Curcumin and Ethanol Effects in Trembler-J Schwann Cell Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Primary Culture Enriched in SCs
2.3. Curcumin
2.4. Determination of Curcumin Concentration
2.5. Curcumin Treatment
2.6. Immunolabeling in Enriched SC Cultures
2.7. Confocal Microscopy
2.8. Fluorescence Image Analysis
2.9. Statistical Analysis
3. Results
3.1. Morphological Differences between FBs and SCs
3.2. Obtaining of Cultures Enriched in +/+ and TrJ/+ SCs
3.3. Determination of Suitable Curcumin Concentration to Be Used in Enriched +/+ and TrJ/+ SC Cultures
3.4. Effect of Curcumin Treatment on SC +/+ and TrJ/+ Enriched Cultures
3.4.1. Alterations in PMP22 Expression
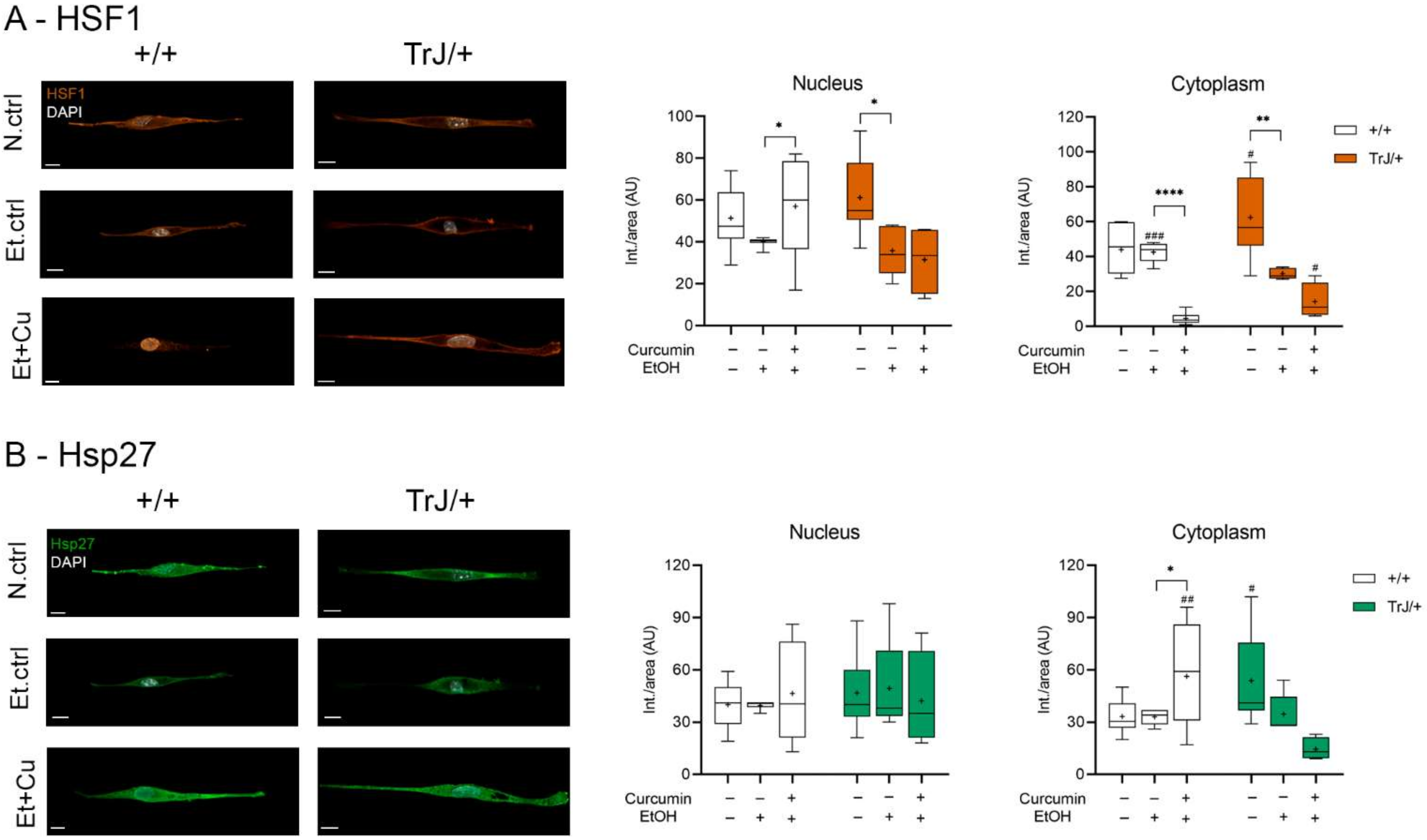
3.4.2. Effect of Curcumin on Heat Shock Response: Expression of HSF1 and Hsp27
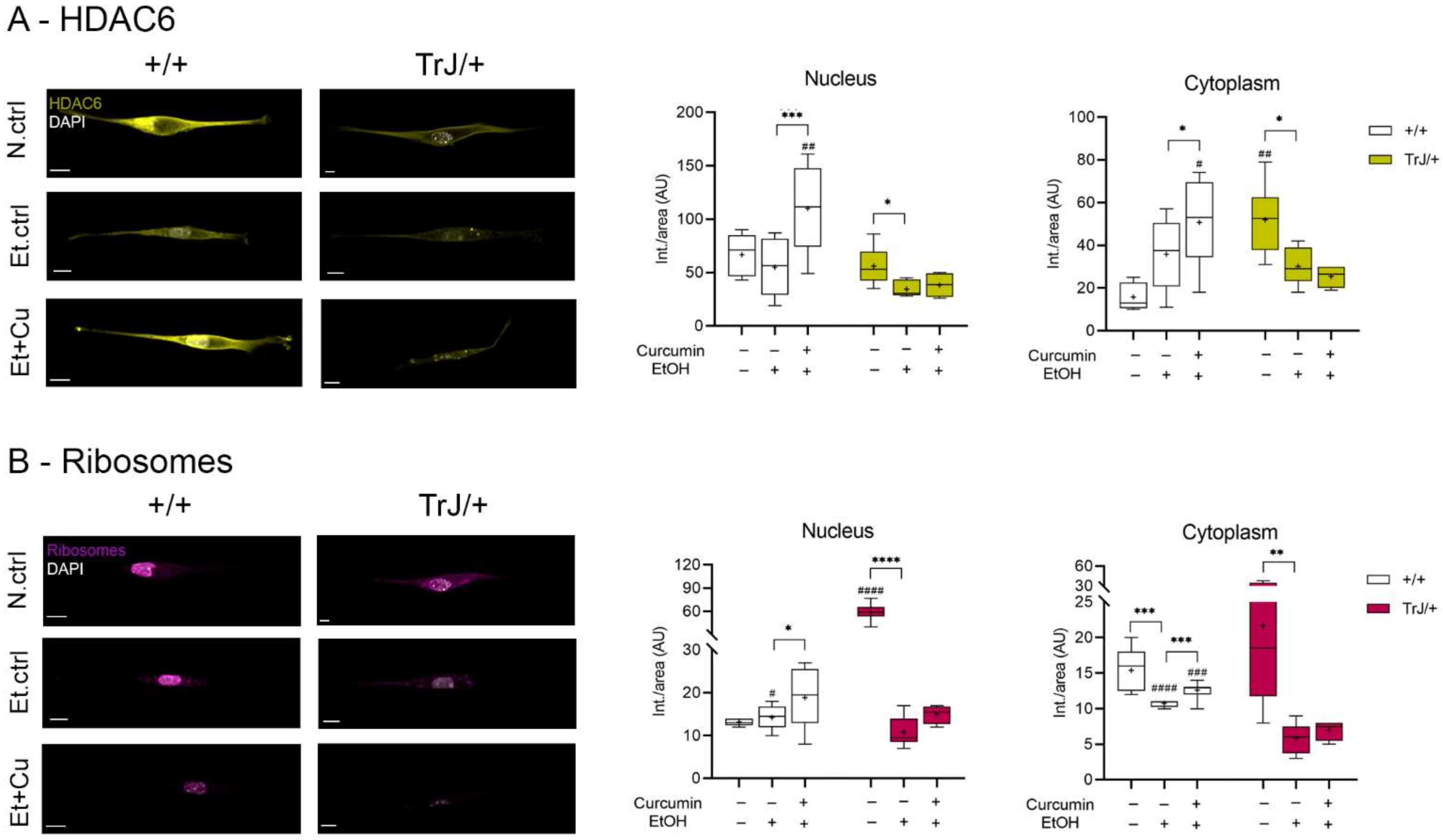
3.4.3. Effect of Curcumin on Autophagy/mTOR: Expression of HDAC6 and Ribosomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skre, H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin. Genet. 1974, 6, 98–118. [Google Scholar] [CrossRef]
- Vallat, J.M. Dominantly inherited peripheral neuropathies. J. Neuropathol. Exp. Neurol. 2003, 62, 699–714. [Google Scholar] [CrossRef][Green Version]
- Sereda, M.W.; Nave, K.A. Animal models of Charcot-Marie-Tooth disease type 1A. Neuromolecular Med. 2006, 8, 205–215. [Google Scholar] [CrossRef]
- Li, J.; Parker, B.; Martyn, C.; Natarajan, C.; Guo, J. The PMP22 gene and its related diseases. Mol. Neurobiol. 2013, 47, 673–698. [Google Scholar] [CrossRef]
- Parmantier, E.; Braun, C.; Thomas, J.L.; Peyron, F.; Martinez, S.; Zalc, B. PMP-22 expression in the central nervous system of the embryonic mouse defines potential transverse segments and longitudinal columns. J. Comp. Neurol. 1997, 378, 159–172. [Google Scholar] [CrossRef]
- Parmantier, E.; Cabon, F.; Braun, C.; D’Urso, D.; Müller, H.W.; Zalc, B. Peripheral Myelin Protein-22 is Expressed in Rat and Mouse Brain and Spinal Cord Motoneurons. Eur. J. Neurosci. 1995, 7, 1080–1088. [Google Scholar] [CrossRef]
- Chanson, J.B.; Echaniz-Laguna, A.; Blanc, F.; Lacour, A.; Ballonzoli, L.; Kremer, S.; Namer, I.J.; Lannes, B.; Tranchant, C.; Vermersch, P.; et al. Central nervous system abnormalities in patients with PMP22 gene mutations: A prospective study. J. Neurol. Neurosurg. Psychiatry 2013, 84, 392–397. [Google Scholar] [CrossRef]
- Damián, J.P.; Vázquez Alberdi, L.; Canclini, L.; Rosso, G.; Bravo, S.O.; Martínez, M.; Uriarte, N.; Ruiz, P.; Calero, M.; Di Tomaso, M.V.; et al. Central Alteration in Peripheral Neuropathy of Trembler-J Mice: Hippocampal pmp22 Expression and Behavioral Profile in Anxiety Tests. Biomolecules 2021, 11, 601. [Google Scholar] [CrossRef]
- Di Tomaso, M.V.; Vázquez Alberdi, L.; Olsson, D.; Cancela, S.; Fernández, A.; Rosillo, J.C.; Reyes Ábalos, A.L.; Álvarez Zabaleta, M.; Calero, M.; Kun, A. Colocalization Analysis of Peripheral Myelin Protein-22 and Lamin-B1 in the Schwann Cell Nuclei of Wt and TrJ Mice. Biomolecules 2022, 12, 456. [Google Scholar] [CrossRef]
