Abstract
Despite substance use disorders (SUD) being one of the leading causes of disability and mortality globally, available therapeutic approaches remain ineffective. The difficulty in accurately characterizing the neurobiological mechanisms involved with a purely qualitative diagnosis is an obstacle to improving the classification and treatment of SUD. In this regard, identifying central and peripheral biomarkers is essential to diagnosing the severity of drug dependence, monitoring therapeutic efficacy, predicting treatment response, and enhancing the development of safer and more effective pharmacological tools. In recent years, the crucial role that the endocannabinoid system (ECS) plays in regulating the reinforcing and motivational properties of drugs of abuse has been described. This has led to studies characterizing ECS alterations after exposure to various substances to identify biomarkers with potential diagnostic, prognostic, or therapeutic utility. This review aims to compile the primary evidence available from rodent and clinical studies on how the ECS components are modified in the context of different substance-related disorders, gathering data from genetic, molecular, functional, and neuroimaging experimental approaches. Finally, this report concludes that additional translational research is needed to further characterize the modifications of the ECS in the context of SUD, and their potential usefulness in the necessary search for biomarkers.
1. Introduction
According to the latest epidemiological data, the estimated number of past-year users of any drug globally stands at 275 million people, increasing by 22 percent between 2010 and 2019. From this population, almost 36.3 million (13%) suffer from diagnosed substance use disorders (SUD) [1]. SUD is a chronic, relapsing clinical condition characterized by compulsive drug-seeking and use despite harmful consequences, constituting one of the leading causes of disability and mortality. Indeed, deaths directly related to SUD amounted to 167,750 in 2015, representing a 60% increase over the previous figure of 2000. In short, opioids produce the highest morbidity and mortality among the drugs consumed, alcohol constitutes the most consumed legal drug globally, and cannabis is the illegal drug with the highest number of users [1,2].
Unfortunately, despite the devastating worldwide impact of SUD, available pharmacological treatments remain insufficiently effective for most people, probably due to the difficulties of characterizing and elucidating the underlying neurobiological mechanisms. While clinically useful, commonly used psychosocial and symptomatic criteria are not enough to accurately capture the vast heterogeneity of SUD, depending on factors such as genetics, age, gender, polydrug use, or psychiatric comorbidities. Furthermore, the current diagnosis of SUD relies on several qualitative outcome measures. For this reason, identifying central and peripheral biomarkers applying a multidisciplinary approach is an urgent need to diagnose the severity of drug dependence, monitor therapeutic efficacy, predict treatment response, or enhance the development of safer and more effective therapeutics for SUD [3,4,5,6].
In recent years, compelling advances have been acquired in understanding how addictive drugs affect the brain initially, particularly in the brain’s reward system, and induce longer-lasting neuroadaptive changes after repeated exposure, leading to the compulsive seeing and taking of drugs that define addiction [7]. The acute administration of drugs of abuse enhances the activity of the mesolimbic dopamine (DA) system [8], increasing DA release in the nucleus accumbens (NAcc) shell through the stimulation of DA neurons from the ventral tegmental area (VTA). This neurochemical response has been related to the rewarding effect [9], producing a hedonic experience that is crucial for the initiation and maintenance of drug consumption [10]. Repeated drug exposure causes adaptive changes in the circuitry of the extended amygdala, resulting in enhanced reactivity to stress [11] and the emergence of negative emotions [12]. Furthermore, chronic drug consumption also leads to profound impairments in decision-making that are closely related to functional and morphological changes occurring in the prefrontal cortex (PFC) [13]. The PFC and other frontal regions play a crucial role in executive processes, including self-regulation, behavioral control, flexibility, and threat conditioning [14,15,16,17]. A down-regulation of DA signaling occurs in the PFC and their associated circuits after repeated drug exposure. These changes are linked to neuroplastic and morphological changes in the PFC glutamatergic neurons [18]. In addition, it is relevant to note that, apart from disturbances mainly in dopaminergic and excitatory neurotransmission systems, the phenomena encompassing drug addiction are also associated with neuroinflammatory processes that have recently received particular attention [19], and are related to the activation of metabolic systems, such as the tryptophan-kynurenine pathway [20,21,22]. Overall, these alterations weaken the inhibitory control that cortical areas exert on the mesolimbic system, impairing the capability to control the decisions related to drug consumption, finally leading to the loss of control, compulsive drug use and relapse that characterize addictive disorders.
The endocannabinoid system (ECS) has received substantial attention, and accumulated evidence points out its crucial role in the neuromodulation of the rewarding and neurophysiological actions of drugs of abuse [23,24]. ECS is a ubiquitous lipid signaling system distributed throughout the organism that participates in multiple intracellular signaling pathways [25,26]. It regulates several physiological functions and mediates the crosstalk between different neurotransmitter systems, therefore representing a key player in controlling behavioral responses [27,28]. In short, cannabinoid receptors (CB1R and CB2R), endogenous ligands or endocannabinoids (eCBs; anandamide (AEA) and 2-arachidonoylglycerol (2-AG)), and the major enzymes responsible for the synthesis (N-acylphosphatidylethanolamine specific phospholipase D (NAPE-PLD) and diacylglycerol lipase (DAGL)) and degradation (fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MAGL)) of eCBs are the main components of the ECS, present in the central and peripheral nervous system [27,29] (see Figure 1), and in many other peripheral tissues regulating distinct functions [30].
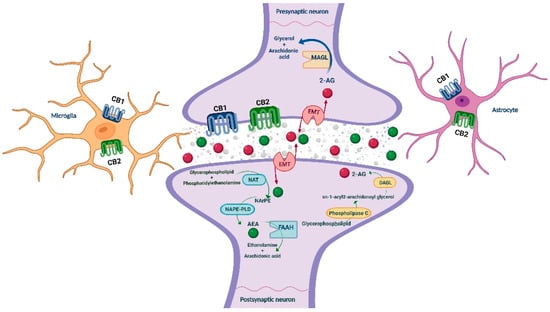
Figure 1.
Schematic representation of the main ECS components, including the metabolizing routes of the eCBs. AEA: anandamide; CB1/CB2: cannabinoid receptors 1 and 2; DAGL: diacylglycerol lipase; EMT: endocannabinoid membrane transporter; FAAH: fatty acid amide hydrolase; MAGL: monoacylglycerol lipase; NAPE-PLD: N-acylphosphatidylethanolamine specific phospholipase D; NArPE: N-arachidonoyl phosphatidylethanolamine; NAT: N-acyl transferase; 2-AG: 2-arachidonoylglycerol. Image created with BioRender.
The connection between the ECS and drug addiction emerges from the very well-known rewarding effects and abuse potential of cannabis preparations. Notably, the phytocannabinoid delta-9-tetrahydrocannabinol (THC) accounts for typical cannabis effects. In addition, ECS components are expressed in the brain regions that make up the mesocorticolimbic pathway, including the VTA, NAcc, or PFC. Notably, the dopaminergic neurons of this brain reward circuitry are controlled by excitatory and inhibitory inputs that are, in turn, modulated by the ECS [31,32,33].
Thus, identifying drug-induced changes in the targets comprising this neuromodulatory system has attracted increasing attention in recent years to discover new biomarkers with diagnostic, prognostic, or therapeutic potential. The present review compiles the primary evidence on the genetic (polymorphisms), molecular, functional and neuroimaging alterations in the components of the ECS (receptors, endocannabinoids and enzymes) that occur as a consequence of exposure to different drugs of abuse (alcohol, cannabis, opioids, stimulants, nicotine, and hallucinogens), from a translational approach that integrates clinical and animal studies.
2. Methods
The literature search for this narrative review was performed in the Medline database (PubMed) employing medical subject headings (MeSH). Specific keywords were employed according to the substance-related disorders included in the review: Alcohol (“ethanol” [MeSH]), cannabis [MeSH], analgesics, opioid [MeSH], N-Methyl-3,4-methylenedioxyamphetamine [MeSH], methamphetamine [MeSH], cocaine [MeSH], nicotine [MeSH], tobacco [MeSH], hallucinogens [MeSH]. These terms were combined with “cannabinoids” [MeSH] by the Boolean operator “AND.” All the authors critically analyzed all the results for each search to decide the selection of each reference according to the adequacy of its content with the subject matter of the study. No PubMed filters were applied to maximize the selection of all the available and appropriate information. All original articles, systematic reviews, or meta-analyses on identifying ECS components alterations in drug addiction were accepted. Those articles not related to the topic of interest, not written in English, or to which access was not possible were discarded.
3. Endocannabinoid Components as Potential Biomarkers in SUD
This section aims to gather evidence about how the ECS is impaired due to acute or chronic drug consumption or to specific stages of drug-related disorders (alcohol, cannabis, opioids, stimulants, tobacco, and hallucinogens), mainly intoxication, withdrawal, dishabituation, or relapse. For this purpose, a translational approach has been applied to combine animal and human studies providing relevant information from a multidisciplinary point of view that includes genomics, epigenetics, genetics, proteomics, or neuroimaging.
3.1. Alcohol-Related Disorders
Alcohol use disorder (AUD) is one of the most common addictive disorders, with significant health and socioeconomic impacts. The search for adequate biomarkers allowing the development of preventive and therapeutic strategies is a significant challenge to improve the clinical management of alcoholic patients. In this regard, ECS components have attracted great interest because of their potential to serve as biomarkers in AUD. This section gathers the most recent studies developed to detect and describe changes in the ECS in patients with AUD and different animal models of ethanol exposure, providing valuable information about its potential diagnostic, prognostic, or therapeutic usefulness (Table 1).

Table 1.
Main findings from human and animal studies aimed to identify alterations of ECS components in alcohol-related disorders.
3.1.1. Gene Polymorphisms of ECS Components
Promising clinical results have been obtained by evaluating the involvement of different single-nucleotide polymorphisms (SNPs) of genes encoding distinct cannabinoid components in AUD. Considering the close relationship between the ECS and dopaminergic system, Hoenicka et al. aimed to analyze genetic factors, underlying the dual pathology of AUD and antisocial personality disorder. The authors evaluated gene variants involved in both systems, analyzing SNPs near the D2 dopamine receptor gene (D2). More specifically, a 10-repeat allele of a variable number tandem repeats (VNTR) of the SLC6A3 gene, the C385A FAAH SNP and the 3′-UTR microsatellites of the CNR1 gene were analyzed. In a sample of 137 alcoholic patients, a close correlation was found in CNR1 and FAAH genes. This result suggests the interaction of the dopaminergic and ECS systems in developing the comorbidity of alcohol misuse and antisocial behavior [34]. However, the interaction between CNR1 (rs2023239) and the D4 dopamine receptor gene (DRD4) did not provide similar results regarding the interconnections between the dopaminergic and cannabinoid systems. The CNR1 C allele group presented more increased alcohol craving than the T allele group, but no correlations were found between DRD4 and CNR1 gene variants [35].
Seven SNPs were significantly associated with Alcohol Use Disorder Identification Test (AUDIT) scores in a European adolescent population of alcohol consumers. Two of them, CNR1 (rs9343525) and monoacylglycerol lipase (MGLL; rs507961) genes, remain significant after statistical corrections for multiple covariables [36]. When evaluating FAAH and MGLL genes SNPs in an alcoholic population of Japanese patients, despite covering most regions of the genes encoding these endocannabinoid metabolic enzymes, no associations were observed with the increased susceptibility to develop alcoholism [37]. However, the polymorphism Q63R of the CNR2 gene was associated with high ethanol consumption in another mixed sample (including males and females) of the Japanese population [38].
Two polymorphisms of CNR1, rs6454674 (SNP3) and rs806368 (SNP8), have been strongly associated with both alcohol dependence and drug dependence. Interestingly, the interaction between both SNPs significantly increased the risk of developing drug or alcohol dependence [39]. The influence of these two allelic variants of CNR1 (rs6454674 and rs806368) with an additional one (rs1049353) was evaluated in another study developed by Marcos and colleagues [40]. The authors analyzed single markers and haplotypes and conducted interaction analyses, concluding that the haplotype TGT (corresponding to rs6454674-rs1049353-rs806368) decreases the susceptibility to develop alcohol dependence. However, an interaction was found between the G allele of rs6454674 SNP and the C allele of rs806368 SNP, supporting the involvement of the ECS in alcohol dependence [40]. The polymorphism rs1049353 (1359 G/A, Thr453Thr) was also evaluated in a Caucasian population, comparing the frequency of its presentation in alcohol and control subjects. In this study, the authors showed that the homozygous genotype CNR1 1359 A/A of the polymorphism increased the vulnerability to develop alcohol withdrawal-induced delirium [41]. Thus, identifying this genetic variant could be an exciting strategy in alcoholic patients to prevent the development of delirium.
Previous studies pointed out that the A or G allele of a CNR1 polymorphism could not be associated with a history of alcohol withdrawal-induced seizures [42]. Nevertheless, post-mortem studies showed a strong association between the C allele of the CNR1 rs2023239 polymorphism, with increased CB1R binding in the prefrontal cortex, alcohol cue-elicited brain activation, and subjective alcohol reward. Thus, patients with the C allele are more susceptible to changes in the mesocorticolimbic circuitry, and more prone to developing alcohol dependence [43].
The SNP of the FAAH gene, C385A (FAAH Pro129Thr, rs324420), is associated with a decreased enzymatic activity of FAAH, resulting in increased anandamide levels and activity. This FAAH SNP has been related to alterations in alcohol consumption. In a study developed by Bühler and colleagues, authors evaluated the involvement of 10 SNPs of different targets, two of them corresponding to the FAAH gene. The positive rating of alcohol-related pictures was associated with the homozygous CC C38A SNP genotype, indicating that this SNP is a candidate for screening patients with a higher risk of alcohol-related problems, such as sleep disturbances [44,45]. This correlation appears to be different depending on the ethnic factors. Indeed, the Thr129 allele frequency is higher in European American participants with current alcohol dependence than in non-dependent controls. Thr129 carriers reported a median of 10 fewer abstinent days and 13 more binge drinking days than controls. Nevertheless, there were no significant differences between Thr129 allele frequency in African American participants in alcoholic patients and controls [46].
The minor allele (A) of the SNP C385A has been associated with reduced FAAH enzyme activity and increased risk for substance use disorders in adults. However, its implication for young people still has to be elucidated. For this purpose, Best and colleagues determined the FAAH C385A genotype in a group of heavy-drinking youths. Authors showed that people with the FAAH minor allele (AC or AA genotype) present significantly more drinking days, heavy episodic drinking, and higher alcohol-related problems and consumption patterns [47]. These findings suggested that the reduced endocannabinoid metabolism induced by the SNP affecting the FAAH gene may be related to heavier alcohol use in the young population before the onset of chronic drinking problems. Thus, identifying this SNP may be a potential marker of possible development of alcohol dependence, allowing the implementation of preventive strategies.
Previous results in humans were confirmed in a preclinical study using genetic knock-in mice containing the human FAAH SNP C385A. These mutant FAAH (A/A) mice developed a greater alcohol intake and preference in the drinking-in-the-dark paradigm (DID), supporting the idea about the involvement of this SNP in alcohol dependence [48].
3.1.2. Gene and Protein Function/Expression Changes of ECS Components
The first evidence suggesting the involvement of ECS in the regulation of ethanol effects was that acute or chronic ethanol exposure modifies the gene expression of the CB1R. Acute ethanol exposure induced neuroplastic alterations in the CNR1 gene expression in different brain areas such as caudate-putamen (CPu), central amygdala (CeA), and ventromedial hypothalamic nucleus [49]. These results were extended by Ortiz and colleagues, evaluating the effect of chronic ethanol exposure on CNR1 gene expression. In this study, the consumption of ethanol for a 52-day period reduced the gene expression of CNR1 in the CPu, the ventromedial nucleus of the hypothalamus, CA1 and CA2 fields of the hippocampus (Hipp), and increased gene levels in the dental gyrus in rats [50]. Furthermore, other authors showed that chronic ethanol vapor exposure increases MGLL and DAGLB mRNA and MAGL protein levels. These changes were accompanied by a significant decrease in CNR1 mRNA and CB1R protein levels in alcohol-exposed rats [51]. The implication of CNR1 genes in alcohol intake was also evaluated in a study using the CRISP/CAS9 method. Hay and colleagues disrupted a highly conserved regulatory sequence in the CNR1 gene in this study. This procedure reduced CNR1 gene expression, causing a subsequent reduction in alcohol intake in the two-bottle choice paradigm, supporting the involvement of ECS in alcohol dependence [52].
