; Anwar, D.; Shibata, M.; Tomomori-Sato, C.; Sato, S.; Ranjan, A.; Seidel, C.W.; Tsukiyama, T.; et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 2015, 6, 5941. [Google Scholar] [CrossRef]
- Malik, S.; Roeder, R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010, 11, 761–772. [Google Scholar] [CrossRef]
- Allen, B.L.; Taatjes, D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef]
- Takahashi, H.; Ranjan, A.; Chen, S.; Suzuki, H.; Shibata, M.; Hirose, T.; Hirose, H.; Sasaki, K.; Abe, R.; Chen, K.; et al. The role of Mediator and Little Elongation Complex in transcription termination. Nat. Commun. 2020, 11, 1063–1082. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.R.; Lin, C.; Garrett, A.S.; Thornton, J.; Mohaghegh, N.; Hu, D.; Jackson, J.; Saraf, A.; Swanson, S.K.; Seidel, C.; et al. The little elongation complex regulates small nuclear RNA transcription. Mol. Cell 2011, 44, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Smith, E.R.; Garruss, A.S.; Mohaghegh, N.; Varberg, J.M.; Lin, C.; Jackson, J.; Gao, X.; Saraf, A.; Florens, L.; et al. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol. Cell 2013, 51, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Yamamoto, J.; Hagiwara, Y.; Chiba, K.; Isobe, T.; Narita, T.; Handa, H.; Yamaguchi, Y. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat. Commun. 2014, 5, 4263. [Google Scholar] [CrossRef] [PubMed]
- Egloff, S.; Al-Rawaf, H.; O’Reilly, D.; Murphy, S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol. Cell. Biol. 2009, 29, 4002–4013. [Google Scholar] [CrossRef]
- Laitem, C.; Zaborowska, J.; Tellier, M.; Yamaguchi, Y.; Cao, Q.; Egloff, S.; Handa, H.; Murphy, S. CTCF regulates NELF, DSIF and P-TEFb recruitment during transcription. Transcription 2015, 6, 79–90. [Google Scholar] [CrossRef]
- Medlin, J.E.; Uguen, P.; Taylor, A.; Bentley, D.L.; Murphy, S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003, 22, 925–934. [Google Scholar] [CrossRef]
- Medlin, J.; Scurry, A.; Taylor, A.; Zhang, F.; Peterlin, B.M.; Murphy, S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005, 24, 4154–4165. [Google Scholar] [CrossRef]
- Egloff, S.; O’Reilly, D.; Chapman, R.D.; Taylor, A.; Tanzhaus, K.; Pitts, L.; Eick, D.; Murphy, S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007, 318, 1777–1779. [Google Scholar] [CrossRef]
- Egloff, S.; Szczepaniak, S.A.; Dienstbier, M.; Taylor, A.; Knight, S.; Murphy, S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 2010, 285, 20564–20569. [Google Scholar] [CrossRef] [PubMed]
- Glover-Cutter, K.; Larochelle, S.; Erickson, B.; Zhang, C.; Shokat, K.; Fisher, R.P.; Bentley, D.L. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter- proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 2009, 29, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.S.; Heidemann, M.; Tietjen, J.R.; Zhang, D.W.; Chapman, R.D.; Eick, D.; Ansari, A.Z. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 2009, 34, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Egloff, S.; Zaborowska, J.; Laitem, C.; Kiss, T.; Murphy, S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell 2012, 45, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, N.; Weiner, A.M. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 1986, 47, 249–258. [Google Scholar] [CrossRef]
- De Vegvar, H.E.; Lund, E.; Dahlberg, J.E. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 1986, 47, 259–266. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Zhou, F.; Li, K.; Cao, H.; Ni, M.; Liu, Y.; Gi, Z.; et al. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017, 170, 1028–1043. [Google Scholar] [CrossRef]
- Westin, G.; Zabielski, J.; Hammarström, K.; Monstein, H.J.; Bark, C.; Pettersson, U. Clustered genes for human U2 RNA. Proc. Natl. Acad. Sci. USA 1984, 81, 3811–3815. [Google Scholar] [CrossRef]
- Van Arsdell, S.W.; Weiner, A.M. Human genes for U2 small nuclear RNA are tandemly repeated. Mol. Cell. Biol. 1984, 4, 492–499. [Google Scholar]