- Hou, J.; Wang, L.; Zhao, J.; Zhuo, H.; Cheng, J.; Chen, X.; Zheng, W.; Hong, Z.; Cai, J. Inhibition of protein PMP22 enhances etoposide-induced cell apoptosis by p53 signaling pathway in Gastric Cancer. Int. J. Biol. Sci. 2021, 17, 3145–3157. [Google Scholar] [CrossRef]
- Sociali, G.; Visigalli, D.; Prukop, T.; Cervellini, I.; Mannino, E.; Venturi, C.; Bruzzone, S.; Sereda, M.W.; Schenone, A. Tolerability and efficacy study of P2X7 inhibition in experimental Charcot-Marie-Tooth type 1A (CMT1A) neuropathy. Neurobiol. Dis. 2016, 95, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Simske, J.S. Vertebrate Claudin/PMP22/EMP22/MP20 family protein TMEM47 regulates epithelial cell junction maturation and morphogenesis. Dev. Dyn. 2016, 245, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Heid, H.; Zimbelmann, R.; Kuhn, C.; Winter-Simanowski, S.; Dörflinger, Y.; Grund, C.; Rickelt, S. Transmembrane protein PERP is a component of tessellate junctions and of other junctional and non-junctional plasma membrane regions in diverse epithelial and epithelium-derived cells. Cell Tissue Res. 2013, 353, 99–115. [Google Scholar] [CrossRef]
- Sereda, M.W.; Meyer Zu Hörste, G.; Suter, U.; Uzma, N.; Nave, K.A. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A). Nat. Med. 2003, 9, 1533–1537. [Google Scholar] [CrossRef]
- Maeda, Y.; Kataoka, Y.; Sugaya, A.; Kariya, S.; Kobayashi, K.; Nishizaki, K. Steroid-dependent sensorineural hearing loss in a patient with Charcot-Marie-Tooth disease showing auditory neuropathy. Auris. Nasus. Larynx 2015, 42, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Zu Horste, G.M.; Prukop, T.; Liebetanz, D.; Mobius, W.; Nave, K.A.; Sereda, M.W. Antiprogesterone therapy uncouples axonal loss from demyelination in a transgenic rat model of CMT1A neuropathy. Ann. Neurol. 2007, 61, 61–72. [Google Scholar] [CrossRef]
- Attardi, L.D.; Reczek, E.E.; Cosmas, C.; Demicco, E.G.; McCurrach, M.E.; Lowe, S.W.; Jacks, T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000, 14, 1835. [Google Scholar] [CrossRef]
- Bird, T.D. Charcot-Marie-Tooth (CMT) Hereditary Neuropathy Overview; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Fortun, J.; Dunn, W.A.; Joy, S.; Li, J.; Notterpek, L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 2003, 23, 10672–10680. [Google Scholar] [CrossRef]
- Okamoto, Y.; Pehlivan, D.; Wiszniewski, W.; Beck, C.R.; Snipes, G.J.; Lupski, J.R.; Khajavi, M. Curcumin facilitates a transitory cellular stress response in trembler-J mice. Hum. Mol. Genet. 2013, 22, 4698–4705. [Google Scholar] [CrossRef]
- Suter, U.; Scherer, S.S. Disease mechanisms in inherited neuropathies. Nat. Rev. Neurosci. 2003, 4, 714–726. [Google Scholar] [CrossRef]
- Jaradeh, S.S. Hereditary Neuropathies. J. Clin. Neuromuscul. Dis. 2003, 5, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Valentijn, L.J.; Baas, F.; Wolterman, R.A.; Hoogendijk, J.E.; van den Bosch, N.H.A.; Zorn, I.; Gabreëls-Festen, A.A.W.M.; de Visser, M.; Bolhuis, P.A. Identical point mutations of PMP-22 in Trembler-J mouse and Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992, 2, 288–291. [Google Scholar] [CrossRef]