The alcohol intake in rats exposed to a protocol of maternal separation during its postnatal period correlated with the increased protein expression of CB1R in the ventral striatum, and decreased in the frontal cortex [53]. These findings were similar to those obtained by Hansson and colleagues regarding FAAH levels in alcohol-preferring rats. In this study, the increased preference strongly correlated with decreased expression of FAAH in the prefrontal cortex (PFC), inducing reduced enzyme activity in this brain region. These results suggest an overactive endocannabinoid transmission of alcohol-preferring rats and a compensatory down-regulation of CB1R signaling [54]. On the other hand, mice with high alcohol preference showed reduced gene expression of CNR2 in the ventral midbrain, supporting the idea of the involvement of this gene in alcohol consumption [38].
ECS changes were also reported in rodents exposed to alcohol withdrawal paradigms. Acute alcohol withdrawal reduced AEA content in the basolateral amygdala (BLA) and 2-AG concentrations in the PFC. Interestingly, authors observed these alterations in male rats but not in females, suggesting sex-dependent changes in this animal model [55]. In addition, acute ethanol withdrawal was also associated with FAAH, MGLL, CNR1, CNR2 and GPR55 gene expression reduction. These changes were more pronounced than those obtained after intermittent alcohol exposure [56]. The intermittent alcohol exposure increased the mRNA levels of the enzyme of synthesis of AEA and 2-AG in the PFC and reduced it in the amygdala (Amy). On the other hand, this type of alcohol exposure was associated with decreased mRNA levels of CNR1 and CNR2 in the striatum [57].
Ventral striatum post-mortem samples of alcohol-dependent patients were analyzed to evaluate changes in the CB1R protein levels, enzymatic activity, and protein levels of FAAH. The results revealed reduced CB1R levels, CB1R-mediated signaling, and FAAH levels compared with the control group. Authors suggested that alterations in FAAH activity modified AEA concentrations, CB1R protein levels, and receptor activity [58]. However, further studies are required to explore the relationship between CB1R in the ventral striatum of alcoholic patients and the development of alcohol dependence. CB1R, CB2R and GPR55 gene and protein levels were also evaluated using human monocyte-derived dendritic cells from alcohol users. Gene expression studies revealed an increase in CNR2 and GPR55 gene expressions without changes in CNR1. Interestingly, both receptors increased after in vitro treatment, with different concentrations of ethanol in cell cultures [59].
3.1.3. Neuroimaging of ECS Components
The first human studies showing the critical role of the endocannabinoid system (ECS) were performed using the CB1R binding radiotracer [11C]OMAR [61]. This study was conducted in a relatively limited group of men (8 healthy controls and 8 with alcohol dependence, 4 weeks after the last drink). Interestingly, the volume of distribution (VT) of [11C]OMAR in patients with alcohol dependence was approximately 20% (p = 0.023) higher than in healthy controls in the Amy, Hipp, putamen, insula, anterior and posterior cingulate cortices and orbitofrontal cortex (OFC). Age, body mass index or smoking status did not influence the outcome. In a similar but amplified study with the radiotracer [18F]FMPEP-d2, Hirvonen et al. scanned 18 male in-patients with alcohol dependence twice, within 3–7 days of admission from ongoing drinking, and after 2–4 weeks of supervised abstinence, and compared them to 19 healthy males. On the first scan, CB1R binding was 20–30% lower in patients with alcohol dependence than in control subjects in all brain regions, and was negatively correlated with years of alcohol abuse. Indeed, this result was in agreement with data from the previous study. In the second scan after 2–4 weeks of abstinence, the CB1R levels did not show significant recovery in those patients, indicating the long-term effects of alcohol dependence and the involvement of CB1R [62]. In a posterior study, Ceccarini et al. [63] investigated the changes in CB1R availability after acute and chronic alcohol abuse. The 20 healthy subjects that received an intravenous ethanol injection to study the acute effects showed a global brain increase in CB1R availability (+15.8%) measured with [18F]MK-9470. In contrast, the chronic drinking patients showed a global decrease (−16.1%). After 1 month of abstinence, the decreased availability of CB1R persisted, according to Hirvonen et al., 2013.
Studies in rodents using PET also investigated the effect of alcohol in the ECS. Ceccarini et al. showed that acute alcohol administration in male rats increased [18F]MK-9470 binding to CB1R in the NAcc, while chronic ethanol consumption decreased [18F]MK-9470 in the Hipp and CPu. In contrast to human studies, the [18F]MK-9470 levels recovered the baseline levels in rats and increased after 7 and 14 days of abstinence [60].
In addition to radiotracers targeting CB1R and CB2R, others targeting the two main hydrolases that metabolize AEA and 2-AG (FAAH and MAGL, respectively) are under development. [11C]CURB was the first PET radiotracer developed to image FAAH, and evaluated first in animals [65], and later in humans [66]. Best et al. studied brain FAAH levels in AUD patients 3–7 days and 2–4 weeks after abstinence using [11C]CURB. They found that brain FAAH levels were lower in alcoholic patients than in controls during early abstinence, while there were no differences at 2–4 weeks [64].
3.1.4. Concluding Remarks—ECS and Alcohol-Related Disorders
Studies developed to date showed the strong involvement of ECS in regulating alcohol effects in humans and animal models. Some relevant conclusions can be drawn from the available evidence: (1) specific CNR1 polymorphisms have been widely associated with AUD; (2) FAAH C385A rs324420 polymorphism seems to be related to increased risk of alcohol misuse and addiction; and (3) chronic alcohol exposure commonly decreases FAAH (gene expression and binding) associated with an increase in eCBs levels and reduced CB1R (gene expression and binding) in several brain regions. Further studies are required to explore, discover, and characterize specific ECS biomarkers with diagnostic or therapeutic potential in alcohol-related disorders.
3.2. Cannabis-Related Disorders
Much research on cannabis has focused on studying dynamic changes in the gene and protein expression of key targets within the ECS, induced by THC or cannabinoids, mainly in rodents, and to a lesser extent, in humans. This review summarizes the most promising results, revealing significant alterations that may serve as potential biomarkers for cannabis use disorders (CUD) (Table 2).

Table 2.
Main findings from human and animal studies aimed to identify alterations of ECS components in cannabis-related disorders.
3.2.1. Gene Polymorphisms of ECS Components
In humans, genetic variants of different critical elements of the ECS have been proposed as a risk factor for cannabis dependence, withdrawal, craving, and cue-elicited brain activation.
A significant correlation was found between the CNR1 SNPs rs1049353, rs806368, and rs806380 and CUD [67,69,70]. The polymorphism CNR1 rs2023239, allele G, is associated with more significant withdrawal and craving for cannabis after short-term abstinence, greater craving after cannabis cue exposure [71], and more significant brain response to cannabis cue in several areas, including the OFC, inferior frontal gyrus (IFG), and anterior cingulate gyrus (ACG) [72]. It has been found that allele G affects CNR1 mRNA expression; in human postmortem brain tissues, a reduction of CNR1 gene expression was detected in the PFC. Moreover, cannabis users’ carriers of the G allele showed smaller hippocampal volume, suggesting that cannabis exposure may interact with CB1R altered by the polymorphism, in such a way that reduces hippocampal volume [73].
Colizzi et al. performed a functional MRI study in cannabis users, demonstrating that G carriers of the CNR1 SNP rs1406977 had superior functional connectivity in the left ventrolateral PFC, and reduced working memory accuracy during the 2-back task. They concluded that the adverse effects of cannabis use are enhanced on a specific genetic background related to CNR1 variations [74]. A longitudinal structural MRI study on regular users of cannabis found an association between reduced volume in the anterior cingulum and CNR1 SNPs 1049353 and rs806368. Authors proposed an association between the level of cannabis exposure, the decreased volume of the brain mentioned above the area, and CNR1 haplotypes variation [68].
Similarly, Ho et al. found a correlation between the CNR1 SNP rs12720071 and smaller parietal white matter volume in schizophrenic patients with CUD [75]. Interestingly, a recent publication linked the polymorphisms CNR1 rs12720071 (G-allele-carriers), the mitogen-activated protein kinase 14 (MAPK14) rs12199654 (A-allele carriers), and smaller cerebral and lobar white matter volumes in schizophrenic patients with heavy marijuana use [76].
In the case of the CB2R, the SNP rs2501431 (CNR2) has been associated with CUD [77]. Recently, the SNPs rs12744386 and rs35761398 (CNR2) have been linked with a high risk for developing schizophrenia in patients with CUD [78].
The human polymorphism FAAH C385A (rs324420) is one of the most explored SNPs. This polymorphism results in a mutant form of FAAH, with reduced expression and cellular stability [79,110]. The A allele has been associated with street drug use [79] and allele C with an increased risk for cannabis dependence progression, although with inconsistent results. On the one hand, allele A has been linked with increased ventral striatum reactivity, brain area closely related with reward, higher impulsivity [80], and more significant bias to appetite stimuli [81]. In other studies, the allele C was associated with the increased activity of brain reward areas in regular marijuana users [72].
Similarly, opposite results were found in the role of alleles A and C in the progression from sporadic cannabis users to cannabis dependents. Some authors found a link between allele A and an increased risk for CUD [79,82,111,112], while others revealed that allele C confers more risk for THC dependence [71,78,83]. Recently, allele C has been associated with craving during marijuana abstinence [71].
Recent research has deeply explored the role of C385A polymorphism in cannabis dependence by using a knock-in mouse model (FAAHC/A) that biologically recapitulates the human polymorphism. Interestingly, the authors found a modified CNR1 gene expression with higher levels at GABAergic terminals and lower levels at glutamatergic terminals in the VTA, which may affect the development of the mesolimbic reward pathway. This study supports the fact that this polymorphism C/A contributes to a preference for THC in adolescent female mice that persists into adulthood compared to female FAAHC/C, which find THC aversive [99]. Furthermore, the SNP rs604300 within the MGLL has been associated with stress adaptation related to cannabis dependence [84]. A recent study showed changes in white matter bundles innervating posterior cingulate and parietal cortex, basal ganglia, and temporal cortex in patients with cannabis dependence, and a significantly lower gray matter thickness and density in the precuneus. These brain structural alterations seemed to be significantly associated with regional differences in MAGL expression [102].
3.2.2. Gene and Protein Function/Expression Changes of ECS Components
Most of the results come from studies in rodents focused on identifying the effects of chronic cannabinoid agonists administration to clarify the mechanisms underlying cannabis dependence, craving, and, more recently, cannabinoid withdrawal.
Chronic cannabinoid administration induced a dynamic and time-dependent reduction of CNR1 mRNA levels in the different brain regions of rodents, such as the CPu. During the first days (5–7 days) of chronic administration, there was no alteration in CNR1 gene expression, but it was markedly reduced at 11 days of administration. These changes occur parallel to motor tolerance, supporting the involvement of CNR1 mRNA reduction in the development of cannabis tolerance [85,86]. However, in an additional study, chronic THC administration during 5 days induced an opposite effect, since CNR1 mRNA levels were increased in the striatum of male Wistar rats [87]. The discrepancy in the results may be due to the different cannabinoid agonists used (CP-55,940 and THC), the pattern of administration (doses, duration), and the strain of rodents used (rats).
Male rats perinatally exposed to chronic THC showed reduced CNR1 gene expression in the PFC and lower 2-AG brain concentrations; changes persist until adulthood. The alterations in CNR1 gene expression have been associated with changes in dopamine D2 receptor (DRD2) gene expression and, even more critically, with a significant hypomethylation at the DRD2 regulatory region [113]. Altogether, these data emphasize the involvement of CB1R in cannabis dependence and its close interaction with the dopamine reward system [114,115].
In humans, increased the CB1R gene expression and hypomethylation of the promotor were detected in the peripheral blood cells of THC-dependent patients [100]. More importantly, CNR1 mRNA levels correlate with a lower score on the Satisfaction With Life Scale (SWLS) and with craving and cannabis dependence, results in agreement with previous animal studies showing a relationship between craving and CNR1 gene expression [116].
In mice models of cannabinoid withdrawal, CNR1 gene expression was increased in the NAcc, the ventromedial hypothalamic nucleus, CeA, and CA1 field of the Hipp [89,90,91,117]. This up-regulation may be due to a compensatory neuroadaptive response to reducing CB1R found after repeated treatment with a cannabinoid receptor agonist [86,118]. Besides, CB2R gene expression has been down-regulated in the NAcc of C57BL/6J mice exposed to cannabinoid withdrawal [91].
Autoradiographic studies revealed that the chronic administration of THC or CB1R agonists, such as the CP-55,940, significantly reduced CB1R binding and led to tolerance in most responses (e.g., motor tolerance) [92,93,96,118,119,120]. The down-regulation and desensitization of CB1R are time-dependent and region-specific, and occur prior to gene expression alterations [94]. Thus, the reduction of CB1R binding may represent the first step in the biochemical mechanisms underlying tolerance to cannabinoids [95,121].
In rats, the acute administration of THC induced an upward shift of AEA concentrations in the NAcc, though there was still a trend towards increased levels after complete chronic treatment [88]. Previous studies found increased concentrations of AEA in limbic forebrain areas after chronic THC administration [97,98]. Thus, cannabinoid tolerance is accompanied by region-dependent changes in the concentrations of AEA and 2-AG [97,98]. In healthy volunteers, a single oral dose of THC increased circulating concentrations of AEA, 2-AG, and other endocannabinoids 2 and 3 h after administration compared with placebo [101].
3.2.3. Neuroimaging of ECS Components
The effects of the chronic use of cannabis on ECS brain components were investigated using PET imaging. Hirvonen et al. investigated the changes in CB1R availability with [18F]FMPEP-d2 in 30 chronic daily cannabis smokers and 28 control subjects. They imaged the chronic cannabis smokers, the day after admission, and approximately 4 weeks after abstinence. At baseline, the VT of [18F]FMPEP-d2 was lower in cannabis smokers than in control subjects in neocortex and limbic areas. After the abstinence period, the VT of [18F]FMPEP-d2 showed significant increases in regions, with reduced VT at baseline [103]. Ceccarini et al. imaged 10 chronic cannabis users and 10 healthy controls using [18F]MK-9470 to measure CB1R availability. The results showed a global decrease in CB1R receptor availability (−11.7%), the most significant changes in the temporal lobe, anterior and posterior cingulate cortex and NAcc [104]. Bhattacharyya et al. investigated the effect of THC on anxiety and amygdala response in humans using the CB1R binding radiotracer [11C]MePPEP. The THC treatment-induced anxiety modulated Amy activation and increased the availability of CB1R [105]. D’Souza et al. investigate the time course of changes in CB1R availability following short and intermediate-term abstinence in a group (n = 11) of cannabis-dependent subjects, imaging them with [11C]OMAR just before starting abstinence and 2 and 28 days after. VT was lower in cannabis-dependent subjects than the healthy controls at baseline. However, this difference was no longer evident after 2 or 28 days of abstinence [106].
Changes in FAAH were investigated in cannabis use patients. Boileau et al. studied the FAAH alterations using [11C]CURB in a small group of chronic cannabis users (n = 10) during early abstinence, compared to a group of healthy controls (n = 22). They showed decreased [11C]CURB binding in the chronic cannabis users from −20% (Amy and cingulate) to −14% (Hipp), and in most of the cortical areas studied [107]. In a more recent study from the same researchers, the results showed lower [11C]CURB binding among a group (n = 14) of young chronic cannabis users (23 ± 5 years of age) compared to healthy controls during early abstinence throughout the whole neocortex and striatum, Amy, Hipp, thalamus and cerebellum [108]. Another study using [11C]OMAR studied the changes in CB1R availability in women with CUD (n = 10), who displayed significantly lower VT than healthy female controls (n = 10) in the Hipp, Amy, cingulate, and insula, similar to previous studies in men [109]. Interestingly, a recent review on psychiatric disorders also evaluated PET studies in CUD and AUD [122].