- Bylund, L.; Kytölä, S.; Lui, W.O.; Larsson, C.; Weber, G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet. Genome Res. 2004, 106, 28–32. [Google Scholar] [CrossRef]
- McCullough, L.; Connell, Z.; Petersen, C.; Formosa, T. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 2015, 201, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Tellier, M.; Foxwell, J.; de Almeida, C.R.; Tan-Wong, S.M.; Dhir, S.; Dujardin, G.; Dhir, A.; Murphy, S.; Proudfoot, N.J. Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol. Cell 2018, 72, 970–984. [Google Scholar] [CrossRef] [PubMed]
- Tellier, M.; Tellier, M.; Zaborowska, J.; Caizzi, L.; Mohammad, E.; Velychko, T.; Schwalb, B.; Ferrer-Vicens, I.; Blears, D.; Nojima, T.; et al. CDK12 globally stimulates RNA polymerase II transcription elongation and carboxyl-terminal domain phosphorylation. Nucl. Acids Res. 2020, 48, 7712–7727. [Google Scholar] [CrossRef] [PubMed]
- Narain, A.; Bhandare, P.; Adhikari, B.; Backes, S.; Eilers, M.; Dölken, L.; Schlosser, A.; Erhard, F.; Baluapuri, A.; Wolf, E. Targeted protein degradation reveals a direct role of SPT6 in RNAPII elongation and termination. Mol. Cell 2021, 81, 3110–3127. [Google Scholar] [CrossRef]
- Kohoutek, J.; Blazek, D. Cyclin K goes with Cdk12 and Cdk13. Cell Div. 2012, 7, 12–21. [Google Scholar] [CrossRef]
- Cenik, B.K.; Shilatifard, A. COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 2021, 22, 38–58. [Google Scholar] [CrossRef]
- O’Reilly, D.; Kuznetsova, O.V.; Laitem, C.; Zaborowska, J.; Dienstbier, M.; Murphy, S. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucl. Acids Res. 2014, 42, 264–275. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Egloff, S.; Vitali, P.; Tellier, M.; Raffel, R.; Murphy, S.; Kiss, T. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 2017, 36, 934–948. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Tellier, M.; Murphy, S. Incomplete removal of ribosomal RNA can affect chromatin RNA-seq data analysis. Transcription 2020, 11, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology resource: Enriching a GOld mine. Nucl. Acids Res. 2021, 49, D325–D334. [CrossRef]
- Fong, N.; Brannan, K.; Erickson, B.; Kim, H.; Cortazar, M.A.; Sheridan, R.M.; Nguyen, T.; Karp, S.; Bentley, D.L. Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition. Mol. Cell 2015, 60, 256–267. [Google Scholar] [CrossRef]
- Gonzalez, M.N.; Blears, D.; Svejstrup, J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2021, 22, 3–21. [Google Scholar] [CrossRef]
- Formosa, T.; Winston, F. The role of FACT in managing chromatin: Disruption, assembly, or repair? Nucl. Acids Res. 2020, 48, 11929–11941. [Google Scholar] [CrossRef]
- Zhang, Z.; Will, C.L.; Bertram, K.; Dybkov, O.; Hartmuth, K.; Agafonov, D.E.; Hofele, R.; Urlaub, H.; Kastner, B.; Lührmann, R.; et al. Molecular architecture of the human 17S U2 snRNP. Nature 2020, 583, 310–313. [Google Scholar] [CrossRef]
- Ni, Z.; Xu, C.; Guo, X.; Hunter, G.O.; Kuznetsova, O.V.; Tempel, W.; Marcon, E.; Zhong, G.; Guo, H.; Kuo, W.W.; et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat. Struct. Mol. Biol. 2014, 21, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.; Rondón, A.G.; Pérez-Calero, C.; Salas-Armenteros, I.; Aguilera, A. The THO Complex as a Paradigm for the Prevention of Cotranscriptional R-Loops. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2019; Volume 84, pp. 105–114. [Google Scholar]
- Friend, K.; Lovejoy, A.F.; Steitz, J.A. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol. Cell 2007, 28, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.W.; Ma, B.; Sadowski, C.L.; Kobayashi, R.; Hernandez, N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996, 15, 7129–7136. [Google Scholar] [CrossRef]
- Fan, Z.; Devlin, J.R.; Hogg, S.J.; Doyle, M.A.; Harrison, P.F.; Todorovski, I.; Cluse, L.A.; Knight, D.A.; Sandow, J.J.; Gregory, G.; et al. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci. Adv. 2020, 6, eaaz5041. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, M.; Dries, R.; Grassetti, A.V.; Dust, S.; Gao, Y.; Huang, H.; Sharma, B.; Day, D.S.; Kwiatkowski, N.; Pomaville, M.; et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 2019, 10, 1757. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.C.; Pombo, A.; Murphy, S. 2003 Interaction of proteins with promoter elements of the human U2 snRNA genes in vivo. Gene 2003, 315, 103–112. [Google Scholar] [CrossRef]