- Notterpek, L.; Ryan, M.C.; Tobler, A.R.; Shooter, E.M. PMP22 accumulation in aggresomes: Implications for CMT1A pathology. Neurobiol. Dis. 1999, 6, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Notterpek, L.; Snipes, G.J.; Naef, R.; Sossin, W.; Laliberté, J.; Iacampo, S.; Suter, U.; Shooter, E.M.; Murphy, R.A. Neurons promote the translocation of peripheral myelin protein 22 into myelin. J. Neurosci. 1997, 17, 7754–7762. [Google Scholar] [CrossRef] [PubMed]
- Kun, A.; Rosso, G.; Canclini, L.; Bresque, M.; Romeo, C.; Cal, K.; Calliari, A.; Hanuz, A.; Roberto, J.; Roberto, J. The Schwann Cell-Axon Link in Normal Condition or Neuro-Degenerative Diseases: An Immunocytochemical Approach. In Applications of Immunocytochemistry; Dehghani, H., Ed.; InTech: London, UK, 2012; pp. 249–266. ISBN 978-953-51-5235-4. [Google Scholar]
- Myers, J.K.; Mobley, C.K.; Sanders, C.R. The Peripheral Neuropathy-Linked Trembler and Trembler-J Mutant Forms of Peripheral Myelin Protein 22 are Folding-Destabilized. Biochemistry 2008, 47, 10620–10629. [Google Scholar] [CrossRef]
- Pantera, H.; Shy, M.E.; Svaren, J. Regulating PMP22 expression as a dosage sensitive neuropathy gene. Brain Res. 2020, 1726, 146491. [Google Scholar] [CrossRef]
- Kun, A.; Canclini, L.; Rosso, G.; Bresque, M.; Romeo, C.; Hanusz, A.; Cal, K.; Calliari, A.; Sotelo Silveira, J.; Sotelo, J.R. F-actin distribution at nodes of Ranvier and Schmidt-Lanterman incisures in mammalian sciatic nerves. Cytoskeleton 2012, 69, 486–495. [Google Scholar] [CrossRef]
- Rosso, G.; Negreira, C.; Sotelo, J.R.; Kun, A. Myelinating and demyelinating phenotype of Trembler-J mouse (a model of Charcot-Marie-Tooth human disease) analyzed by atomic force microscopy and confocal microscopy. J. Mol. Recognit. 2012, 25, 247–255. [Google Scholar] [CrossRef]
- Notterpek, L.; Shooter, E.M.; Snipes, G.J. Upregulation of the endosomal-lysosomal pathway in the Trembler-J neuropathy. J. Neurosci. 1997, 17, 4190–4200. [Google Scholar] [CrossRef]
- Khalil, A.A.; Kabapy, N.F.; Deraz, S.F.; Smith, C. Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets? Biochim. Biophys. Acta-Rev. Cancer 2011, 1816, 89–104. [Google Scholar] [CrossRef]
- Borges, T.J.; Wieten, L.; Van Herwijnen, M.J.C.; Broere, F.; Van der Zee, R.; Bonorino, C.; Van Eden, W. The anti-inflammatory mechanisms of Hsp70. Front. Immunol. 2012, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.D.; Prince, T.; Gong, J.; Calderwood, S.K. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS ONE 2012, 7, e39679. [Google Scholar] [CrossRef]
- Morimoto, R.I. Regulation of the heat-shock transcriptional response: Cross talk between a family of heat-shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Chang, C.W.; Róna, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.S.; Fontes, M.R.M.; Stewart, M.; Vértessy, B.G.; Forwood, J.K.; et al. Structural Biology and Regulation of Protein Import into the Nucleus. J. Mol. Biol. 2016, 428, 2060–2090. [Google Scholar] [CrossRef]