The effects of ECS genetic polymorphisms in the binding of radiotracers were described in recent studies. Boileau et al. studied FAAH changes using [11C]CURB in a small group of chronic cannabis users, showing that the brain binding of the [11C]CURB is dependent on the genetic FAAH polymorphism rs324420 (C385A) [107].
3.2.4. Concluding Remarks—ECS and Cannabis-Related Disorders
The evidence suggests a strong involvement of ECS in regulating cannabis effects in humans and animal models. Some relevant conclusions can be drawn from the available evidence: (1) specific CNR1 polymorphisms have been associated with significant cannabis withdrawal syndrome and craving; (2) the consequences of FAAH C385A rs324420 polymorphism on cannabis effects and the development of cannabis dependence vary according to the carrying allele (C or A); and (3) cannabis chronic exposure induces a down-regulation of CB1R (gene expression and binding) that is normalized or even increased during withdrawal. Further studies are required to explore, discover, and characterize specific ECS biomarkers with diagnostic or therapeutic potential in cannabis-related disorders.
3.3. Opioid-Related Disorders
The crosstalk between the endogenous opioid and cannabinoid systems has a pivotal role in regulating the rewarding and neurophysiological actions of several abuse drugs, including opioids (i.e., morphine, heroin) [123,124,125]. This section contains the available data from animal and clinical studies about the genetic, molecular and neuroimaging modifications in ECS components related to opioid addiction (Table 3).

Table 3.
Main findings from human and animal studies aimed to identify alterations of ECS components in opioid-related disorders.
3.3.1. Gene Polymorphisms of ECS Components
There is very scarce information about ECS polymorphisms related to opioid addiction. One study aimed to genotype 6 polymorphisms of the CNR1 gene and the 385C>A SNP of the FAAH gene in former heroin addicts and their corresponding controls distinguishing between ethnicities (Caucasians, Hispanics, African Americans and Asians). Long repeats of the triplet polymorphism of CNR1 18087-18131(TAA)8–17 were significantly associated with vulnerability to develop heroin addiction. Conversely, the allele 1359A and the genotype 1359AA of CNR1 were associated with a protective effect, particularly in Caucasians. However, no association was found with the missense substitution 385C>A in the FAAH gene [126]. Another work focused on evaluating the association of the CNR1 SNP rs2023239 and the development of major depressive disorder (MDD) and/or suicidal behavior in opiate-dependent outpatients under stable methadone treatment. Interestingly, after regression analysis adjusting for covariables, the C allele of CNR1 rs2023239 was closely related with a lower prevalence of lifetime MDD, but not with suicidal behavior. Although authors acknowledged that replication studies were necessary, these results were encouraging, since the early detection of opioid addicts at risk of major depression was crucial to better implementing psychiatric care in this population [127].
3.3.2. Gene and Protein Function/Expression Changes of ECS Components
One of the first steps to elucidate the involvement of ECS in opioid addiction was the evaluation of the effects of acute or chronic morphine administration on the expression or function of distinct ECS components. Romero and colleagues published the first study showing changes in CB1R binding in different brain regions under basal ([3H]CP-55,940) or stimulated ([35S]GTPγS + WIN-55,212-2) conditions in Swiss mice exposed to morphine (5 days, twice daily, 8–45 mg/kg, s.c.). Although CB1R binding levels were very similar between morphine-dependent and control mice, a small but significant increase was found in the globus pallidus. In addition, the WIN-55,212-2-stimulated [35S]GTPγS binding to CB1R was significantly higher in the substantia nigra and central gray substance. These data suggested that morphine might alter the coupling to intracellular mechanisms rather than cannabinoid receptor binding [128].
Subsequent studies continued to evaluate the effects of repeated morphine exposure (6 days, twice daily, 10–100 mg/kg, i.p.) on CB1R binding and gene expression in different brain regions of Wistar rats. Morphine administration increased CB1R binding in medial CPu, septum and NAcc while decreasing it in the dental gyrus and BLA. Furthermore, CNR1 mRNA levels measured by in situ hybridization were significantly reduced by morphine exposure in the CPu and cerebellum. In contrast, an up-regulation was found in the CA2 region of the Hipp and septum [129]. The authors of the previous study employed the same procedure of morphine exposure to further characterize the changes in eCBs and their receptors in different brain regions involved in drug reward. AEA levels were similar between morphine-dependent and control rats, while CB1R binding was reduced in the cerebral cortex and midbrain of rats exposed to morphine. Moreover, CB1R-stimulated [35S]GTPγS binding increased in the cerebral cortex and decreased in the brainstem. These alterations in ECS components suggested that its pharmacological manipulation might represent a novel tool to manage the negative consequences of opioid addiction [130].
Additional studies also aimed to fully characterize the alterations of ECS components in the reward system after sustained morphine treatment. Vigano et al. chronically treated Sprague–Dawley rats with morphine (4.5 days, twice daily, 5 mg/kg, s.c.) and evaluated CB1R binding, CB1R-stimulated [35S]GTPγS binding, and eCBs levels. Morphine-tolerant rats showed reduced CB1R binding in the cerebellum and Hipp, decreased CB1R function in the NAcc, and lower 2-AG levels in limbic regions such as NAcc and Hipp [131]. Furthermore, repeated morphine treatment (5 days, twice daily, 5 mg/kg, i.p.) in the same rat strain induced a reduction of MAGL protein and CNR2 gene expression in the VTA, whereas AEA or 2-AG levels were unaltered in this brain region. However, the authors also explored the effects of a single morphine injection and obtained a significant decrease in AEA levels [132]. Indeed, Jin and colleagues evaluated the differences between the administration of morphine under acute (10 mg/kg, s.c.) or chronic (12 days, twice daily, 10 mg/kg, s.c.) conditions on CB1R in several brain areas of Wistar rats. Prolonged morphine administration significantly increased CB1R gene and protein expression in the cortex, cerebellum, and Hipp. Acute treatment did not modify CB1R protein levels in these regions, and decreased CNR1 gene expression in the cerebellum. Interestingly, acute and particularly repeated morphine treatment induced an up-regulation of CNR1 gene expression in peripheral blood mononuclear cells (PBMCs). This study provided relevant evidence about the effects of morphine on CB1R in the central nervous system and peripheral immune system cells, adding new clues to the involvement of the ECS in opioid addiction [133]. Moreover, the effects of acute (5 and 10 mg/kg, s.c.) and chronic (5 days, 10–40 mg/kg, s.c.) morphine treatment on AEA and 2-AG levels were also evaluated in the NAcc (shell subregion) of Wistar rats, obtaining an increase in AEA and a decrease in 2-AG [134].
The conditioned place preference (CPP) paradigm has been employed to analyze morphine-induced changes in ECS components in its different experimental phases (conditioning, extinction or withdrawal, and reinstatement). In Sprague–Dawley rats, 5 days of conditioning with morphine (10 mg/kg, s.c.) induced an up-regulation of CNR2 gene expression in the cortex, spleen, and PBMCs, while decreasing in the brainstem. Notably, this study also included clinical samples from morphine abusers (n = 8) and their corresponding controls (n = 5). Despite the limited number of patients, higher levels of cannabinoid receptors gene expression were found in the PBMCs of subjects with at least 1 year of morphine overuse history compared with controls [135]. Later, Yuan et al. evaluated the effects of morphine withdrawal at different phases (acute, latent, and chronic) on CB1R protein expression after 5 days of conditioning (10 mg/kg, s.c.) in the CPP. Regardless of the phase, CB1R was elevated in the NAcc of rats undergoing morphine withdrawal, a result that was accompanied by an increase of CB1R-positive inhibitory terminals [136].
Moreover, short-term morphine withdrawal evaluated 72 h after the 7-days of conditioning (10 mg/kg, s.c.) in the CPP was associated with increased DAGLα in the NAcc [137]. These findings led authors to hypothesize the involvement of 2-AG on the CB1R-mediated modulation of inhibitory synaptic transmission in the NAcc under morphine withdrawal conditions, which may be one of the mechanisms associated with opioid relapse. Another study performed in C57BL/6J mice intended to characterize the gene expression alterations of ECS components (cannabinoid receptors, synthesizing and degradative enzymes) in several brain regions at different phases of the CPP paradigm (expression, extinction, and reinstatement) with morphine. Interestingly, opposite gene expression changes were found between expression and reinstatement phases, whereas no alterations were associated with extinction [138].
Other experimental designs were employed to investigate alterations of the ECS after opioid exposure. Vigan et al. analyzed the modifications of AEA and 2-AG levels in the PFC, NAcc, CPu, and Hipp of Sprague–Dawley rats subjected to a morphine behavioral sensitization procedure, divided into three phases (induction, withdrawal, and expression). The results pointed out that the ECS underwent significant changes during the different phases of sensitization to morphine, motivating further studies to address if the functional manipulation of ECS could play a therapeutic role in drug-seeking behavior, particularly in opioid addiction [139]. Besides, the effects of intravenous heroin self-administration on CB1R binding and function were evaluated. There was an enhancement of CB1R density in the VTA and Amy and increased CB1R coupling in the PFC, NAcc, CPu, Hipp, and Amy. These findings suggested that the voluntary chronic intake of opioids regulates CB1R expression and activity in reward-related brain structures, which could represent long-term neuroadaptations, contributing to the development of drug addiction and dependence [140].
Finally, brain endocannabinoid levels (i.e., AEA and 2-AG) were significantly changed in the NAcc, CPu, and mesencephalon of adolescent and adult Long-Evans rats exposed to a postnatal maternal deprivation model (3 h daily from postnatal day 1–14). Interestingly, the vulnerability to morphine reinforcing and motivational actions was significantly higher in deprived rats. Thus, it could be argued that the alterations in brain endocannabinoid levels may be related to the escalation behavior in a self-administration paradigm [141].
3.3.3. Concluding Remarks—ECS and Opioid-Related Disorders
The results compiled suggest a significant involvement of ECS in regulating opioid effects in humans and animal models. However, the available evidence is minimal, and there are many controversies, probably due to differences in experimental designs, particularly in animal model studies. Despite this, it could be proposed that repeated administration of opioids (e.g., heroin, morphine) leads to increased levels and function of CB1R. Better knowledge about the opioid-induced alterations on ECS components and its involvement in the reinforcing and motivational actions of opioids are crucial steps to identify biomarkers with potential usefulness to improve the diagnosis and therapeutics of opioid-related disorders.
3.4. Stimulant-Related Disorders
Several studies highlight the functional importance of the ECS in the regulation of the actions produced by psychostimulant drugs, such as amphetamines and cocaine. The importance of studying components of the ECS as a consequence of consumption or exposure to different psychostimulant drugs is fundamental to finding more personalized drugs for the treatment of this type of addiction. In this section, the principal results found in this area are gathered (Table 4).

Table 4.
Main findings from human and animal studies aimed to identify alterations of ECS components in stimulant-related disorders.
3.4.1. Gene Polymorphisms of ECS Components
Cocaine-dependent individuals are at high risk of relapsing into heavy use, even after a period of abstinence. There may be a genetic contribution to cocaine dependence and relapse risk. To date, it has been confirmed that few genetic variants contribute to this vulnerability. Two independent CNR1 variants, the G allele genotypes of rs6454674 (SNP3^G+) and the T/T genotype of rs806368 (SNP8^T/T), have significant interaction effects on the risk of cocaine dependence in European Americans (EA), as well as in African Americans (AA) with these genotypes [142]. Similarly, cocaine-dependent individuals and controls of African ancestry were genotyped for two SNP in the CNR1 gene (rs6454674, rs806368). A significant difference in genotype frequencies between cases and controls was observed for both SNP [143].
On the other hand, the triplet repeat (AAT)n polymorphism near the CNR1 gene has been examined in the Afro-Caribbean population among cocaine dependents, with schizophrenia or not. In this case, the frequency of the repeated allele (AAT)12 was increased in both cocaine-dependent non-schizophrenics and schizophrenics compared to the controls [144]. Therefore, all these studies confirm the association between CNR1 variants and cocaine dependence in different ethnicities.
A variant (rs324420) within the FAAH gene on chromosome 1 encodes a substitution (Pro129Thr), which results in decreased FAAH activity, and consequently, increased AEA levels. This has been linked to alterations in mood and stress reactivity and an increased risk of addiction. In a recent study, 70 participants with cocaine use disorder received intravenous doses of saline and cocaine (20, 40 mg). In this study, it was found that the prevalence of the variant allele was higher in the cocaine use disorder group, and also, the drug effect (high and depression) was even higher compared with patients without this polymorphism [145]. In another similar study, this polymorphism was evaluated with methamphetamine (METH)-dependent patients and controls in a Chinese Han population. In these patients, the A allele of the 385C/A (rs324420) polymorphism significantly increased the risk of METH dependence [146].
Similarly, the association of the FAAH Pro129Thr polymorphism with METH dependence, METH-induced psychosis, manic episodes, and panic disorder was investigated in a Malaysian population. This polymorphism was significantly associated with the risk of METH dependence and METH-induced mania in this ethnicity [147]. In short, FAAH Pro129Thr polymorphism may contribute to an increased predisposition to cocaine and METH dependence.
3.4.2. Gene and Protein Function/Expression Changes of ECS Components
METH induces specific damage in the dopaminergic neurotransmitter system and is associated with cell death. A single administration of METH (30 mg/kg, i.p.) alters the concentrations of the endocannabinoids AEA and 2-AG in the striatum of adult male mice, suggesting that the ECS may be involved in brain responses to this drug [148].
Ecstasy, 3,4-methylenedioxymethamphetamine (MDMA), is one of the most widely used psychedelic drugs worldwide. Colado and colleagues revealed that MDMA administration (12.5 mg/kg, i.p.) in adult Dark Agouti rats increased CNR2 expression, mainly in the microglia of the PFC and hypothalamus [149]. On the other hand, AEA and 2-AG plasma concentrations were not affected by MDMA in a double-blind placebo-controlled study, with 20 healthy recreational polydrug users receiving a pre-treatment with a 5-HT2 receptor blocker (ketanserin, 40 mg) before MDMA (75 mg) [162].
Cocaine is an addictive drug that disrupts mesolimbic dopaminergic neurons by inhibiting dopamine uptake, leading to compulsive behavior and relapse. Acute exposure to cocaine decreased DAGLA gene expression and increased tyrosine hydroxylase (TH) in the cerebellum of male C57BL/6J mice. Likewise, chronic cocaine administration decreased FAAH and DAGLβ protein expressions in cerebellar Purkinje cells [150]. Repeated cocaine administration (5 days pretreatment, 20 mg/kg, i.p.) and treatment (10 mg/kg after 6 days of extinction) in the same mouse strain increased CNR1 gene expression and decreased NAPE-PLD and DAGLα in the hippocampus [151]. However, mice subjected to two sessions of crack cocaine inhalation per day for 11 days reduced mRNA expression levels of ECS components such as FAAH, MGLL and CNR1 in the PFC, whereas mRNA levels of the synthesis enzymes NAPE-PLD and DAGLA were not altered in this brain area [152]. Apparent discrepancies in the results found in these studies may be due to differences in the brain areas (Hipp and PFC), the route of administration (i.p. and inhaled), or the treatment duration (13 and 11 days).
On the other hand, the effects of cocaine sensitization regimen (15 mg/kg, i.p.) for 7 consecutive days during different windows of adolescent vulnerability (PND 33–39, PND 40–46, PND 47–53) on CB1R and CB2R protein levels were evaluated in the PFC and Hipp in male Sprague–Dawley rats. The results revealed that CB1R and CB2R are differentially modulated in PFC and Hipp during adolescence. CB1R protein expression increased, while CB2R decreased in both areas during adolescence compared to adulthood. However, cocaine only altered CB1R and CB2R expression in PND33 to PND39 in the PFC, an effect that did not persist into adulthood, identifying a period of vulnerability during adolescence [153]. Indeed, in a study evaluating CB1R and CB2R status in the cerebral cortex of cocaine-treated adult rodents (mice and rats), CB1R protein was reduced in the PFC. However, CB2R protein was not significantly altered in the PFC of cocaine-dependent rodents [154].