- Stunkel, W.; Kober, I.; Seifart, K.H. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol. Cell. Biol. 1997, 17, 4397–4405. [Google Scholar] [CrossRef][Green Version]
- Boyd, D.C.; Greger, I.H.; Murphy, S. In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene 2000, 247, 33–44. [Google Scholar] [CrossRef]
- Zhao, X.; Pendergrast, P.S.; Hernandez, N. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell 2001, 7, 539–549. [Google Scholar] [CrossRef]
- Tsui, C.; Inouye, C.; Levy, M.; Lu, A.; Florens, L.; Washburn, M.P.; Tjian, R. dCas9-targeted locus-specific protein isolation method identifies histone gene regulators. Proc. Natl. Acad. Sci. USA 2018, 115, E2734–E2741. [Google Scholar] [CrossRef]
| Name | Forward Primer | Reverse Primer |
|---|---|---|
| dCas9 EcoVR | GAATGATATCGTGCTGACCCTGACAC | GTCCACATCGTAGTCGGACAG |
| dCas9 FseI | GCCATCGTGCCTCAGAGCTTTC | GAACGGCCGGCCTTTTTCGTGG |
| dCas9 ExF Seq | GGGCACATACCACGATCTGCTG | |
| dCas9 Mut Seq | GGCAAGAGCGACAACGTGCCCTC | |
| dCas9 ExR Seq | GGATTCTCCTCGACGTCACCGC | |
| G1 | CACCCCGCCCTTTCACAGAGGGCG | AAACCGCCCTCTGTGAAAGGGCGG |
| G2 | CACCATGCCCCGCCCTTTCACAGA | AAACTCTGTGAAAGGGCGGGGCAT |
| G3 | CACCAAATGAAAGCCCGGGAACGC | AAACGCGTTCCCGGGCTTTCATTT |
| G4 | CACCTCTCATCCACATTCAAGTCG | CACCTCTCATCCACATTCAAGTCG |
| G5 | CACCGACGGTGACGGCGGGCGCGA | AAACTCGCGCCCGCCGTCACCGTC |
| G6 | CACCAAGGCGAGCGCATCGCTTCT | CACCAAGGCGAGCGCATCGCTTCT |
| G7 | CACCCCGTCTACCGCCCGCACATC | AAACGATGTGCGGGCGGTAGACGG |
| G8 | CACCGCACGCTGTCGTTTCCACCG | CACCGCACGCTGTCGTTTCCACCG |
| G9 | CACCCCGGAAGGGTTTCGCGTCAT | CACCCCGGAAGGGTTTCGCGTCAT |
| G10 | CACCAGTGGGTGGCGACCTTTTAA | CACCAGTGGGTGGCGACCTTTTAA |
| U2 N | GGAGCGGAGCGTTCTCTGTCTCCCC | AGAGTGTGAGCCCTCATTCACGCCC |
| U2 DSE | TGGCTCGATACGAACAAGGAAG | GTTCCCGGGCTTTCATTTCG |
| U2 UpPSE | GGGAACGCCGAAGAAGCACGGG | CCCCAGCCTCGCTCCTTGCCC |
| U2 PSE | ATGAGAGTGGGACGGTGA | CACTTGATCTTAGCCAAAAGG |
| U2 3′ box | ACGAGTCCTGTGACGCGCCGGCTTG | CTCCGGGTGGGTCCCATTCCTTTAA |
| U2 CT-rich | CCTCCCCGCCTCTCCCTCGCTC | GGACAAATAGCCAACGCATGCGG |
| U1 PSE | GGAAAGGGCTCGGGAGTGCGCG | CAGGTAAGTATGAGAGCTTGGGC |
| Chr6 off target | AGACTACACGATACAACATCCAC | AGCAGGAATCAGAACTCCCATC |
| Chr7 off target | AGCAGGAATCAGAACTCCCATC | ACCAAGGAGGAAAGGTAGTAGC |
| Antibodies | Reference |
|---|---|
| AffiniPure Donkey Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch 711-005-152 |
| Anti-CRISPR-Cas9 | Abcam ab191468 |
| CDK12 | LS Bio LS-C288466-100 |
| CDK12 | Novus Biologicals NB100-87011 |
| Cyclin K | Bethyl Laboratories A301-939A |
| Oct-1 | Bethyl Laboratories A301-717A |
| RNA Polymerase II | Novus Biologicals NBP2-32080 |
| SPT6 | Cell Signalling 15616 |
| Streptavidin (HRP) | Abcam ab7403 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guiro, J.; Fagbemi, M.; Tellier, M.; Zaborowska, J.; Barker, S.; Fournier, M.; Murphy, S. CAPTURE of the Human U2 snRNA Genes Expands the Repertoire of Associated Factors. Biomolecules 2022, 12, 704. https://doi.org/10.3390/biom12050704
Guiro J, Fagbemi M, Tellier M, Zaborowska J, Barker S, Fournier M, Murphy S. CAPTURE of the Human U2 snRNA Genes Expands the Repertoire of Associated Factors. Biomolecules. 2022; 12(5):704. https://doi.org/10.3390/biom12050704
Chicago/Turabian StyleGuiro, Joana, Mathias Fagbemi, Michael Tellier, Justyna Zaborowska, Stephanie Barker, Marjorie Fournier, and Shona Murphy. 2022. "CAPTURE of the Human U2 snRNA Genes Expands the Repertoire of Associated Factors" Biomolecules 12, no. 5: 704. https://doi.org/10.3390/biom12050704
APA StyleGuiro, J., Fagbemi, M., Tellier, M., Zaborowska, J., Barker, S., Fournier, M., & Murphy, S. (2022). CAPTURE of the Human U2 snRNA Genes Expands the Repertoire of Associated Factors. Biomolecules, 12(5), 704. https://doi.org/10.3390/biom12050704