- Home, T.; Jensen, R.A.; Rao, R. Heat shock factor 1 in protein homeostasis and oncogenic signal integration. Cancer Res. 2015, 75, 907–912. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsujimura, A.; Taguchi, K.; Tanaka, M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 2017, 13, 133–148. [Google Scholar] [CrossRef]
- Schäfer, C.; Seeliger, H.; Bader, D.C.; Assmann, G.; Buchner, D.; Guo, Y.; Ziesch, A.; Palagyi, A.; Ochs, S.; Laubender, R.P.; et al. Heat shock protein 27 as a prognostic and predictive biomarker in pancreatic ductal adenocarcinoma. J. Cell. Mol. Med. 2012, 16, 1776–1791. [Google Scholar] [CrossRef]
- Garrido, C.; Brunet, M.; Didelot, C.; Zermati, Y.; Schmitt, E.; Kroemer, G. Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties. Cell Cycle 2006, 5, 2592–2601. [Google Scholar] [CrossRef]
- Arrigo, A.-P.; Gibert, B. HspB1, HspB5 and HspB4 in Human Cancers: Potent Oncogenic Role of Some of Their Client Proteins. Cancers 2014, 6, 333–365. [Google Scholar] [CrossRef]
- Choi, S.; Lee, H.-J.; Jin, Y.B.; Jang, J.; Kang, G.; Lee, M.; Kim, C.-H.; Kim, J.; Yoon, S.S.; Lee, Y.; et al. MMP9 processing of HSPB1 regulates tumor progression. PLoS ONE 2014, 9, e85509. [Google Scholar] [CrossRef]
- Vidyasagar, A.; Wilson, N.A.; Djamali, A. Heat shock protein 27 (HSP27): Biomarker of disease and therapeutic target. Firogenes Tissue Repairs 2012, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Matsuzaki, H.; Tanaka, M.; Takemura, Y.; Kuroda, S.; Ono, Y.; Kikkawa, U. Activation of protein kinase B (Akt/RAC-protein kinase) by cellular stress and its association with heat shock protein Hsp27. FEBS Lett. 1997, 410, 493–498. [Google Scholar] [CrossRef]
- Wu, R.; Kausar, H.; Johnson, P.; Montoya-Durango, D.E.; Merchant, M.; Rane, M.J. Hsp27 regulates Akt activation and polymorphonuclear leukocyte apoptosis by scaffolding MK2 to Akt signal complex. J. Biol. Chem. 2007, 282, 21598–21608. [Google Scholar] [CrossRef] [PubMed]
- Rane, M.J.; Pan, Y.; Singh, S.; Powell, D.W.; Wu, R.; Cummins, T.; Chen, Q.; McLeish, K.R.; Klein, J.B. Heat shock protein 27 controls apoptosis by regulating Akt activation. J. Biol. Chem. 2003, 278, 27828–27835. [Google Scholar] [CrossRef]
- Chittoor-Vinod, V.G.; Lee, S.; Judge, S.M.; Notterpek, L. Inducible HSP70 is critical in preventing the aggregation and enhancing the processing of PMP22. ASN Neuro 2015, 7, 1759091415569909. [Google Scholar] [CrossRef] [PubMed]
- Fortun, J.; Verrier, J.D.; Go, J.C.; Madorsky, I.; Dunn, W.A.; Notterpek, L. The formation of peripheral myelin protein 22 aggregates is hindered by the enhancement of autophagy and expression of cytoplasmic chaperones. Neurobiol. Dis. 2007, 25, 252–265. [Google Scholar] [CrossRef]
- Benoy, V.; Vanden Berghe, P.; Jarpe, M.; Van Damme, P.; Robberecht, W.; Van Den Bosch, L. Development of Improved HDAC6 Inhibitors as Pharmacological Therapy for Axonal Charcot–Marie–Tooth Disease. Neurotherapeutics 2017, 14, 417–428. [Google Scholar] [CrossRef]