In addition, changes in the expression of CB1R and CB2R in Wistar rats exposed to maternal deprivation (24 h, PND1 and 9) were also evaluated. Overall, maternal deprivation induced sex-dependent and long-lasting alterations in the CB1R expression. Both control and maternally deprived animals received cocaine (8 mg/kg/day) or saline during adolescence (PND 28–48). Adolescent cocaine administration in control animals increased CB1R expression in the Hipp, whereas the opposite was found in maternally deprived animals. Furthermore, cocaine administration during adolescence increased CB1R function in the Hipp exclusively in males in both groups [155].
Bystrowska et al. used male Wistar rats to evaluate the changes induced by cocaine after two types of experimental designs: (1) Intravenous self-administration for 14 days, and (2) Intravenous self-administration for 14 days, followed by 10 days of extinction [156]. Immunohistochemical techniques in the brain samples analyzed the levels of CB1R and CB2R. After the self-administration procedure, the results revealed a significant decrease in CB1R expression in the PFC, Amy, and VTA. An increase of CB1R in the Amy was also found in the extinction period. On the other hand, CB2R expression decreased in the PFC and NAcc in any of the methods of cocaine exposure. In another study from the same group, a priming dose of cocaine (10 mg/kg, i.p.) increased the concentrations of AEA in the Hipp and cortex, 2-AG in the Hipp and NAcc, and CB1R and CB2R in the PFC and the lateral septal nuclei only in animals that had self-administered cocaine previously.
Interestingly, drug-induced relapse resulted in an increase in N-Acylphosphatidylethanolamines (NAPEs) levels in cortical areas and striatum and a decrease in oleoylethanolamide (OEA) and palmitoylethanolamide (PEA) levels in the NAcc, cerebellum, and Hipp. Conversely, a down-regulation of CB1R protein expression in the VTA was found. Therefore, all these changes indicate that the ECS is involved in cocaine-reinforced behaviors and drug-induced relapse [157].
The results of the ex vivo recordings of neurons in the VTA revealed that cocaine self-administration abolished the mechanisms of ECS-mediated long-term depression (LTD) in glutamatergic synapsis by an altered function of presynaptic CB1R [124]. In another study using the cocaine self-administration paradigm after a prolonged period of abstinence (30 days), increased and reduced DAGL and MAGL levels in the NAcc, respectively, were found during abstinence [123].
In a fascinating comparative study between different rat species, Lewis and Fischer 344 (F344) investigated, displaying a differential sensitivity to cocaine-induced reinstatement, the effects of cocaine self-administration (1 mg/kg per infusion, 21 days) on ECS components expression in the Hipp, which were measured by immunohistochemistry. In this study, Lewis rats showed lower CB1R and higher CB2R expression than Fisher rats measured 24 h after the last self-administration session [160].
Three CB2-specific probes were employed in quantitative real-time PCR (qPCR) and in situ hybridization (ISH) assays to evaluate how cocaine alters CB2R expression in striatal medial spiny neurons expressing dopamine D1 or D2 receptors (D1-MSNs, D2-MSNs) and in microglia. A single injection of cocaine does not alter its expression; however, repeated cocaine injections or self-administration up-regulated CB2R gene expression in both the brain (cortex and striatum) and periphery (spleen). In the striatum, CB2R up-regulation occurred mainly in the D1-MSNs of the NAcc. Brain CB2R could modulate cocaine action through D1-MSNs, considering their significant role in brain reward [161].
Free N-acyl-ethanolamines (NAEs) and 2-acylglycerols levels were analyzed in abstinent cocaine addicts from outpatient treatment programs diagnosed with cocaine use disorder, and in healthy control volunteers. In cocaine use disorder subjects, the NAEs were increased, and 2-acylglycerols decreased compared to controls. In addition, NAEs were significantly elevated in cocaine use disorder patients diagnosed with anxiety compared to cocaine use disorder subjects without this comorbidity. Notably, NAEs concentrations were increased by comorbid mood and anxiety disorders in cocaine addicts. Hence, these results suggest that NAE changes may be biomarkers of cocaine use disorder with psychiatric comorbidity [163].
Recently, hair concentrations of glucocorticoids (cortisone, cortisol), 2-AG, AEA, OEA, and PEA were compared among 48 recreational cocaine users, 25 cocaine-dependent users, and 67 non-stimulant controls. Significantly higher hair cortisone concentrations were found in recreational cocaine users and cocaine-dependent users compared to controls. OEA and PEA hair concentrations were substantially lower in cocaine-dependent users than recreational cocaine users and controls. No significant differences in AEA and 2-AG concentrations were found among the studied groups. Based on these results, the HPA axis and ECS appear to be critical regulators for cocaine use disorders [164].
3.4.3. Concluding Remarks—ECS and Stimulant-Related Disorders
Alterations in the different components of the ECS were found at central and peripheral levels in animals and humans exposed to psychostimulant drugs such as amphetamines and cocaine. Specific polymorphisms of the CNR1 gene or the FAAH rs324420 SNP have been associated with the development of dependence on stimulants (i.e., cocaine and METH). Moreover, multiple changes of ECS targets were detected mainly in studies with cocaine, revealing the importance of this system in regulating the different aspects of stimulant addiction, such as reward, motivation, withdrawal, or relapse. Even though more studies in both animals and humans are necessary, the current evidence suggests that ECS components may become promising biomarkers in stimulant-related disorders, and may reach a relevant implication in improving their therapeutic approach.
3.5. Tobacco-Related Disorders
According to the World Health Organization (WHO), the tobacco epidemic is one of the biggest public health threats worldwide. Tobacco kills more than 8 million people each year, primarily due to its consumption [165]. In that sense, developing new preventive and therapeutic avenues is crucial to lessen tobacco-related disorders’ impact. Several lines of evidence have shown that the ECS regulates nicotine’s reinforcing and motivational actions, with a particular focus on the therapeutic opportunities related to the pharmacological manipulation of CB1R [166,167]. The close overlap of cannabinoid and nicotinic acetylcholine receptors in reward-related brain areas suggests the interaction between both systems [168] (Table 5).

Table 5.
Main findings from human and animal studies aiming to identify the alterations of ECS components in tobacco-related disorders.
3.5.1. Gene Polymorphisms of ECS Components
Distinct CNR1 gene allelic variants have been studied in patients with nicotine dependence. The first one, published in 2008, genotyped 10 SNPs in the CNR1 in 2 independent samples (Virginia Study of Nicotine Dependence (VAND) and Virginia Study of Anxiety and Neuroticism (VAANX)) to test for association with smoking initiation and nicotine dependence. Concretely, 2 out of 10 markers (SNPs rs6928499 and rs2023239) were significantly associated with both measured parameters. Besides, the authors performed haplotype analyses, determining that the haplotype 1-1-2 of SNPs rs2023239-rs12720071-rs806368 was associated with nicotine dependence in both independent samples. Importantly, all these associations were female-specific according to the stratification by sex [169]. Another study investigated the influence of CNR1 SNP rs2023239 in nicotine reinforcement, and cue-elicited craving in regular tobacco smokers. The C allele variant of this SNP was associated with reduced nicotine reinforcement, whereas no changes were detected concerning craving measures. These results pointed out the involvement of CB1R in nicotine addiction, although further studies are necessary to improve the understanding of CB1R-dependent mechanisms underlying the regulation of nicotine rewarding properties [170].
In addition, Evans and colleagues evaluated if specific CNR1 polymorphisms comprising the TAG haplotype (SNPs rs806379, rs1535255, and rs2023239) could be associated with cognitive impairment during nicotine withdrawal. Interestingly, the analyses revealed that tobacco smokers homozygous for the major allele of the CNR1 SNP rs806379 attenuated the cognitive disruption induced by nicotine withdrawal. This result suggested the therapeutic potential of CB1R blockade for smoking cessation, particularly in individuals with more significant nicotine withdrawal-related cognitive alterations [171].
3.5.2. Gene and Protein Function/Expression Changes of ECS Components
Among the first approaches to elucidate the involvement of ECS components on nicotine-induced effects are studies evaluating the consequences of repeated exposure to nicotine in rodents. One of the first reports demonstrates that nicotine produces significant modifications of different ECS components, suggesting its pivotal role in nicotine addiction [172]. Thus, a 7-day administration of nicotine to Wistar rats induced significant changes in the levels of AEA and 2-AG in several brain areas (cerebral cortex, limbic forebrain, striatum, Hipp, and brainstem), as well as concrete alterations of CB1R gene or protein expression in the septum and cerebral cortex, respectively.
Early-life exposure to nicotine is a critical concern considering the high vulnerability to induce long-term alterations in the normal brain development during infancy and/or adolescence, including those related to disturbances in the modulatory role of the ECS. In this regard, various rodent studies have focused on evaluating how nicotine administration during postnatal or adolescence may impair ECS components’ function or expression. Torres et al. exposed C57BL/6J mouse pups from postnatal day 3 (PND3) to PND14 to tobacco smoke (two 1 h exposures per day, 3R4F reference cigarettes). At different periods (infancy, adolescence, and adulthood), protein expression changes in CB1R, CB2R, NAPE-PLD, DAGL, FAAH, and MAGL in the brainstem and striatum of mice were analyzed. Curiously, differential alterations occurred depending on the brain region and age, although infancy was the most critical period for changes in the ECS components. Authors concluded that exposure to tobacco smoke during critical early life periods could disturb the ECS in regions involved in sudden infant death syndrome, emotional regulation, and vulnerability to drug addiction [173].
Likewise, a subchronic nicotine treatment’s short- and long-term effects (0.4 mg/kg/day, i.p.) during adolescence (from PND34 to PND43) on hippocampal and striatal CB1R were analyzed in Wistar rats of both sexes. Interestingly, whereas no short-term changes on CB1R protein expression were observed, there were very significant long-term CB1R density alterations in the hippocampus and striatum, with a similar effect in male and female rats. Furthermore, these modifications were accompanied by a significant down-regulation of mu-opioid receptors in both brain regions and sexes. These results suggested that juvenile nicotine exposure induces profound and long-lasting disturbances in the endocannabinoid and opioid systems, which may account for higher vulnerability to developing drug addiction later in life [174]. In addition, Mateos and colleagues also evaluated the long-term effects of adolescent exposure to nicotine (1.4 mg/kg/day, i.p., from PND28 to PND43) on recognition memory, emotional behavior, and CB1R activity in cingulated cortex and Hipp. Surprisingly, nicotine-induced memory impairments were present only in females, whereas an increase in CB1R function (CP-55,940-stimulated GTPγS binding) was only found in male rats [175].
Consequences of nicotine exposure (7 days, 0.4 mg/kg/day, i.p.) on CB1R density during the adolescence (PND30) or adulthood (PND60) of Sprague–Dawley rats were notably different. In several regions of the PFC and Hipp and the VTA, quantitative autoradiography revealed a significant increase in CB1R binding, while no changes were detected in adult-treated rats. These changes were associated with a higher susceptibility to the locomotor-decreasing effects of both THC and CP-55,940, only in nicotine-treated rats during the adolescent period. These findings confirm the close functional interaction between nicotinic and cannabinoid systems, pointing out that early nicotine exposure could heighten the vulnerability to cannabis use [176].
Behavioral sensitization induced by juvenile exposure to nicotine in adolescent Sprague–Dawley rats (PND28) was helpful to classify the animals as low or high responders (depending on the nicotine-induced hyperlocomotion effects). Following a 1-week or 3-weeks injection-free period, all the animals received a low dose nicotine challenge (0.1 mg/kg, s.c.) to evaluate locomotor sensitization, social interaction, and CNR1 gene expression in the BLA and CeA. Only high responders showed nicotine-induced locomotor sensitization and decreased social interaction, together with a significant reduction of CNR1 mRNA expression in BLA and CeA after a 1-week injection-free period [177].
In another study, nicotine was combined with a high-fat diet (HFD) for 4 weeks to evaluate the consequences on CNR1 gene expression in hypothalamic nuclei (arcuate, paraventricular, ventromedial, lateral) and Hipp. The authors demonstrated that a significant up-regulation of CNR1 mRNA levels was found when combining nicotine and HFD. These results suggest enhanced endocannabinoid response in diet-induced obesity combined with nicotine exposure [178].
To date, two publications have specifically evaluated the involvement of the ECS in the regulation of nicotine withdrawal. First, a 7-day nicotine dependence procedure (5.2 mg/kg/day, transdermal patch) was performed in Wistar rats to finally measure several spontaneous somatic withdrawal signs, including locomotor activity and anxiety-like behavior, at 16 h and 34 h after patch removal. AEA levels were significantly lower in the Hipp of abstinent rats at 16 h, whereas they were higher in the Amy and hypothalamus at 34 h of nicotine withdrawal. However, the levels of 2-AG did not differ between nicotine withdrawal and control groups [179]. Second, precipitated nicotine withdrawal was induced in C57BL/6J mice (25 mg/kg/day, 14 days, Alzet minipump), showing episodic memory deficits evaluated in an object recognition task. Interestingly, this cognitive impairment was associated with a significant increase of 2-AG levels and a reduction in MAGL protein expression [180]. Altogether, these findings emphasize how ECS components are significantly modified during nicotine withdrawal, providing essential clues for developing cannabinoid-based pharmacological strategies to maintain abstinence and avoid nicotine relapse.
In a self-administration paradigm, Buczynski et al. compared the effects of volitional intake and forced administration of nicotine on levels of eCBs detected by in vivo microdialysis in the VTA of Wistar rats. Interestingly, nicotine self-administration (volitional), but not yoked administration (forced), reduced oleoylethanolamine (OEA) and increased AEA levels in the VTA in comparison with control animals. However, 2-AG levels increased regardless of the nicotine exposure paradigm, and FAAH activity was not affected. These results demonstrated for the first time that nicotine-induced modulation of eCBs levels was influenced by the volitional nature of drug exposure [181].
3.5.3. Neuroimaging of ECS Components
Very few PET imaging studies have linked tobacco consumption and cannabinoid receptors. A PET imaging study using the CB1R selective ligand [18F]FMPEP revealed a 20% lower total brain CB1R density in tobacco smokers who were otherwise healthy than the non-smoking group [183]. In a study performed in patients with schizophrenia and healthy controls, tobacco consumption was also studied. The results showed that [11C]OMAR VT was lower in the Amy, caudate, Hipp, hypothalamus, insula, putamen pallidum, frontal, parietal, occipital, and posterior cingulate cortices of schizophrenic patients than in healthy controls. In contrast, there were no significant differences in [11C]OMAR VT between smoker schizophrenic patients and healthy controls. Significantly lower [11C]OMAR VT was detected in non-smoker versus smoker schizophrenics in the parietal and occipital cortices [184]. Interestingly, while in healthy controls, tobacco consumption decreases the availability of CB1R, in patients with schizophrenia, a slight increase was found.
In an earlier study in female rats by Gérard et al., they scanned the rats at baseline and after chronic administration of nicotine using [18F]MK-9470. Their results showed that chronic nicotine administration did not significantly change CB1R availability in rats [182].
3.5.4. Concluding Remarks—ECS- and Tobacco-Related Disorders
Although there is limited evidence regarding the role of the ECS components in tobacco addiction, some interesting conclusions can be extracted from the previously reviewed studies. Some polymorphisms of the CNR1 gene have been suggested to be associated with nicotine dependence, and, interestingly, the rs806379 SNP appears to be a protective factor, since it has been related to an attenuated cognitive disruption during nicotine withdrawal. On the other hand, exposure to nicotine at early life stages is closely related to long-term disturbances in specific ECS markers. Furthermore, animal models evaluating nicotine withdrawal or motivation have revealed differential changes in the levels of eCBs. Finally, very scarce neuroimaging data suggest a reduction of CB1R availability in tobacco smokers. Additional research is essential to improve the understanding of ECS involvement in nicotine addiction and identify potential biomarkers.
3.6. Hallucinogen-Related Disorders
According to the National Institute on Drug Abuse (NIDA), hallucinogens can be classified into two categories: (1) Classical hallucinogens (D-lysergic acid diethylamide (LSD), 4-phosphoryloxy-N,N-dimethyltryptamine (psilocybin), mescaline, N,N-dimethyltryptamine (DMT), and 251-NBOMe), and (2) Dissociative hallucinogens (Phencyclidine (PCP), ketamine, dextromethorphan or Salvia divinorum). Both hallucinogens can generate visual and auditory sensations that are not real. At the same time, dissociative drugs, additionally, can make people who consume these substances feel out of control or disconnected from their body and environment [185]. These substances are primarily used recreationally. Indeed, 7.5% of adolescents consumed hallucinogens at some time in their lives, according to data reported in 2020 [186]. Therefore, due to the potential health impact of hallucinogen use, it is necessary to look for potential biomarkers to design better preventive and therapeutic strategies (Table 6).

Table 6.
Main findings from human and animal studies aiming to identify alterations of ECS components in hallucinogen-related disorders.
3.6.1. Gene and Protein Function/Expression Changes of ECS Components
A study carried out by Guimarães et al. in a cohort of healthy volunteers showed that oral administration of Ayahuasca, a tisane containing alkaloids such as DMT, reduced plasma concentrations of AEA at 4 h, while 2-AG was slightly reduced at one and a half hours, followed by an increase at 4 h [187].
On the other hand, several studies evaluated the effect of dissociative drugs on the endogenous cannabinoid system. The subchronic administration of ketamine in mice increased the concentrations of AEA in the CPu, Amy, and PFC. However, 2-AG concentrations were increased in CPu and NAcc, but decreased in PFC. Moreover, MGLL mRNA levels were reduced in CPu and PFC, although only MAGL protein expression in CPu was reduced [188]. Acute postnatal administration of PCP in mice reduced CB1R levels in the PFC, while increasing it in the dental gyrus of Hipp [189].
Furthermore, in two sub-chronic studies in rats, it was found that AEA levels were increased in the NAcc after a motor activity test [190] or social interaction [191]. However, in the social interaction study, 2-AG levels increased in the NAcc and CPu, AEA decreased in the medial PFC (mPFC) and Amy, and NAPE-PLD expression increased in the mPFC [191]. Interestingly, Vigano et al. showed that the chronic administration of PCP in rats increased 2-AG levels in PFC, together with an increase in CB1R density in Amy and VTA [192].
3.6.2. Concluding Remarks—ECS and Hallucinogen-Related Disorders
Although classical and dissociative hallucinogens produce alterations in the ECS, very scarce information is available, so further studies are needed to analyze their potential use as biomarkers in hallucinogen-related disorders.
4. Conclusions and Future Perspectives
The evidence from rodent and human studies gathered in this narrative review highlights the alterations that occur in the main components of the ECS upon exposure to drugs of abuse, especially at early life stages or associated with distinct addictive phases (acute/chronic exposure, dependence, withdrawal, or relapse). It is important to note that the currently available information supports the potential usefulness of identifying changes of cannabinoid receptors, ligands, or enzymes as biomarkers to improve the diagnostic classification of patients with SUD, and increase the success of their pharmacological treatment. Nonetheless, the results included in the present review should be interpreted with caution due to some limitations. For instance, several reviewed studies did not consider differences in gender, age, or associated comorbidities such as polyconsumption, a prevalent circumstance in addicted individuals. Furthermore, there is a great variety in the experimental designs employed, particularly with animal models regarding drug exposure duration, doses, and administration patterns. Thus, one of the biggest challenges in future studies is replicating the available results by applying similar procedures. Only in this way will it be possible to be sure about the direction and magnitude of the changes in the different components of the ECS, which is essential in the search for reliable biomarkers with potential application in the clinical setting. In this regard, a multidisciplinary and translational approach combining cutting-edge technologies (i.e., omics) in biological samples from animal models and patients is crucial to rapidly understanding the complex role that the ECS plays in drug addiction. Finally, well-designed clinical studies employing low invasive methods (e.g., neuroimaging) and accessible biological samples (e.g., blood), as well as specific selection criteria, are mandatory to explore further how ECS components could serve as potential diagnostic, prognostic, monitoring, or therapeutic biomarkers in substance-related disorders.
Author Contributions
F.N. and J.M. designed, coordinated, and reviewed the sections and contents of this manuscript and oversaw the organization to distribute the writing tasks among the authors. F.N., M.S.G.-G., A.G., D.N., F.L.-P. and Á.M. performed the literature searches. F.N., M.S.G.-G., A.G., D.N., F.L.-P., Á.M., T.F. and J.M. participated in the manuscript writing. All the authors critically reviewed, and J.M. approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The preparation of the manuscript was supported by “Ministerio de Sanidad, Delegación del Gobierno para el Plan Nacional Sobre Drogas” (PNSD, 2019I012 to J.M.), “ISCIII-Redes de Investigación Cooperativa Orientadas a Resultados en Salud (RICORS), Red de Investigación en Atención Primaria de Adicciones (RIAPAd)” (grant number RD21/0009/0008 to J.M.), “Ministerio de Ciencia e Innovación programa Ramón y Cajal” (RYC201722666 to T.F.), and “Turku University hospital” (grant to F.L.-P.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- UNODC. World Drug Report; UNODC: Vienna, Austria, 2021. [Google Scholar]
- WHO. Global Status Report on Alcohol and Health; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Bough, K.J.; Pollock, J.D. Defining Substance Use Disorders: The Need for Peripheral Biomarkers. Trends Mol. Med. 2018, 24, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Carrison, K.A.; Potenza, M.N. Neuroimaging and biomarkers in addiction treatment. Curr. Psychiatry Rep. 2014, 16, 513. [Google Scholar] [CrossRef] [PubMed]
- Kwako, L.E.; Bickel, W.K.; Goldman, D. Addiction Biomarkers: Dimensional Approaches to Understanding Addiction. Trends Mol. Med. 2018, 24, 121–128. [Google Scholar] [CrossRef]
- Volkow, N.D.; Koob, G.; Baler, R. Biomarkers in substance use disorders. ACS Chem. Neurosci. 2015, 6, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J.; Luscher, C. The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 2019, 102, 48–59. [Google Scholar] [CrossRef]
- Maldonado, R. The neurobiology of addiction. In Addiction Mechanisms, Phenomenology and Treatment; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar] [CrossRef]
- Di Chiara, G.; Bassareo, V.; Fenu, S.; De Luca, M.A.; Spina, L.; Cadoni, C.; Acquas, E.; Carboni, E.; Valentini, V.; Lecca, D. Dopamine and drug addiction: The nucleus accumbens shell connection. Neuropharmacology 2004, 47 (Suppl. S1), 227–241. [Google Scholar] [CrossRef] [PubMed]
- Koob, G.F.; Volkow, N.D. Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry 2016, 3, 760–773. [Google Scholar] [CrossRef]
- Battaglia, S.; Serio, G.; Scarpazza, C.; D’Ausilio, A.; Borgomaneri, S. Frozen in (e)motion: How reactive motor inhibition is influenced by the emotional content of stimuli in healthy and psychiatric populations. Behav. Res. Ther. 2021, 146, 103963. [Google Scholar] [CrossRef] [PubMed]
- Jennings, J.H.; Sparta, D.R.; Stamatakis, A.M.; Ung, R.L.; Pleil, K.E.; Kash, T.L.; Stuber, G.D. Distinct extended amygdala circuits for divergent motivational states. Nature 2013, 496, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Belin, D.; Belin-Rauscent, A.; Murray, J.E.; Everitt, B.J. Addiction: Failure of control over maladaptive incentive habits. Curr. Opin. Neurobiol. 2013, 23, 564–572. [Google Scholar] [CrossRef]
- Battaglia, S.; Harrison, B.J.; Fullana, M.A. Does the human ventromedial prefrontal cortex support fear learning, fear extinction or both? A commentary on subregional contributions. Mol. Psychiatry 2021, 1–3. [Google Scholar] [CrossRef]
- Battaglia, S.; Garofalo, S.; di Pellegrino, G.; Starita, F. Revaluing the Role of vmPFC in the Acquisition of Pavlovian Threat Conditioning in Humans. J. Neurosci. 2020, 40, 8491–8500. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, R.Z.; Volkow, N.D. Dysfunction of the prefrontal cortex in addiction: Neuroimaging findings and clinical implications. Nat. Rev. Neurosci. 2011, 12, 652–669. [Google Scholar] [CrossRef] [PubMed]
- Vitale, F.; Monti, I.; Padron, I.; Avenanti, A.; de Vega, M. The neural inhibition network is causally involved in the disembodiment effect of linguistic negation. Cortex 2022, 147, 72–82. [Google Scholar] [CrossRef]
- Volkow, N.D.; Koob, G.F.; McLellan, A.T. Neurobiologic Advances from the Brain Disease Model of Addiction. N. Engl. J. Med. 2016, 374, 363–371. [Google Scholar] [CrossRef]
- Morcuende, A.; Navarrete, F.; Nieto, E.; Manzanares, J.; Femenia, T. Inflammatory Biomarkers in Addictive Disorders. Biomolecules 2021, 11, 1824. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Toth, F.; Polyak, H.; Szabo, A.; Mandi, Y.; Vecsei, L. Immune Influencers in Action: Metabolites and Enzymes of the Tryptophan-Kynurenine Metabolic Pathway. Biomedicines 2021, 9, 734. [Google Scholar] [CrossRef]
- Morales-Puerto, N.; Gimenez-Gomez, P.; Perez-Hernandez, M.; Abuin-Martinez, C.; Gil de Biedma-Elduayen, L.; Vidal, R.; Gutierrez-Lopez, M.D.; O’Shea, E.; Colado, M.I. Addiction and the kynurenine pathway: A new dancing couple? Pharmacol. Ther. 2021, 223, 107807. [Google Scholar] [CrossRef]
- Tanaka, M.; Vecsei, L. Monitoring the kynurenine system: Concentrations, ratios or what else? Adv. Clin. Exp. Med. 2021, 30, 775–778. [Google Scholar] [CrossRef]
- Maldonado, R.; Valverde, O.; Berrendero, F. Involvement of the endocannabinoid system in drug addiction. Trends Neurosci. 2006, 29, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Manzanares, J.; Cabanero, D.; Puente, N.; Garcia-Gutierrez, M.S.; Grandes, P.; Maldonado, R. Role of the endocannabinoid system in drug addiction. Biochem. Pharmacol. 2018, 157, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Piomelli, D. The molecular logic of endocannabinoid signalling. Nat. Rev. Neurosci. 2003, 4, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Kumar, U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. [Google Scholar] [CrossRef] [PubMed]
- Katona, I.; Freund, T.F. Multiple functions of endocannabinoid signaling in the brain. Annu. Rev. Neurosci. 2012, 35, 529–558. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.L.; Abbott, J.K. Cannabinoids and the brain: The effects of endogenous and exogenous cannabinoids on brain systems and function. In The Complex Connection between Cannabis and Schizophrenia; Academic Press: Cambridge, MA, USA, 2018; pp. 37–74. [Google Scholar]
- Mackie, K. Distribution of cannabinoid receptors in the central and peripheral nervous system. Handb. Exp. Pharmacol. 2005, 299–325. [Google Scholar] [CrossRef]
- Rodriguez de Fonseca, F.; Del Arco, I.; Bermudez-Silva, F.J.; Bilbao, A.; Cippitelli, A.; Navarro, M. The endocannabinoid system: Physiology and pharmacology. Alcohol Alcohol. 2005, 40, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Asth, L.; Santos, A.C.; Moreira, F.A. The endocannabinoid system and drug-associated contextual memories. Behav. Pharmacol. 2021, 621. [Google Scholar] [CrossRef]
- Parolaro, D.; Vigano, D.; Realini, N.; Rubino, T. Role of endocannabinoids in regulating drug dependence. Neuropsychiatr. Dis. Treat. 2007, 3, 711–721. [Google Scholar] [CrossRef][Green Version]
- Parsons, L.H.; Hurd, Y.L. Endocannabinoid signalling in reward and addiction. Nat. Rev. Neurosci. 2015, 16, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Hoenicka, J.; Ponce, G.; Jimenez-Arriero, M.A.; Ampuero, I.; Rodriguez-Jimenez, R.; Rubio, G.; Aragues, M.; Ramos, J.A.; Palomo, T. Association in alcoholic patients between psychopathic traits and the additive effect of allelic forms of the CNR1 and FAAH endocannabinoid genes, and the 3′ region of the DRD2 gene. Neurotox. Res. 2007, 11, 51–60. [Google Scholar] [CrossRef]
- van den Wildenberg, E.; Janssen, R.G.; Hutchison, K.E.; van Breukelen, G.J.; Wiers, R.W. Polymorphisms of the dopamine D4 receptor gene (DRD4 VNTR) and cannabinoid CB1 receptor gene (CNR1) are not strongly related to cue-reactivity after alcohol exposure. Addict. Biol. 2007, 12, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Elkrief, L.; Spinney, S.; Vosberg, D.E.; Banaschewski, T.; Bokde, A.L.W.; Quinlan, E.B.; Desrivieres, S.; Flor, H.; Garavan, H.; Gowland, P.; et al. Endocannabinoid Gene x Gene Interaction Association to Alcohol Use Disorder in Two Adolescent Cohorts. Front. Psychiatry 2021, 12, 645746. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Ishiguro, H.; Higuchi, S.; Onaivi, E.S.; Arinami, T. Association study between alcoholism and endocannabinoid metabolic enzyme genes encoding fatty acid amide hydrolase and monoglyceride lipase in a Japanese population. Psychiatr. Genet. 2007, 17, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, H.; Iwasaki, S.; Teasenfitz, L.; Higuchi, S.; Horiuchi, Y.; Saito, T.; Arinami, T.; Onaivi, E.S. Involvement of cannabinoid CB2 receptor in alcohol preference in mice and alcoholism in humans. Pharm. J. 2007, 7, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Kranzler, H.R.; Luo, X.; Covault, J.; Gelernter, J. CNR1 variation modulates risk for drug and alcohol dependence. Biol. Psychiatry 2007, 62, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Marcos, M.; Pastor, I.; de la Calle, C.; Barrio-Real, L.; Laso, F.J.; Gonzalez-Sarmiento, R. Cannabinoid receptor 1 gene is associated with alcohol dependence. Alcohol. Clin. Exp. Res. 2012, 36, 267–271. [Google Scholar] [CrossRef]
- Schmidt, L.G.; Samochowiec, J.; Finckh, U.; Fiszer-Piosik, E.; Horodnicki, J.; Wendel, B.; Rommelspacher, H.; Hoehe, M.R. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002, 65, 221–224. [Google Scholar] [CrossRef]