- Novelle, M.G.; Davis, A.; Price, N.L.; Ali, A.; Fürer-Galvan, S.; Zhang, Y.; Becker, K.; Bernier, M.; de Cabo, R. Caloric restriction induces heat shock response and inhibits B16F10 cell tumorigenesis both in vitro and in vivo. Aging (Albany. N. Y.) 2015, 7, 233–240. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Zhang, C.-X.; Liu, Y.-M.; Chen, K.-L.; Chen, G. A comparative study of anti-aging properties and mechanism: Resveratrol and caloric restriction. Oncotarget 2017, 8, 65717–65729. [Google Scholar] [CrossRef]
- Soh, J.W.; Marowsky, N.; Nichols, T.J.; Rahman, A.M.; Miah, T.; Sarao, P.; Khasawneh, R.; Unnikrishnan, A.; Heydari, A.R.; Silver, R.B.; et al. Curcumin is an early-acting stage-specific inducer of extended functional longevity in Drosophila. Exp. Gerontol. 2013, 48, 229–239. [Google Scholar] [CrossRef]
- Sinclair, D.A. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005, 126, 987–1002. [Google Scholar] [CrossRef]
- Kume, S.; Uzu, T.; Kashiwagi, A.; Koya, D. SIRT1, a Calorie Restriction Mimetic, in a New Therapeutic Approach for Type 2 Diabetes Mellitus and Diabetic Vascular Complications. Endocr. Metab. Immune Disord. Drug Targets 2012, 10, 16–24. [Google Scholar] [CrossRef]
- Willcox, B.J.; Willcox, D.C. Caloric restriction, caloric restriction mimetics, and healthy aging in Okinawa: Controversies and clinical implications. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Garg, G.; Singh, A.K.; Bissoyi, A.; Rizvi, S.I. Fisetin, a potential caloric restriction mimetic, attenuates senescence biomarkers in rat erythrocytes. Biochem. Cell Biol. 2019, 97, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Ingram, D.K.; Zhu, M.; Mamczarz, J.; Zou, S.; Lane, M.A.; Roth, G.S.; deCabo, R. Calorie restriction mimetics: An emerging research field. Aging Cell 2006, 5, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Pietrocola, F.; Madeo, F.; Kroemer, G. Caloric restriction mimetics: Natural/physiological pharmacological autophagy inducers. Autophagy 2014, 10, 1879–1882. [Google Scholar] [CrossRef]
- Madeo, F.; Pietrocola, F.; Eisenberg, T.; Kroemer, G. Caloric restriction mimetics: Towards a molecular definition. Nat. Rev. Drug Discov. 2014, 13, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, P.; Alam, A.; Choksi, A.; Chattopadhyay, S.; Poddar, P. Retention of Anticancer Activity of Curcumin after Conjugation with Fluorescent Gold Quantum Clusters: An in Vitro and in Vivo Xenograft Study. ACS Omega 2018, 3, 4776–4785. [Google Scholar] [CrossRef]
- Beevers, C.S.; Li, F.; Liu, L.; Huang, S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int. J. Cancer 2006, 119, 757–764. [Google Scholar] [CrossRef]
- Watson, J.L.; Hill, R.; Yaffe, P.B.; Greenshields, A.; Walsh, M.; Lee, P.W.; Giacomantonio, C.A.; Hoskin, D.W. Curcumin causes superoxide anion production and p53-independent apoptosis in human colon cancer cells. Cancer Lett. 2010, 297, 1–8. [Google Scholar] [CrossRef]