- Preuss, U.W.; Koller, G.; Zill, P.; Bondy, B.; Soyka, M. Alcoholism-related phenotypes and genetic variants of the CB1 receptor. Eur. Arch. Psychiatry Clin. Neurosci. 2003, 253, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, K.E.; Haughey, H.; Niculescu, M.; Schacht, J.; Kaiser, A.; Stitzel, J.; Horton, W.J.; Filbey, F. The incentive salience of alcohol: Translating the effects of genetic variant in CNR1. Arch. Gen. Psychiatry 2008, 65, 841–850. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buhler, K.M.; Huertas, E.; Echeverry-Alzate, V.; Gine, E.; Molto, E.; Montoliu, L.; Lopez-Moreno, J.A. Risky alcohol consumption in young people is associated with the fatty acid amide hydrolase gene polymorphism C385A and affective rating of drug pictures. Mol. Genet. Genom. 2014, 289, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Kazmi, N.; Brooks, A.T.; Krumlauf, M.; Schwandt, M.L.; George, D.T.; Hodgkinson, C.A.; Wallen, G.R.; Ramchandani, V.A. FAAH and CNR1 Polymorphisms in the Endocannabinoid System and Alcohol-Related Sleep Quality. Front. Psychiatry 2021, 12, 712178. [Google Scholar] [CrossRef] [PubMed]
- Sloan, M.E.; Gowin, J.L.; Yan, J.; Schwandt, M.L.; Spagnolo, P.A.; Sun, H.; Hodgkinson, C.A.; Goldman, D.; Ramchandani, V.A. Severity of alcohol dependence is associated with the fatty acid amide hydrolase Pro129Thr missense variant. Addict. Biol. 2018, 23, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Best, L.M.; Wardell, J.D.; Tyndale, R.F.; McPhee, M.D.; Le Foll, B.; Kish, S.J.; Boileau, I.; Hendershot, C.S. Association of the Fatty Acid Amide Hydrolase C385A Polymorphism with Alcohol Use Severity and Coping Motives in Heavy-Drinking Youth. Alcohol. Clin. Exp. Res. 2021, 45, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, T.; Lee, F.; Kreek, M.J. Involvement of Endocannabinoids in Alcohol “Binge” Drinking: Studies of Mice with Human Fatty Acid Amide Hydrolase Genetic Variation and After CB1 Receptor Antagonists. Alcohol. Clin. Exp. Res. 2016, 40, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.M.; Ortiz, S.; Perez-Rial, S.; Manzanares, J. Time dependent alterations on tyrosine hydroxylase, opioid and cannabinoid CB1 receptor gene expressions after acute ethanol administration in the rat brain. Eur. Neuropsychopharmacol. 2008, 18, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, S.; Oliva, J.M.; Perez-Rial, S.; Palomo, T.; Manzanares, J. Chronic ethanol consumption regulates cannabinoid CB1 receptor gene expression in selected regions of rat brain. Alcohol Alcohol. 2004, 39, 88–92. [Google Scholar] [CrossRef][Green Version]
- Fu, R.; Tang, Y.; Li, W.; Ren, Z.; Li, D.; Zheng, J.; Zuo, W.; Chen, X.; Zuo, Q.K.; Tam, K.L.; et al. Endocannabinoid signaling in the lateral habenula regulates pain and alcohol consumption. Transl. Psychiatry 2021, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Hay, E.A.; McEwan, A.; Wilson, D.; Barrett, P.; D’Agostino, G.; Pertwee, R.G.; MacKenzie, A. Disruption of an enhancer associated with addictive behaviour within the cannabinoid receptor-1 gene suggests a possible role in alcohol intake, cannabinoid response and anxiety-related behaviour. Psychoneuroendocrinology 2019, 109, 104407. [Google Scholar] [CrossRef] [PubMed]
- Romano-Lopez, A.; Mendez-Diaz, M.; Ruiz-Contreras, A.E.; Carrisoza, R.; Prospero-Garcia, O. Maternal separation and proclivity for ethanol intake: A potential role of the endocannabinoid system in rats. Neuroscience 2012, 223, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Hansson, A.C.; Bermudez-Silva, F.J.; Malinen, H.; Hyytia, P.; Sanchez-Vera, I.; Rimondini, R.; Rodriguez de Fonseca, F.; Kunos, G.; Sommer, W.H.; Heilig, M. Genetic impairment of frontocortical endocannabinoid degradation and high alcohol preference. Neuropsychopharmacology 2007, 32, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Henricks, A.M.; Berger, A.L.; Lugo, J.M.; Baxter-Potter, L.N.; Bieniasz, K.V.; Petrie, G.; Sticht, M.A.; Hill, M.N.; McLaughlin, R.J. Sex- and hormone-dependent alterations in alcohol withdrawal-induced anxiety and corticolimbic endocannabinoid signaling. Neuropharmacology 2017, 124, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.; Rivera, P.; Pavon, F.J.; Decara, J.; Suarez, J.; Rodriguez de Fonseca, F.; Parsons, L.H. Differential effects of single versus repeated alcohol withdrawal on the expression of endocannabinoid system-related genes in the rat amygdala. Alcohol. Clin. Exp. Res. 2012, 36, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Marin, L.; Pavon, F.J.; Decara, J.; Suarez, J.; Gavito, A.; Castilla-Ortega, E.; Rodriguez de Fonseca, F.; Serrano, A. Effects of Intermittent Alcohol Exposure on Emotion and Cognition: A Potential Role for the Endogenous Can.n.n.n.nnabinoid System and Neuroinflammation. Front. Behav. Neurosci. 2017, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Vinod, K.Y.; Kassir, S.A.; Hungund, B.L.; Cooper, T.B.; Mann, J.J.; Arango, V. Selective alterations of the CB1 receptors and the fatty acid amide hydrolase in the ventral striatum of alcoholics and suicides. J. Psychiatr. Res. 2010, 44, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Agudelo, M.; Yndart, A.; Morrison, M.; Figueroa, G.; Munoz, K.; Samikkannu, T.; Nair, M.P. Differential expression and functional role of cannabinoid genes in alcohol users. Drug Alcohol Depend. 2013, 133, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Ceccarini, J.; Casteels, C.; Koole, M.; Bormans, G.; Van Laere, K. Transient changes in the endocannabinoid system after acute and chronic ethanol exposure and abstinence in the rat: A combined PET and microdialysis study. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Neumeister, A.; Normandin, M.D.; Murrough, J.W.; Henry, S.; Bailey, C.R.; Luckenbaugh, D.A.; Tuit, K.; Zheng, M.Q.; Galatzer-Levy, I.R.; Sinha, R.; et al. Positron emission tomography shows elevated cannabinoid CB1 receptor binding in men with alcohol dependence. Alcohol. Clin. Exp. Res. 2012, 36, 2104–2109. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, J.; Zanotti-Fregonara, P.; Umhau, J.C.; George, D.T.; Rallis-Frutos, D.; Lyoo, C.H.; Li, C.T.; Hines, C.S.; Sun, H.; Terry, G.E.; et al. Reduced cannabinoid CB1 receptor binding in alcohol dependence measured with positron emission tomography. Mol. Psychiatry 2013, 18, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Ceccarini, J.; Hompes, T.; Verhaeghen, A.; Casteels, C.; Peuskens, H.; Bormans, G.; Claes, S.; Van Laere, K. Changes in cerebral CB1 receptor availability after acute and chronic alcohol abuse and monitored abstinence. J. Neurosci. 2014, 34, 2822–2831. [Google Scholar] [CrossRef] [PubMed]
- Best, L.M.; Williams, B.; Le Foll, B.; Mansouri, E.; Bazinet, R.P.; Lin, L.; De Luca, V.; Lagzdins, D.; Rusjan, P.; Tyndale, R.F.; et al. Lower brain fatty acid amide hydrolase in treatment-seeking patients with alcohol use disorder: A positron emission tomography study with [C-11]CURB. Neuropsychopharmacology 2020, 45, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.A.; Garcia, A.; Parkes, J.; Houle, S.; Tong, J.; Vasdev, N. [11C]CURB: Evaluation of a novel radiotracer for imaging fatty acid amide hydrolase by positron emission tomography. Nucl. Med. Biol. 2011, 38, 247–253. [Google Scholar] [CrossRef]
- Rusjan, P.M.; Wilson, A.A.; Mizrahi, R.; Boileau, I.; Chavez, S.E.; Lobaugh, N.J.; Kish, S.J.; Houle, S.; Tong, J. Mapping human brain fatty acid amide hydrolase activity with PET. J. Cereb. Blood Flow Metab. 2013, 33, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Hartman, C.A.; Hopfer, C.J.; Haberstick, B.; Rhee, S.H.; Crowley, T.J.; Corley, R.P.; Hewitt, J.K.; Ehringer, M.A. The association between cannabinoid receptor 1 gene (CNR1) and cannabis dependence symptoms in adolescents and young adults. Drug Alcohol Depend. 2009, 104, 11–16. [Google Scholar] [CrossRef]
- Hill, S.Y.; Sharma, V.; Jones, B.L. Lifetime use of cannabis from longitudinal assessments, cannabinoid receptor (CNR1) variation, and reduced volume of the right anterior cingulate. Psychiatry Res. Neuroimaging 2016, 255, 24–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agrawal, A.; Wetherill, L.; Dick, D.M.; Xuei, X.; Hinrichs, A.; Hesselbrock, V.; Kramer, J.; Nurnberger, J.I., Jr.; Schuckit, M.; Bierut, L.J.; et al. Evidence for association between polymorphisms in the cannabinoid receptor 1 (CNR1) gene and cannabis dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009, 150B, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Hopfer, C.J.; Young, S.E.; Purcell, S.; Crowley, T.J.; Stallings, M.C.; Corley, R.P.; Rhee, S.H.; Smolen, A.; Krauter, K.; Hewitt, J.K.; et al. Cannabis receptor haplotype associated with fewer cannabis dependence symptoms in adolescents. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141B, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Haughey, H.M.; Marshall, E.; Schacht, J.P.; Louis, A.; Hutchison, K.E. Marijuana withdrawal and craving: Influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction 2008, 103, 1678–1686. [Google Scholar] [CrossRef] [PubMed]
- Filbey, F.M.; Schacht, J.P.; Myers, U.S.; Chavez, R.S.; Hutchison, K.E. Individual and additive effects of the CNR1 and FAAH genes on brain response to marijuana cues. Neuropsychopharmacology 2010, 35, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Schacht, J.P.; Hutchison, K.E.; Filbey, F.M. Associations between cannabinoid receptor-1 (CNR1) variation and hippocampus and amygdala volumes in heavy cannabis users. Neuropsychopharmacology 2012, 37, 2368–2376. [Google Scholar] [CrossRef] [PubMed]
- Colizzi, M.; Fazio, L.; Ferranti, L.; Porcelli, A.; Masellis, R.; Marvulli, D.; Bonvino, A.; Ursini, G.; Blasi, G.; Bertolino, A. Functional genetic variation of the cannabinoid receptor 1 and cannabis use interact on prefrontal connectivity and related working memory behavior. Neuropsychopharmacology 2015, 40, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.C.; Wassink, T.H.; Ziebell, S.; Andreasen, N.C. Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr. Res. 2011, 128, 66–75. [Google Scholar] [CrossRef][Green Version]
- Onwuameze, O.E.; Nam, K.W.; Epping, E.A.; Wassink, T.H.; Ziebell, S.; Andreasen, N.C.; Ho, B.C. MAPK14 and CNR1 gene variant interactions: Effects on brain volume deficits in schizophrenia patients with marijuana misuse. Psychol. Med. 2013, 43, 619–631. [Google Scholar] [CrossRef]
- Gerra, M.C.; Jayanthi, S.; Manfredini, M.; Walther, D.; Schroeder, J.; Phillips, K.A.; Cadet, J.L.; Donnini, C. Gene variants and educational attainment in cannabis use: Mediating role of DNA methylation. Transl. Psychiatry 2018, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Arias Horcajadas, F.; Davila Piriz, J.R.; Parra Gonzalez, A.; Sanchez Romero, S.; Sanchez-Morla, E.; Ampuero Sanchez, I.; Ramos Atance, J.A. Cannabinoid receptor type 2 gene is associated with comorbidity of schizophrenia and cannabis dependence and fatty acid amide hydrolase gene is associated with cannabis dependence in the Spanish population. Adicciones 2021, 1587. [Google Scholar] [CrossRef]
- Sipe, J.C.; Chiang, K.; Gerber, A.L.; Beutler, E.; Cravatt, B.F. A missense mutation in human fatty acid amide hydrolase associated with problem drug use. Proc. Natl. Acad. Sci. USA 2002, 99, 8394–8399. [Google Scholar] [CrossRef] [PubMed]
- Hariri, A.R.; Gorka, A.; Hyde, L.W.; Kimak, M.; Halder, I.; Ducci, F.; Ferrell, R.E.; Goldman, D.; Manuck, S.B. Divergent effects of genetic variation in endocannabinoid signaling on human threat- and reward-related brain function. Biol. Psychiatry 2009, 66, 9–16. [Google Scholar] [CrossRef]
- Hindocha, C.; Freeman, T.P.; Schafer, G.; Gardner, C.; Bloomfield, M.A.P.; Bramon, E.; Morgan, C.J.A.; Curran, H.V. Acute effects of cannabinoids on addiction endophenotypes are moderated by genes encoding the CB1 receptor and FAAH enzyme. Addict. Biol. 2020, 25, e12762. [Google Scholar] [CrossRef]
- Melroy-Greif, W.E.; Wilhelmsen, K.C.; Ehlers, C.L. Genetic variation in FAAH is associated with cannabis use disorders in a young adult sample of Mexican Americans. Drug Alcohol Depend. 2016, 166, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Tyndale, R.F.; Payne, J.I.; Gerber, A.L.; Sipe, J.C. The fatty acid amide hydrolase C385A (P129T) missense variant in cannabis users: Studies of drug use and dependence in Caucasians. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144, 660–666. [Google Scholar] [CrossRef]
- Carey, C.E.; Agrawal, A.; Zhang, B.; Conley, E.D.; Degenhardt, L.; Heath, A.C.; Li, D.; Lynskey, M.T.; Martin, N.G.; Montgomery, G.W.; et al. Monoacylglycerol lipase (MGLL) polymorphism rs604300 interacts with childhood adversity to predict cannabis dependence symptoms and amygdala habituation: Evidence from an endocannabinoid system-level analysis. J. Abnorm. Psychol. 2015, 124, 860–877. [Google Scholar] [CrossRef] [PubMed]
- Rubino, T.; Massi, P.; Patrini, G.; Venier, I.; Giagnoni, G.; Parolaro, D. Chronic CP-55,940 alters cannabinoid receptor mRNA in the rat brain: An in situ hybridization study. Neuroreport 1994, 5, 2493–2496. [Google Scholar] [CrossRef] [PubMed]
- Romero, J.; Berrendero, F.; Manzanares, J.; Perez, A.; Corchero, J.; Fuentes, J.A.; Fernandez-Ruiz, J.J.; Ramos, J.A. Time-course of the cannabinoid receptor down-regulation in the adult rat brain caused by repeated exposure to delta9-tetrahydrocannabinol. Synapse 1998, 30, 298–308. [Google Scholar] [CrossRef]
- Romero, J.; Garcia-Palomero, E.; Castro, J.G.; Garcia-Gil, L.; Ramos, J.A.; Fernandez-Ruiz, J.J. Effects of chronic exposure to delta9-tetrahydrocannabinol on cannabinoid receptor binding and mRNA levels in several rat brain regions. Brain Res. Mol. Brain Res. 1997, 46, 100–108. [Google Scholar] [CrossRef]
- Ellgren, M.; Artmann, A.; Tkalych, O.; Gupta, A.; Hansen, H.S.; Hansen, S.H.; Devi, L.A.; Hurd, Y.L. Dynamic changes of the endogenous cannabinoid and opioid mesocorticolimbic systems during adolescence: THC effects. Eur. Neuropsychopharmacol. 2008, 18, 826–834. [Google Scholar] [CrossRef]
- Oliva, J.M.; Ortiz, S.; Palomo, T.; Manzanares, J. Spontaneous cannabinoid withdrawal produces a differential time-related responsiveness in cannabinoid CB1 receptor gene expression in the mouse brain. J. Psychopharmacol. 2004, 18, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.M.; Ortiz, S.; Palomo, T.; Manzanares, J. Behavioural and gene transcription alterations induced by spontaneous cannabinoid withdrawal in mice. J. Neurochem. 2003, 85, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, F.; Aracil-Fernandez, A.; Manzanares, J. Cannabidiol regulates behavioural alterations and gene expression changes induced by spontaneous cannabinoid withdrawal. Br. J. Pharmacol. 2018, 175, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Oviedo, A.; Glowa, J.; Herkenham, M. Chronic cannabinoid administration alters cannabinoid receptor binding in rat brain: A quantitative autoradiographic study. Brain Res. 1993, 616, 293–302. [Google Scholar] [CrossRef]