- Caillaud, M.; Msheik, Z.; Ndong-Ntoutoume, G.M.A.; Vignaud, L.; Richard, L.; Favreau, F.; Faye, P.A.; Sturtz, F.; Granet, R.; Vallat, J.M.; et al. Curcumin–cyclodextrin/cellulose nanocrystals improve the phenotype of Charcot-Marie-Tooth-1A transgenic rats through the reduction of oxidative stress. Free Radic. Biol. Med. 2020, 161, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Barzegar, A.; Moosavi-Movahedi, A.A. Intracellular ROS protection efficiency and free radical-scavenging activity of curcumin. PLoS ONE 2011, 6, e26012. [Google Scholar] [CrossRef] [PubMed]
- Perugini, J.; Di Mercurio, E.; Tossetta, G.; Severi, I.; Monaco, F.; Reguzzoni, M.; Tomasetti, M.; Dani, C.; Cinti, S.; Giordano, A. Biological Effects of Ciliary Neurotrophic Factor on hMADS Adipocytes. Front. Endocrinol. (Lausanne) 2019, 10. [Google Scholar] [CrossRef]
- Kang, S.K.; Cha, S.H.; Jeon, H.G. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006, 15, 165–174. [Google Scholar] [CrossRef]
- Liu, X.; You, L.; Tarafder, S.; Zou, L.; Fang, Z.; Chen, J.; Lee, C.H.; Zhang, Q. Curcumin-releasing chitosan/aloe membrane for skin regeneration. Chem. Eng. J. 2019, 359, 1111–1119. [Google Scholar] [CrossRef]
- Thaloor, D.; Miller, K.J.; Gephart, J.; Mitchell, P.O.; Pavlath, G.K. Systemic administration of the NF-κB inhibitor curcumin stimulates muscle regeneration after traumatic injury. Am. J. Physiol.-Cell Physiol. 1999, 277, C320–C329. [Google Scholar] [CrossRef]
- Khajavi, M.; Shiga, K.; Wiszniewski, W.; He, F.; Shaw, C.A.; Yan, J.; Wensel, T.G.; Snipes, G.J.; Lupski, J.R. Oral Curcumin Mitigates the Clinical and Neuropathologic Phenotype of the Trembler-J Mouse: A Potential Therapy for Inherited Neuropathy. Am. J. Hum. Genet. 2007, 81, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Fortun, J.; Go, J.C.; Li, J.; Amici, S.A.; Dunn, W.A.; Notterpek, L. Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiol. Dis. 2006, 22, 153–164. [Google Scholar] [CrossRef]
- Rosso, G.; Liashkovich, I.; Young, P.; Röhr, D.; Shahin, V. Schwann cells and neurite outgrowth from embryonic dorsal root ganglions are highly mechanosensitive. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 493–501. [Google Scholar] [CrossRef]
- Kreider, B.Q.; Messing, A.; Doan, H.; Kim, S.U.; Lisak, R.P.; Pleasure, D.E. Enrichment of Schwann cell cultures from neonatal rat sciatic nerve by differential adhesion. Brain Res. 1981, 3, 54–67. [Google Scholar] [CrossRef]
- Li, R. Culture methods for selective growth of normal rat and human Schwann cells. Methods Cell Biol. 1998, 57, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Mauritz, C.; Grothe, C.; Haastert, K. Comparative study of cell culture and purification methods to obtain highly enriched cultures of proliferating adult rat Schwann cells. J. Neurosci. Res. 2004, 77, 453–461. [Google Scholar] [CrossRef]
- Caillaud, M.; Chantemargue, B.; Richard, L.; Vignaud, L.; Favreau, F.; Faye, P.-A.; Vignoles, P.; Sturtz, F.; Trouillas, P.; Vallat, J.; et al. Local low dose curcumin treatment improves functional recovery and remyelination in a rat model of sciatic nerve crush through inhibition of oxidative stress. Neuropharmacology 2018, 139, 98–116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, K.; Hu, F.; Chen, L.; Zhang, X.; Wang, F.; Yan, B. Protective effects of natural compounds against oxidative stress in ischemic diseases and cancers via activating the Nrf2 signaling pathway: A mini review. J. Biochem. Mol. Toxicol. 2020, 35, 1–13. [Google Scholar] [CrossRef]