- Rodriguez de Fonseca, F.; Gorriti, M.A.; Fernandez-Ruiz, J.J.; Palomo, T.; Ramos, J.A. Downregulation of rat brain cannabinoid binding sites after chronic delta 9-tetrahydrocannabinol treatment. Pharmacol. Biochem. Behav. 1994, 47, 33–40. [Google Scholar] [CrossRef]
- Tai, S.; Hyatt, W.S.; Gu, C.; Franks, L.N.; Vasiljevik, T.; Brents, L.K.; Prather, P.L.; Fantegrossi, W.E. Repeated administration of phytocannabinoid Delta(9)-THC or synthetic cannabinoids JWH-018 and JWH-073 induces tolerance to hypothermia but not locomotor suppression in mice, and reduces CB1 receptor expression and function in a brain region-specific manner. Pharmacol. Res. 2015, 102, 22–32. [Google Scholar] [CrossRef]
- Breivogel, C.S.; Childers, S.R.; Deadwyler, S.A.; Hampson, R.E.; Vogt, L.J.; Sim-Selley, L.J. Chronic delta9-tetrahydrocannabinol treatment produces a time-dependent loss of cannabinoid receptors and cannabinoid receptor-activated G proteins in rat brain. J. Neurochem. 1999, 73, 2447–2459. [Google Scholar] [CrossRef] [PubMed]
- Romero, J.; Garcia, L.; Fernandez-Ruiz, J.J.; Cebeira, M.; Ramos, J.A. Changes in rat brain cannabinoid binding sites after acute or chronic exposure to their endogenous agonist, anandamide, or to delta 9-tetrahydrocannabinol. Pharmacol. Biochem. Behav. 1995, 51, 731–737. [Google Scholar] [CrossRef]
- Gonzalez, S.; Fernandez-Ruiz, J.; Di Marzo, V.; Hernandez, M.; Arevalo, C.; Nicanor, C.; Cascio, M.G.; Ambrosio, E.; Ramos, J.A. Behavioral and molecular changes elicited by acute administration of SR141716 to Delta9-tetrahydrocannabinol-tolerant rats: An experimental model of cannabinoid abstinence. Drug Alcohol Depend. 2004, 74, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Berrendero, F.; Bisogno, T.; Gonzalez, S.; Cavaliere, P.; Romero, J.; Cebeira, M.; Ramos, J.A.; Fernandez-Ruiz, J.J. Enhancement of anandamide formation in the limbic forebrain and reduction of endocannabinoid contents in the striatum of delta9-tetrahydrocannabinol-tolerant rats. J. Neurochem. 2000, 74, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Burgdorf, C.E.; Jing, D.; Yang, R.; Huang, C.; Hill, M.N.; Mackie, K.; Milner, T.A.; Pickel, V.M.; Lee, F.S.; Rajadhyaksha, A.M. Endocannabinoid genetic variation enhances vulnerability to THC reward in adolescent female mice. Sci. Adv. 2020, 6, eaay1502. [Google Scholar] [CrossRef] [PubMed]
- Rotter, A.; Bayerlein, K.; Hansbauer, M.; Weiland, J.; Sperling, W.; Kornhuber, J.; Biermann, T. CB1 and CB2 receptor expression and promoter methylation in patients with cannabis dependence. Eur. Addict. Res. 2013, 19, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Walter, C.; Ferreiros, N.; Bishay, P.; Geisslinger, G.; Tegeder, I.; Lotsch, J. Exogenous delta(9)-tetrahydrocannabinol influences circulating endogenous cannabinoids in humans. J. Clin. Psychopharmacol. 2013, 33, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Manza, P.; Yuan, K.; Shokri-Kojori, E.; Tomasi, D.; Volkow, N.D. Brain structural changes in cannabis dependence: Association with MAGL. Mol. Psychiatry 2020, 25, 3256–3266. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, J.; Goodwin, R.S.; Li, C.T.; Terry, G.E.; Zoghbi, S.S.; Morse, C.; Pike, V.W.; Volkow, N.D.; Huestis, M.A.; Innis, R.B. Reversible and regionally selective downregulation of brain cannabinoid CB1 receptors in chronic daily cannabis smokers. Mol. Psychiatry 2012, 17, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Ceccarini, J.; Kuepper, R.; Kemels, D.; van Os, J.; Henquet, C.; Van Laere, K. [18F]MK-9470 PET measurement of cannabinoid CB1 receptor availability in chronic cannabis users. Addict. Biol. 2015, 20, 357–367. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Egerton, A.; Kim, E.; Rosso, L.; Riano Barros, D.; Hammers, A.; Brammer, M.; Turkheimer, F.E.; Howes, O.D.; McGuire, P. Acute induction of anxiety in humans by delta-9-tetrahydrocannabinol related to amygdalar cannabinoid-1 (CB1) receptors. Sci. Rep. 2017, 7, 15025. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.C.; Cortes-Briones, J.A.; Ranganathan, M.; Thurnauer, H.; Creatura, G.; Surti, T.; Planeta, B.; Neumeister, A.; Pittman, B.; Normandin, M.; et al. Rapid Changes in CB1 Receptor Availability in Cannabis Dependent Males after Abstinence from Cannabis. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2016, 1, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Boileau, I.; Mansouri, E.; Williams, B.; Le Foll, B.; Rusjan, P.; Mizrahi, R.; Tyndale, R.F.; Huestis, M.A.; Payer, D.E.; Wilson, A.A.; et al. Fatty Acid Amide Hydrolase Binding in Brain of Cannabis Users: Imaging With the Novel Radiotracer [(11)C]CURB. Biol. Psychiatry 2016, 80, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.R.; Watts, J.J.; Da Silva, T.; Tyndale, R.F.; Rusjan, P.M.; Houle, S.; Wilson, A.A.; Ross, R.A.; Boileau, I.; Mizrahi, R. Fatty acid amide hydrolase is lower in young cannabis users. Addict. Biol. 2021, 26, e12872. [Google Scholar] [CrossRef] [PubMed]
- Spindle, T.R.; Kuwabara, H.; Eversole, A.; Nandi, A.; Vandrey, R.; Antoine, D.G.; Umbricht, A.; Guarda, A.S.; Wong, D.F.; Weerts, E.M. Brain imaging of cannabinoid type I (CB1 ) receptors in women with cannabis use disorder and male and female healthy controls. Addict. Biol. 2021, 26, e13061. [Google Scholar] [CrossRef]
- Chiang, K.P.; Gerber, A.L.; Sipe, J.C.; Cravatt, B.F. Reduced cellular expression and activity of the P129T mutant of human fatty acid amide hydrolase: Evidence for a link between defects in the endocannabinoid system and problem drug use. Hum. Mol. Genet. 2004, 13, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Zhou, W.Z.; Zhang, P.W.; Johnson, C.; Wei, L.; Uhl, G.R. Meta-analysis and genome-wide interpretation of genetic susceptibility to drug addiction. BMC Genom. 2011, 12, 508. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.M.; Gerber, A.L.; Cadet, J.L.; Beutler, E.; Sipe, J.C. The fatty acid amide hydrolase 385 A/A (P129T) variant: Haplotype analysis of an ancient missense mutation and validation of risk for drug addiction. Hum. Genet. 2006, 120, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Stark, T.; Maurel, O.M.; Iannotti, F.A.; Kuchar, M.; Ruda-Kucerova, J.; Piscitelli, F.; Laudani, S.; Pekarik, V.; Salomone, S.; et al. Crosstalk between the transcriptional regulation of dopamine D2 and cannabinoid CB1 receptors in schizophrenia: Analyses in patients and in perinatal Delta9-tetrahydrocannabinol-exposed rats. Pharmacol. Res. 2021, 164, 105357. [Google Scholar] [CrossRef] [PubMed]
- Zehra, A.; Burns, J.; Liu, C.K.; Manza, P.; Wiers, C.E.; Volkow, N.D.; Wang, G.J. Cannabis Addiction and the Brain: A Review. J. Neuroimmu. Pharmacol. 2018, 13, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.; Weizman, A.; Weinstein, A. Modulatory effects of cannabinoids on brain neurotransmission. Eur. J. Neurosci. 2019, 50, 2322–2345. [Google Scholar] [CrossRef] [PubMed]
- Iversen, L. Cannabis and the brain. Brain 2003, 126, 1252–1270. [Google Scholar] [CrossRef] [PubMed]
- Aracil-Fernandez, A.; Almela, P.; Manzanares, J. Pregabalin and topiramate regulate behavioural and brain gene transcription changes induced by spontaneous cannabinoid withdrawal in mice. Addict. Biol. 2013, 18, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.J.; Hampson, R.E.; Deadwyler, S.A.; Childers, S.R. Effects of chronic treatment with delta9-tetrahydrocannabinol on cannabinoid-stimulated [35S]GTPgammaS autoradiography in rat brain. J. Neurosci. 1996, 16, 8057–8066. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.; Rodriguez de Fonseca, F. Cannabinoid addiction: Behavioral models and neural correlates. J. Neurosci. 2002, 22, 3326–3331. [Google Scholar] [CrossRef] [PubMed]
- Tanda, G.; Goldberg, S.R. Cannabinoids: Reward, dependence, and underlying neurochemical mechanisms—A review of recent preclinical data. Psychopharmacology 2003, 169, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Colizzi, M.; McGuire, P.; Pertwee, R.G.; Bhattacharyya, S. Effect of cannabis on glutamate signalling in the brain: A systematic review of human and animal evidence. Neurosci. Biobehav. Rev. 2016, 64, 359–381. [Google Scholar] [CrossRef] [PubMed]
- Sloan, M.E.; Grant, C.W.; Gowin, J.L.; Ramchandani, V.A.; Le Foll, B. Endocannabinoid signaling in psychiatric disorders: A review of positron emission tomography studies. Acta Pharmacol. Sin. 2019, 40, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Befort, K. Interactions of the opioid and cannabinoid systems in reward: Insights from knockout studies. Front. Pharmacol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Navarro, M.; Carrera, M.R.; Fratta, W.; Valverde, O.; Cossu, G.; Fattore, L.; Chowen, J.A.; Gomez, R.; del Arco, I.; Villanua, M.A.; et al. Functional interaction between opioid and cannabinoid receptors in drug self-administration. J. Neurosci. 2001, 21, 5344–5350. [Google Scholar] [CrossRef]
- Scavone, J.L.; Sterling, R.C.; Van Bockstaele, E.J. Cannabinoid and opioid interactions: Implications for opiate dependence and withdrawal. Neuroscience 2013, 248, 637–654. [Google Scholar] [CrossRef] [PubMed]
- Proudnikov, D.; Kroslak, T.; Sipe, J.C.; Randesi, M.; Li, D.; Hamon, S.; Ho, A.; Ott, J.; Kreek, M.J. Association of polymorphisms of the cannabinoid receptor (CNR1) and fatty acid amide hydrolase (FAAH) genes with heroin addiction: Impact of long repeats of CNR1. Pharmacogenom. J. 2010, 10, 232–242. [Google Scholar] [CrossRef]
- Icick, R.; Peoc’h, K.; Karsinti, E.; Ksouda, K.; Hajj, A.; Bloch, V.; Prince, N.; Mouly, S.; Bellivier, F.; Lepine, J.P.; et al. A cannabinoid receptor 1 polymorphism is protective against major depressive disorder in methadone-maintained outpatients. Am. J. Addict. 2015, 24, 613–620. [Google Scholar] [CrossRef]
- Romero, J.; Fernandez-Ruiz, J.J.; Vela, G.; Ruiz-Gayo, M.; Fuentes, J.A.; Ramos, J.A. Autoradiographic analysis of cannabinoid receptor binding and cannabinoid agonist-stimulated [35S]GTP gamma S binding in morphine-dependent mice. Drug Alcohol Depend. 1998, 50, 241–249. [Google Scholar] [CrossRef]
- Gonzalez, S.; Fernandez-Ruiz, J.; Sparpaglione, V.; Parolaro, D.; Ramos, J.A. Chronic exposure to morphine, cocaine or ethanol in rats produced different effects in brain cannabinoid CB(1) receptor binding and mRNA levels. Drug Alcohol Depend. 2002, 66, 77–84. [Google Scholar] [CrossRef]
- Gonzalez, S.; Schmid, P.C.; Fernandez-Ruiz, J.; Krebsbach, R.; Schmid, H.H.; Ramos, J.A. Region-dependent changes in endocannabinoid transmission in the brain of morphine-dependent rats. Addict. Biol. 2003, 8, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Vigano, D.; Grazia Cascio, M.; Rubino, T.; Fezza, F.; Vaccani, A.; Di Marzo, V.; Parolaro, D. Chronic morphine modulates the contents of the endocannabinoid, 2-arachidonoyl glycerol, in rat brain. Neuropsychopharmacology 2003, 28, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lipinski, A.A.; Liktor-Busa, E.; Smith, A.F.; Moutal, A.; Khanna, R.; Langlais, P.R.; Largent-Milnes, T.M.; Vanderah, T.W. The Effects of Repeated Morphine Treatment on the Endogenous Cannabinoid System in the Ventral Tegmental Area. Front. Pharmacol. 2021, 12, 632757. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Pan, L.; Guo, Y.; Zheng, Y.; Nie, Z.; Zhu, R. Expression and localization of cannabinoid receptor 1 in rats. brain treated with acute and repeated morphine. Acta Neurobiol. Exp. (Wars) 2014, 74, 288–297. [Google Scholar] [PubMed]
- Sustkova-Fiserova, M.; Jerabek, P.; Havlickova, T.; Syslova, K.; Kacer, P. Ghrelin and endocannabinoids participation in morphine-induced effects in the rat nucleus accumbens. Psychopharmacology 2016, 233, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Zhang, M.; Cao, Y. Exposure to morphine affects the expression of endocannabinoid receptors and immune functions. J. Neuroimmunol. 2012, 247, 52–58. [Google Scholar] [CrossRef]
- Yuan, W.X.; Heng, L.J.; Ma, J.; Wang, X.Q.; Qu, L.J.; Duan, L.; Kang, J.J.; Chen, L.W.; Gao, G.D. Increased expression of cannabinoid receptor 1 in the nucleus accumbens core in a rat model with morphine withdrawal. Brain Res. 2013, 1531, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Ma, J.; Cui, W.; Yuan, W.X.; Zhu, G.; Yang, Q.; Heng, L.J.; Gao, G.D. The endocannabinoid system regulates synaptic transmission in nucleus accumbens by increasing DAGL-alpha expression following short-term morphine withdrawal. Br. J. Pharmacol. 2016, 173, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, C.L.; Qiu, Z.G. Differential expression of endocannabinoid system-related genes in the dorsal hippocampus following expression and reinstatement of morphine conditioned place preference in mice. Neurosci. Lett. 2017, 643, 38–44. [Google Scholar] [CrossRef]
- Vigano, D.; Valenti, M.; Cascio, M.G.; Di Marzo, V.; Parolaro, D.; Rubino, T. Changes in endocannabinoid levels in a rat model of behavioural sensitization to morphine. Eur. J. Neurosci. 2004, 20, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Vigano, D.; Fadda, P.; Rubino, T.; Fratta, W.; Parolaro, D. Bidirectional regulation of mu-opioid and CB1-cannabinoid receptor in rats self-administering heroin or WIN 55,212-2. Eur. J. Neurosci. 2007, 25, 2191–2200. [Google Scholar] [CrossRef] [PubMed]
- Naudon, L.; Piscitelli, F.; Giros, B.; Di Marzo, V.; Dauge, V. Possible involvement of endocannabinoids in the increase of morphine consumption in maternally deprived rat. Neuropharmacology 2013, 65, 193–199. [Google Scholar] [CrossRef]
- Zuo, L.; Kranzler, H.R.; Luo, X.; Yang, B.Z.; Weiss, R.; Brady, K.; Poling, J.; Farrer, L.; Gelernter, J. Interaction between two independent CNR1 variants increases risk for cocaine dependence in European Americans: A replication study in family-based sample and population-based sample. Neuropsychopharmacology 2009, 34, 1504–1513. [Google Scholar] [CrossRef]