- Lundvig, D.M.S.; Pennings, S.W.C.; Brouwer, K.M.; Mtaya-Mlangwa, M.; Mugonzibwa, E.A.; Kuijpers-Jagtman, A.M.; Von den Hoff, J.W.; Wagener, F.A.D.T.G. Curcumin induces differential expression of cytoprotective enzymes but similar apoptotic responses in fibroblasts and myofibroblasts. Exp. Cell Res. 2015, 330, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, Q.; Yang, C.; Yang, H.; Rao, J.; Zhang, X. Curcumin exerts anti-tumor effects on diffuse large B cell lymphoma via regulating PPARγ expression. Biochem. Biophys. Res. Commun. 2020, 524, 70–76. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Zhou, Y.; Bazick, H.; Miles, J.R.; Fethiere, A.I.; Al Salihi, M.O.; Fazio, S.; Tavori, H.; Notterpek, L. A neutral lipid-enriched diet improves myelination and alleviates peripheral nerve pathology in neuropathic mice. Exp. Neurol. 2019, 321, 113031. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Fortun, J.; Li, J.; Go, J.C.; Fenstermaker, A.; Fletcher, B.S.; Notterpek, L. Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J. Neurochem. 2005, 92, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Shooter, E.M.; Notterpek, L. Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol. Dis. 2002, 10, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Suter, U.; Snipes, G.J.; Welcher, A.A.; Shooter, E.M.; Murphy, R.A. Detection and processing of peripheral myelin protein PMP22 in cultured Schwann cells. J. Biol. Chem. 1993, 268, 10372–10379. [Google Scholar] [CrossRef]
- Tello Velasquez, J.; Watts, M.E.; Todorovic, M.; Nazareth, L.; Pastrana, E.; Diaz-Nido, J.; Lim, F.; Ekberg, J.A.K.; Quinn, R.J.; St John, J.A. Low-dose curcumin stimulates proliferation, migration and phagocytic activity of olfactory ensheathing cells. PLoS ONE 2014, 9, e111787. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Li, Q. Curcumin accelerates the repair of sciatic nerve injury in rats through reducing Schwann cells apoptosis and promoting myelinization. Biomed. Pharmacother. 2017, 92, 1103–1110. [Google Scholar] [CrossRef]
- Abuelba, H.; Cotrutz, C.E.; Stoica, B.A.; Stoica, L.; Olinici, D.; Petreuş, T. In vitro evaluation of curcumin effects on breast adenocarcinoma 2D and 3D cell cultures. Rom. J. Morphol. Embryol. 2015, 56, 71–76. [Google Scholar] [PubMed]
- Galvao, J.; Davis, B.; Tilley, M.; Normando, E.; Duchen, M.R.; Cordeiro, M.F. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J. 2014, 28, 1317–1330. [Google Scholar] [CrossRef]
- Ilieva, Y.; Dimitrova, L.; Zaharieva, M.M.; Kaleva, M.; Alov, P.; Tsakovska, I.; Pencheva, T.; Pencheva-El Tibi, I.; Najdenski, H.; Pajeva, I. Cytotoxicity and Microbicidal Activity of Commonly Used Organic Solvents: A Comparative Study and Application to a Standardized Extract from Vaccinium macrocarpon. Toxics 2021, 9, 92. [Google Scholar] [CrossRef]
- Adler, S.; Pellizzer, C.; Paparella, M.; Hartung, T.; Bremer, S. The effects of solvents on embryonic stem cell differentiation. Toxicol. Vitr. 2006, 20, 265–271. [Google Scholar] [CrossRef]
- Müller, T.E.; Nunes, M.E.M.; Rodrigues, N.R.; Fontana, B.D.; Hartmann, D.D.; Franco, J.L.; Rosemberg, D.B. Neurochemical mechanisms underlying acute and chronic ethanol-mediated responses in zebrafish: The role of mitochondrial bioenergetics. Neurochem. Int. 2019, 131, 104584. [Google Scholar] [CrossRef]
- Pereira, R.B.; Andrade, P.B.; Valentão, P. A Comprehensive View of the Neurotoxicity Mechanisms of Cocaine and Ethanol. Neurotox. Res. 2015, 28, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Luo, J. Endoplasmic reticulum stress and ethanol neurotoxicity. Biomolecules 2015, 5, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, A.-P. Structure-functions of HspB1 (Hsp27). Methods Mol. Biol. 2011, 787, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Strauch, A.; Haslbeck, M. The function of small heat-shock proteins and their implication in proteostasis. Essays Biochem. 2016, 60, 163–172. [Google Scholar] [CrossRef]
- Huang, C.; Wu, J.; Xu, L.; Wang, J.; Chen, Z.; Yang, R. Regulation of HSF1 protein stabilization: An updated review. Eur. J. Pharmacol. 2018, 822, 69–77. [Google Scholar] [CrossRef]
- Lee, J.Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010, 29, 969–980. [Google Scholar] [CrossRef]
- Richter-Landsberg, C.; Leyk, J. Inclusion body formation, macroautophagy, and the role of HDAC6 in neurodegeneration. Acta Neuropathol. 2013, 126, 793–807. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef]
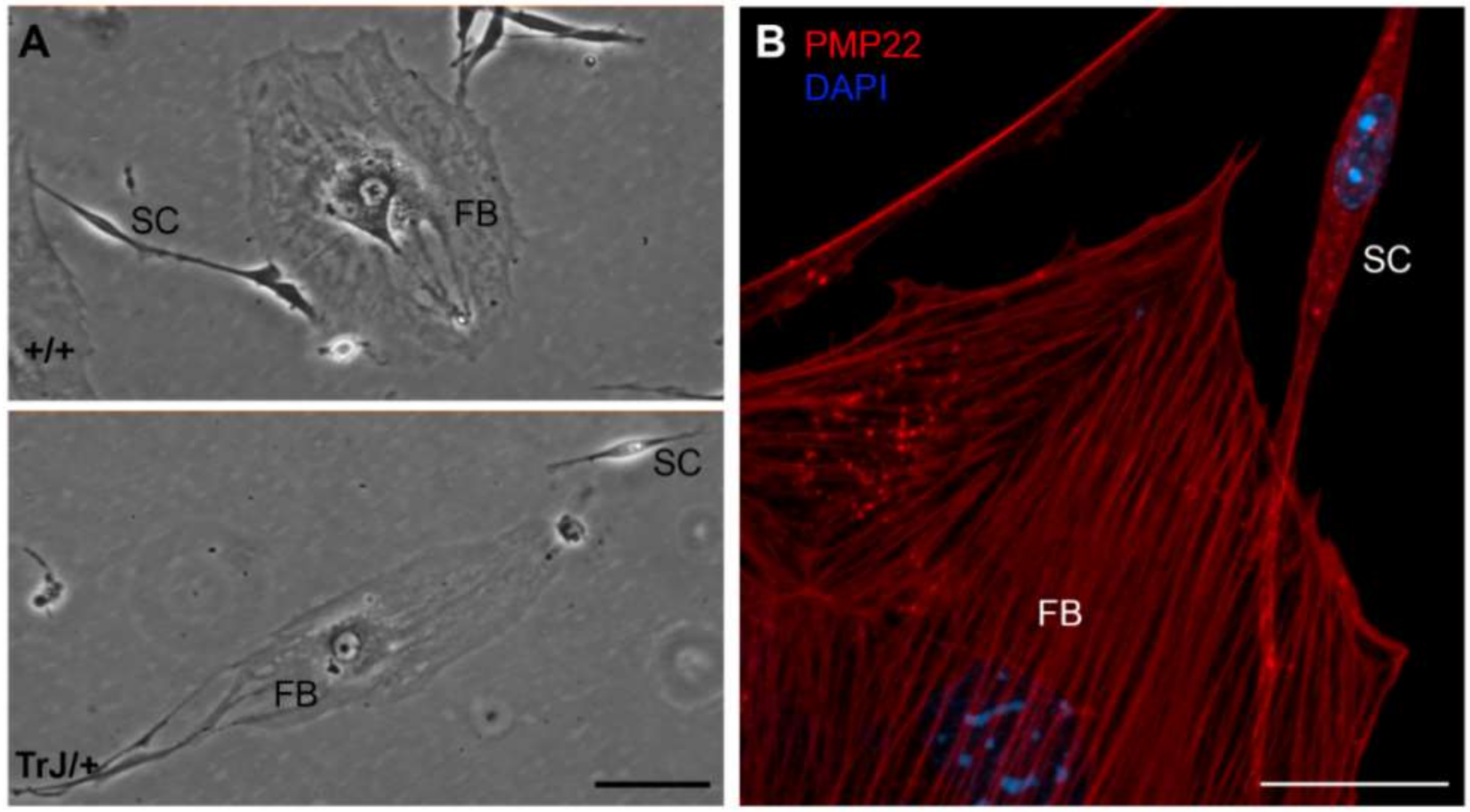
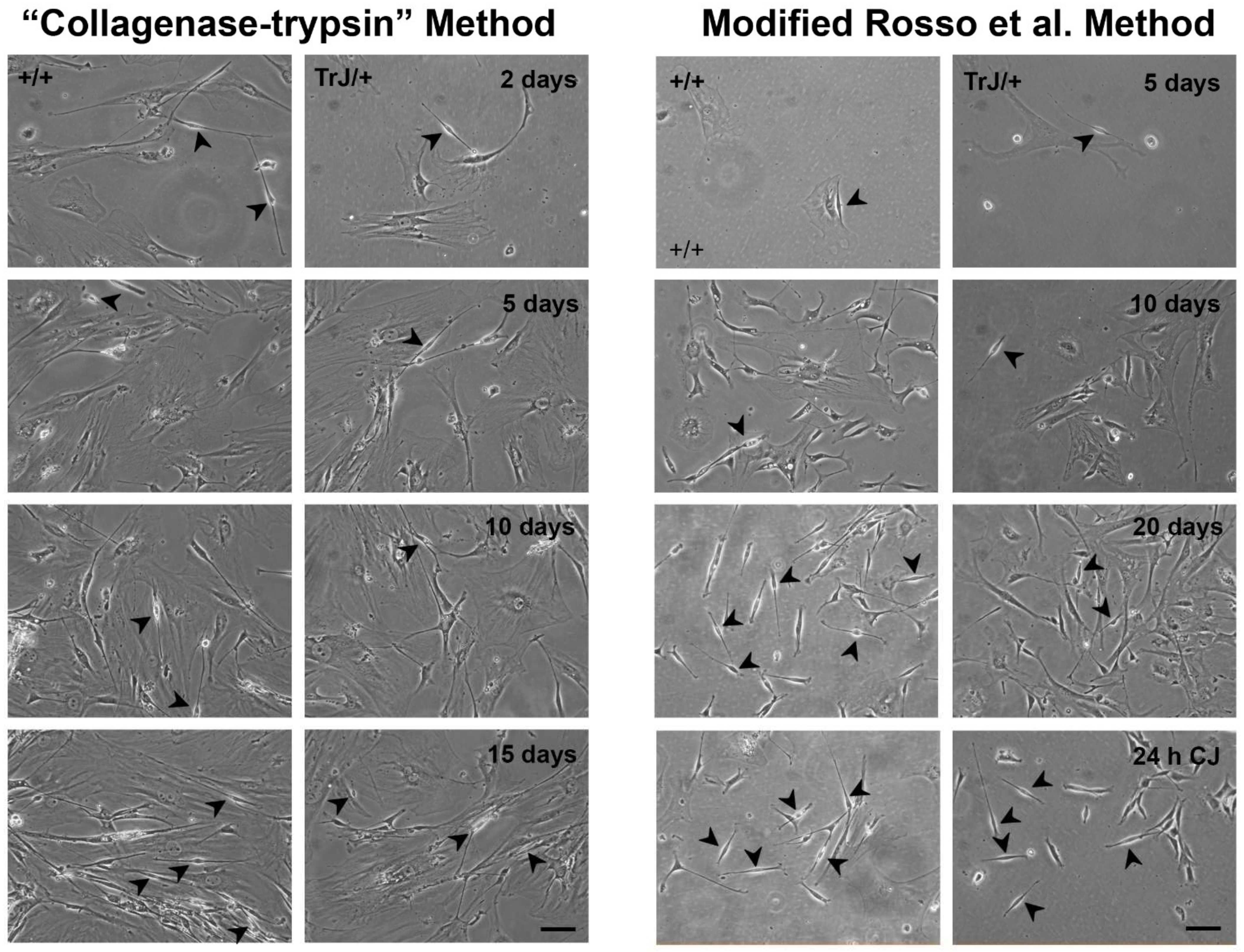
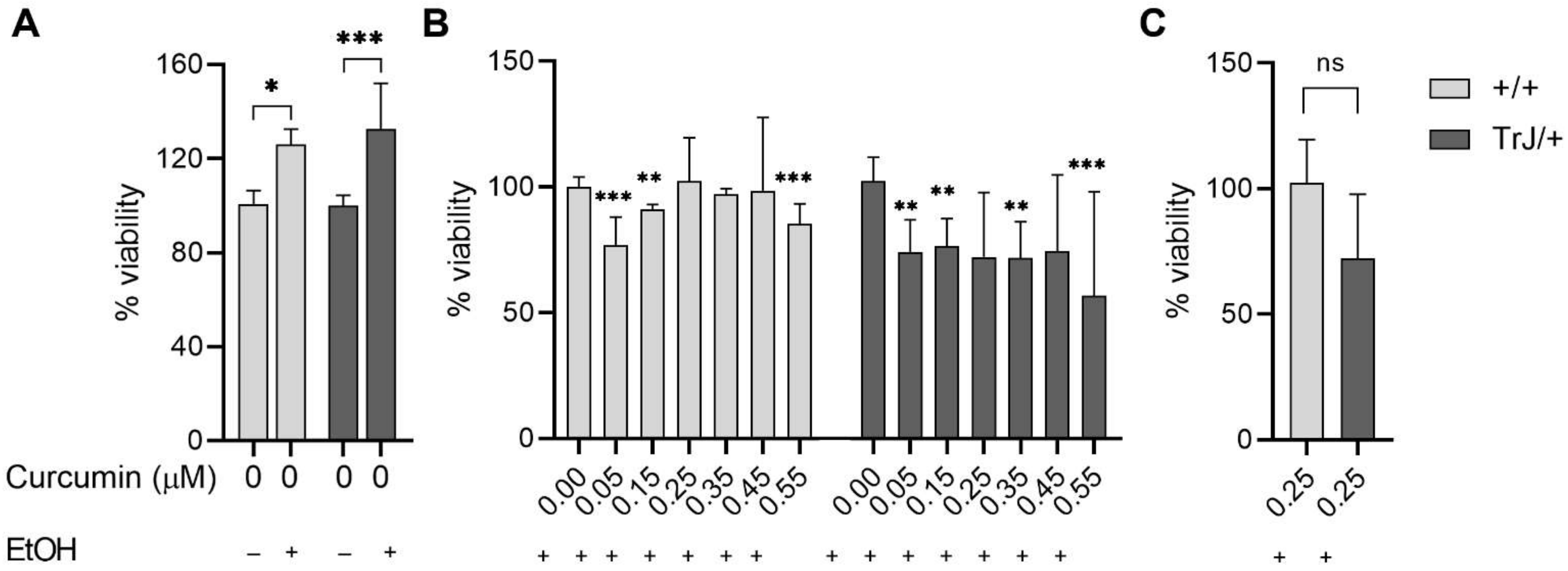
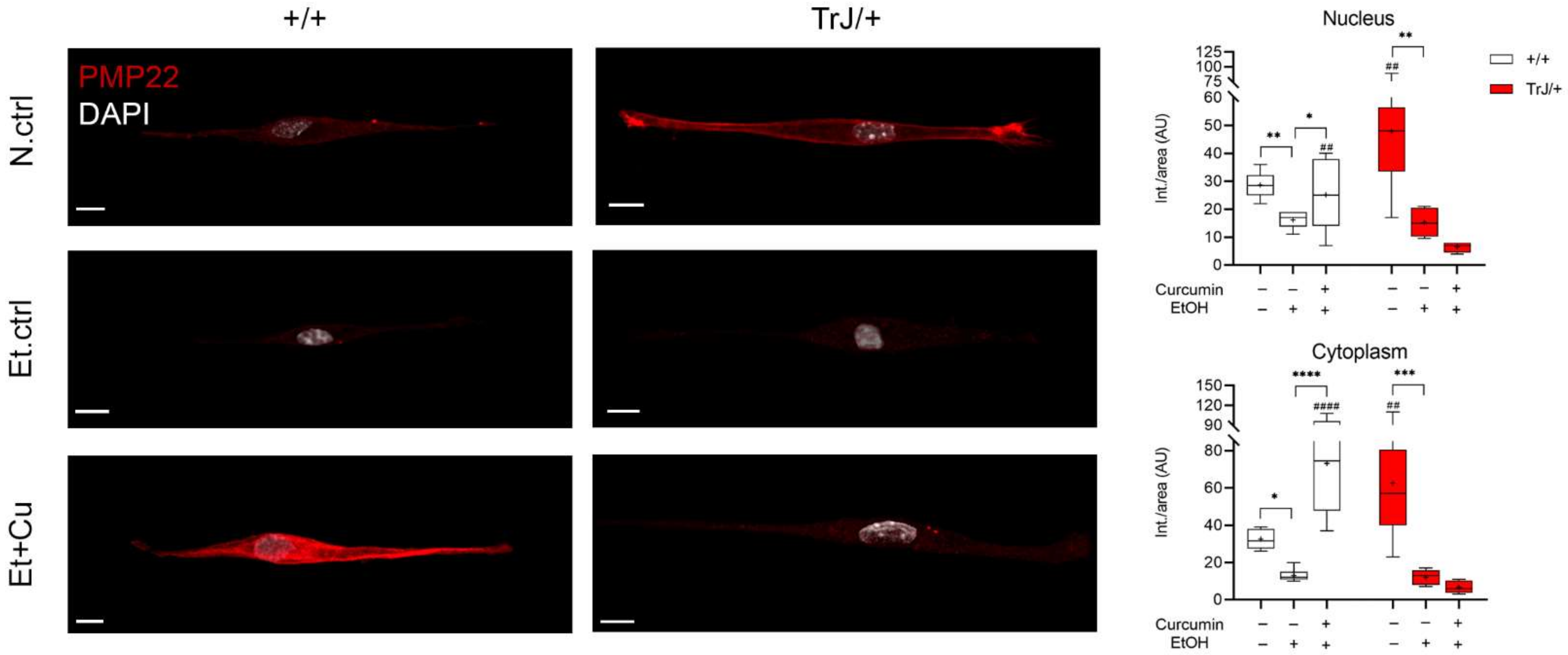
| Cell Type | Area (µm2) | Major Length (µm) | Minor Length (µm) |
|---|---|---|---|
| FB | 180,554 ± 27,043 | 615 ± 61 | 275 ± 41 |
| SC | 6062 ± 699 | 456 ± 51 | 30 ± 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez Alberdi, L.; Rosso, G.; Velóz, L.; Romeo, C.; Farias, J.; Di Tomaso, M.V.; Calero, M.; Kun, A. Curcumin and Ethanol Effects in Trembler-J Schwann Cell Culture. Biomolecules 2022, 12, 515. https://doi.org/10.3390/biom12040515
Vázquez Alberdi L, Rosso G, Velóz L, Romeo C, Farias J, Di Tomaso MV, Calero M, Kun A. Curcumin and Ethanol Effects in Trembler-J Schwann Cell Culture. Biomolecules. 2022; 12(4):515. https://doi.org/10.3390/biom12040515
Chicago/Turabian StyleVázquez Alberdi, Lucia, Gonzalo Rosso, Lucía Velóz, Carlos Romeo, Joaquina Farias, María Vittoria Di Tomaso, Miguel Calero, and Alejandra Kun. 2022. "Curcumin and Ethanol Effects in Trembler-J Schwann Cell Culture" Biomolecules 12, no. 4: 515. https://doi.org/10.3390/biom12040515
APA StyleVázquez Alberdi, L., Rosso, G., Velóz, L., Romeo, C., Farias, J., Di Tomaso, M. V., Calero, M., & Kun, A. (2022). Curcumin and Ethanol Effects in Trembler-J Schwann Cell Culture. Biomolecules, 12(4), 515. https://doi.org/10.3390/biom12040515