- Clarke, T.K.; Bloch, P.J.; Ambrose-Lanci, L.M.; Ferraro, T.N.; Berrettini, W.H.; Kampman, K.M.; Dackis, C.A.; Pettinati, H.M.; O’Brien, C.P.; Oslin, D.W.; et al. Further evidence for association of polymorphisms in the CNR1 gene with cocaine addiction: Confirmation in an independent sample and meta-analysis. Addict. Biol. 2013, 18, 702–708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ballon, N.; Leroy, S.; Roy, C.; Bourdel, M.C.; Charles-Nicolas, A.; Krebs, M.O.; Poirier, M.F. (AAT)n repeat in the cannabinoid receptor gene (CNR1): Association with cocaine addiction in an African-Caribbean population. Pharmacogenom. J. 2006, 6, 126–130. [Google Scholar] [CrossRef][Green Version]
- Patel, M.M.; Nielsen, D.A.; Kosten, T.R.; De La Garza, R., 2nd; Newton, T.F.; Verrico, C.D. FAAH variant Pro129Thr modulates subjective effects produced by cocaine administration. Am. J. Addict. 2018, 27, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.; Deng, X.D.; Ma, Y.; Liu, Y. FAAH levels and its genetic polymorphism association with susceptibility to methamphetamine dependence. Ann. Hum. Genet. 2020, 84, 259–270. [Google Scholar] [CrossRef]
- Sim, M.S.; Hatim, A.; Reynolds, G.P.; Mohamed, Z. Association of a functional FAAH polymorphism with methamphetamine-induced symptoms and dependence in a Malaysian population. Pharmacogenomics 2013, 14, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Nader, J.; Rapino, C.; Gennequin, B.; Chavant, F.; Francheteau, M.; Makriyannis, A.; Duranti, A.; Maccarrone, M.; Solinas, M.; Thiriet, N. Prior stimulation of the endocannabinoid system prevents methamphetamine-induced dopaminergic neurotoxicity in the striatum through activation of CB2 receptors. Neuropharmacology 2014, 87, 214–221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torres, E.; Gutierrez-Lopez, M.D.; Borcel, E.; Peraile, I.; Mayado, A.; O’Shea, E.; Colado, M.I. Evidence that MDMA (‘ecstasy’) increases cannabinoid CB2 receptor expression in microglial cells: Role in the neuroinflammatory response in rat brain. J. Neurochem. 2010, 113, 67–78. [Google Scholar] [CrossRef]
- Palomino, A.; Pavon, F.J.; Blanco-Calvo, E.; Serrano, A.; Arrabal, S.; Rivera, P.; Alen, F.; Vargas, A.; Bilbao, A.; Rubio, L.; et al. Effects of acute versus repeated cocaine exposure on the expression of endocannabinoid signaling-related proteins in the mouse cerebellum. Front. Integr. Neurosci. 2014, 8, 22. [Google Scholar] [CrossRef]
- Blanco, E.; Galeano, P.; Palomino, A.; Pavon, F.J.; Rivera, P.; Serrano, A.; Alen, F.; Rubio, L.; Vargas, A.; Castilla-Ortega, E.; et al. Cocaine-induced behavioral sensitization decreases the expression of endocannabinoid signaling-related proteins in the mouse hippocampus. Eur. Neuropsychopharmacol. 2016, 26, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Areal, L.B.; Rodrigues, L.C.; Andrich, F.; Moraes, L.S.; Cicilini, M.A.; Mendonca, J.B.; Pelicao, F.S.; Nakamura-Palacios, E.M.; Martins-Silva, C.; Pires, R.G. Behavioural, biochemical and molecular changes induced by chronic crack-cocaine inhalation in mice: The role of dopaminergic and endocannabinoid systems in the prefrontal cortex. Behav. Brain Res. 2015, 290, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cabrerizo, R.; Garcia-Fuster, M.J. Opposite regulation of cannabinoid CB1 and CB2 receptors in the prefrontal cortex of rats treated with cocaine during adolescence. Neurosci. Lett. 2016, 615, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Alvaro-Bartolome, M.; Garcia-Sevilla, J.A. Dysregulation of cannabinoid CB1 receptor and associated signaling networks in brains of cocaine addicts and cocaine-treated rodents. Neuroscience 2013, 247, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Llorente-Berzal, A.; Assis, M.A.; Rubino, T.; Zamberletti, E.; Marco, E.M.; Parolaro, D.; Ambrosio, E.; Viveros, M.P. Sex-dependent changes in brain CB1R expression and functionality and immune CB2R expression as a consequence of maternal deprivation and adolescent cocaine exposure. Pharmacol. Res. 2013, 74, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Bystrowska, B.; Frankowska, M.; Smaga, I.; Pomierny-Chamiolo, L.; Filip, M. Effects of Cocaine Self-Administration and Its Extinction on the Rat Brain Cannabinoid CB1 and CB2 Receptors. Neurotox. Res. 2018, 34, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Bystrowska, B.; Frankowska, M.; Smaga, I.; Niedzielska-Andres, E.; Pomierny-Chamiolo, L.; Filip, M. Cocaine-Induced Reinstatement of Cocaine Seeking Provokes Changes in the Endocannabinoid and N-Acylethanolamine Levels in Rat Brain Structures. Molecules 2019, 24, 1125. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Gobira, P.H.; Werner, C.T.; Martin, J.A.; Iida, M.; Thomas, S.A.; Erias, K.; Miracle, S.; Lafargue, C.; An, C.; et al. A role for the endocannabinoid enzymes monoacylglycerol and diacylglycerol lipases in cue-induced cocaine craving following prolonged abstinence. Addict. Biol. 2021, 26, e13007. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hausknecht, K.A.; Gancarz-Kausch, A.M.; Oubraim, S.; Shen, R.Y.; Haj-Dahmane, S. Cocaine self-administration abolishes endocannabinoid-mediated long-term depression of glutamatergic synapses in the ventral tegmental area. Eur. J. Neurosci. 2020, 52, 4517–4524. [Google Scholar] [CrossRef] [PubMed]
- Rivera, P.; Miguens, M.; Coria, S.M.; Rubio, L.; Higuera-Matas, A.; Bermudez-Silva, F.J.; de Fonseca, F.R.; Suarez, J.; Ambrosio, E. Cocaine self-administration differentially modulates the expression of endogenous cannabinoid system-related proteins in the hippocampus of Lewis vs. Fischer 344 rats. Int. J. Neuropsychopharmacol. 2013, 16, 1277–1293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; De Biase, L.; Chandra, R.; Shen, H.; Liu, Q.R.; Gardner, E.; Lobo, M.K.; Xi, Z.X. Repeated cocaine administration upregulates CB2 receptor expression in striatal medium-spiny neurons that express dopamine D1 receptors in mice. Acta Pharmacol. Sin. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Haijen, E.; Farre, M.; de la Torre, R.; Pastor, A.; Olesti, E.; Pizarro, N.; Ramaekers, J.G.; Kuypers, K.P.C. Peripheral endocannabinoid concentrations are not associated with verbal memory impairment during MDMA intoxication. Psychopharmacology 2018, 235, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Pavon, F.J.; Araos, P.; Pastor, A.; Calado, M.; Pedraz, M.; Campos-Cloute, R.; Ruiz, J.J.; Serrano, A.; Blanco, E.; Rivera, P.; et al. Evaluation of plasma-free endocannabinoids and their congeners in abstinent cocaine addicts seeking outpatient treatment: Impact of psychiatric co-morbidity. Addict. Biol. 2013, 18, 955–969. [Google Scholar] [CrossRef]
- Voegel, C.D.; Kroll, S.L.; Schmid, M.W.; Kexel, A.K.; Baumgartner, M.R.; Kraemer, T.; Binz, T.M.; Quednow, B.B. Alterations of stress-related glucocorticoids and endocannabinoids in hair of chronic cocaine users. Int. J. Neuropsychopharmacol. 2021. epub-ahead. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease. Institute of Health Metrics. Available online: https://www.healthdata.org/ (accessed on 17 July 2021).
- Gamaleddin, I.H.; Trigo, J.M.; Gueye, A.B.; Zvonok, A.; Makriyannis, A.; Goldberg, S.R.; Le Foll, B. Role of the endogenous cannabinoid system in nicotine addiction: Novel insights. Front. Psychiatry 2015, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Rezayof, A.; Hashemizadeh, S. Critical role of cannabinoid CB1 receptors in nicotine reward and addiction. In Neuropathology of Drug Addictions and Substance Misuse; Elsevier: Amsterdam, The Netherlands, 2016; Volume 1, pp. 158–167. [Google Scholar]
- Auber, A.; Justinova, Z.; Scherma, M.; Goldberg, S.R. Cannabinoid-nicotine interactions. In Cannabinoid Modulation of Emotion. Memory, and Motivation; Campolongo, P., Fattore, L., Eds.; Springer: New York, NY, USA, 2015. [Google Scholar]
- Chen, X.; Williamson, V.S.; An, S.S.; Hettema, J.M.; Aggen, S.H.; Neale, M.C.; Kendler, K.S. Cannabinoid receptor 1 gene association with nicotine dependence. Arch. Gen. Psychiatry 2008, 65, 816–824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chukwueke, C.C.; Kowalczyk, W.J.; Gendy, M.; Taylor, R.; Tyndale, R.F.; Le Foll, B.; Heishman, S.J. The CB1R rs2023239 receptor gene variant significantly affects the reinforcing effects of nicotine, but not cue reactivity, in human smokers. Brain Behav. 2021, 11, e01982. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.E.; Sutton, S.K.; Jentink, K.G.; Lin, H.Y.; Park, J.Y.; Drobes, D.J. Cannabinoid receptor 1 (CNR1) gene variant moderates neural index of cognitive disruption during nicotine withdrawal. Genes Brain Behav. 2016, 15, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Cascio, M.G.; Fernandez-Ruiz, J.; Fezza, F.; Di Marzo, V.; Ramos, J.A. Changes in endocannabinoid contents in the brain of rats chronically exposed to nicotine, ethanol or cocaine. Brain Res. 2002, 954, 73–81. [Google Scholar] [CrossRef]
- Torres, L.H.; Balestrin, N.T.; Spelta, L.E.W.; Duro, S.O.; Pistis, M.; Marcourakis, T. Exposure to tobacco smoke during the early postnatal period modifies receptors and enzymes of the endocannabinoid system in the brainstem and striatum in mice. Toxicol. Lett. 2019, 302, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Marco, E.M.; Granstrem, O.; Moreno, E.; Llorente, R.; Adriani, W.; Laviola, G.; Viveros, M.P. Subchronic nicotine exposure in adolescence induces long-term effects on hippocampal and striatal cannabinoid-CB1 and mu-opioid receptors in rats. Eur. J. Pharmacol. 2007, 557, 37–43. [Google Scholar] [CrossRef]
- Mateos, B.; Borcel, E.; Loriga, R.; Luesu, W.; Bini, V.; Llorente, R.; Castelli, M.P.; Viveros, M.P. Adolescent exposure to nicotine and/or the cannabinoid agonist CP 55,940 induces gender-dependent long-lasting memory impairments and changes in brain nicotinic and CB(1) cannabinoid receptors. J. Psychopharmacol. 2011, 25, 1676–1690. [Google Scholar] [CrossRef]
- Werling, L.L.; Reed, S.C.; Wade, D.; Izenwasser, S. Chronic nicotine alters cannabinoid-mediated locomotor activity and receptor density in periadolescent but not adult male rats. Int. J. Dev. Neurosci. 2009, 27, 263–269. [Google Scholar] [CrossRef]
- Aydin, C.; Oztan, O.; Isgor, C. Long-term effects of juvenile nicotine exposure on abstinence-related social anxiety-like behavior and amygdalar cannabinoid receptor 1 (CB1R) mRNA expression in the novelty-seeking phenotype. Behav. Brain Res. 2012, 228, 236–239. [Google Scholar] [CrossRef][Green Version]
- Guo, T.; Tanaka, T.; Matsumoto, M.; Kaneko, K.; Unzai, T.; Ogino, Y.; Aotani, D.; Kusakabe, T.; Iwakura, H.; Miyazawa, T.; et al. A combination of dietary fat intake and nicotine exposure enhances CB1 endocannabinoid receptor expression in hypothalamic nuclei in male mice. Neurosci. Lett. 2020, 714, 134550. [Google Scholar] [CrossRef]
- Cippitelli, A.; Astarita, G.; Duranti, A.; Caprioli, G.; Ubaldi, M.; Stopponi, S.; Kallupi, M.; Sagratini, G.; Rodriguez de Fonseca, F.; Piomelli, D.; et al. Endocannabinoid regulation of acute and protracted nicotine withdrawal: Effect of FAAH inhibition. PLoS ONE 2011, 6, e28142. [Google Scholar] [CrossRef] [PubMed]
- Saravia, R.; Flores, A.; Plaza-Zabala, A.; Busquets-Garcia, A.; Pastor, A.; de la Torre, R.; Di Marzo, V.; Marsicano, G.; Ozaita, A.; Maldonado, R.; et al. CB1 Cannabinoid Receptors Mediate Cognitive Deficits and Structural Plasticity Changes During Nicotine Withdrawal. Biol. Psychiatry 2017, 81, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Buczynski, M.W.; Polis, I.Y.; Parsons, L.H. The volitional nature of nicotine exposure alters anandamide and oleoylethanolamide levels in the ventral tegmental area. Neuropsychopharmacology 2013, 38, 574–584. [Google Scholar] [CrossRef]
- Gerard, N.; Ceccarini, J.; Bormans, G.; Vanbilloen, B.; Casteels, C.; Goffin, K.; Bosier, B.; Lambert, D.M.; Van Laere, K. Influence of chronic nicotine administration on cerebral type 1 cannabinoid receptor binding: An in vivo micro-PET study in the rat using [18F]MK-9470. J. Mol. Neurosci. 2010, 42, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, J.; Zanotti-Fregonara, P.; Gorelick, D.A.; Lyoo, C.H.; Rallis-Frutos, D.; Morse, C.; Zoghbi, S.S.; Pike, V.W.; Volkow, N.D.; Huestis, M.A.; et al. Decreased Cannabinoid CB1 Receptors in Male Tobacco Smokers Examined With Positron Emission Tomography. Biol. Psychiatry 2018, 84, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, M.; Cortes-Briones, J.; Radhakrishnan, R.; Thurnauer, H.; Planeta, B.; Skosnik, P.; Gao, H.; Labaree, D.; Neumeister, A.; Pittman, B.; et al. Reduced Brain Cannabinoid Receptor Availability in Schizophrenia. Biol. Psychiatry 2016, 79, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- NIDA. Hallucinogens Drug Facts. Available online: https://www.drugabuse.gov/publications/drugfacts/hallucinogens (accessed on 10 July 2021).
- NIDA. Monitoring the Future Study: Trends in Prevalence of Various Drugs. Available online: https://www.drugabuse.gov/drug-topics/trends-statistics/monitoring-future/monitoring-future-study-trends-in-prevalence-various-drugs (accessed on 11 July 2021).
- Dos Santos, R.G.; Crippa, J.A.; Osorio, F.L.; Rocha, J.M.; Rossi, G.N.; Marchioni, C.; Queiroz, M.E.C.; Silveira, G.O.; Yonamine, M.; Hallak, J.E.C. Possible Interactions Between 5-HT2A Receptors and the Endocannabinoid System in Humans: Preliminary Evidence of Interactive Effects of Ayahuasca and Endocannabinoids in a Healthy Human Subject. J. Clin. Psychopharmacol. 2018, 38, 644–646. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, H.; Wang, L.; Zhang, J.; Liu, C.; Wan, X.; Liu, X.; Hu, Y.; Fang, Q.; Xiao, Y.; et al. Endocannabinoid signaling regulates the reinforcing and psychostimulant effects of ketamine in mice. Nat. Commun. 2020, 11, 5962. [Google Scholar] [CrossRef] [PubMed]
- Lovelace, J.W.; Vieira, P.A.; Corches, A.; Mackie, K.; Korzus, E. Impaired fear memory specificity associated with deficient endocannabinoid-dependent long-term plasticity. Neuropsychopharmacology 2014, 39, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Seillier, A.; Martinez, A.A.; Giuffrida, A. Differential effects of Delta9-tetrahydrocannabinol dosing on correlates of schizophrenia in the sub-chronic PCP rat model. PLoS ONE 2020, 15, e0230238. [Google Scholar] [CrossRef] [PubMed]
- Seillier, A.; Martinez, A.A.; Giuffrida, A. Phencyclidine-induced social withdrawal results from deficient stimulation of cannabinoid CB(1) receptors: Implications for schizophrenia. Neuropsychopharmacology 2013, 38, 1816–1824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vigano, D.; Guidali, C.; Petrosino, S.; Realini, N.; Rubino, T.; Di Marzo, V.; Parolaro, D. Involvement of the endocannabinoid system in phencyclidine-induced cognitive deficits modelling schizophrenia. Int. J. Neuropsychopharmacol. 2009, 12, 599–614. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).