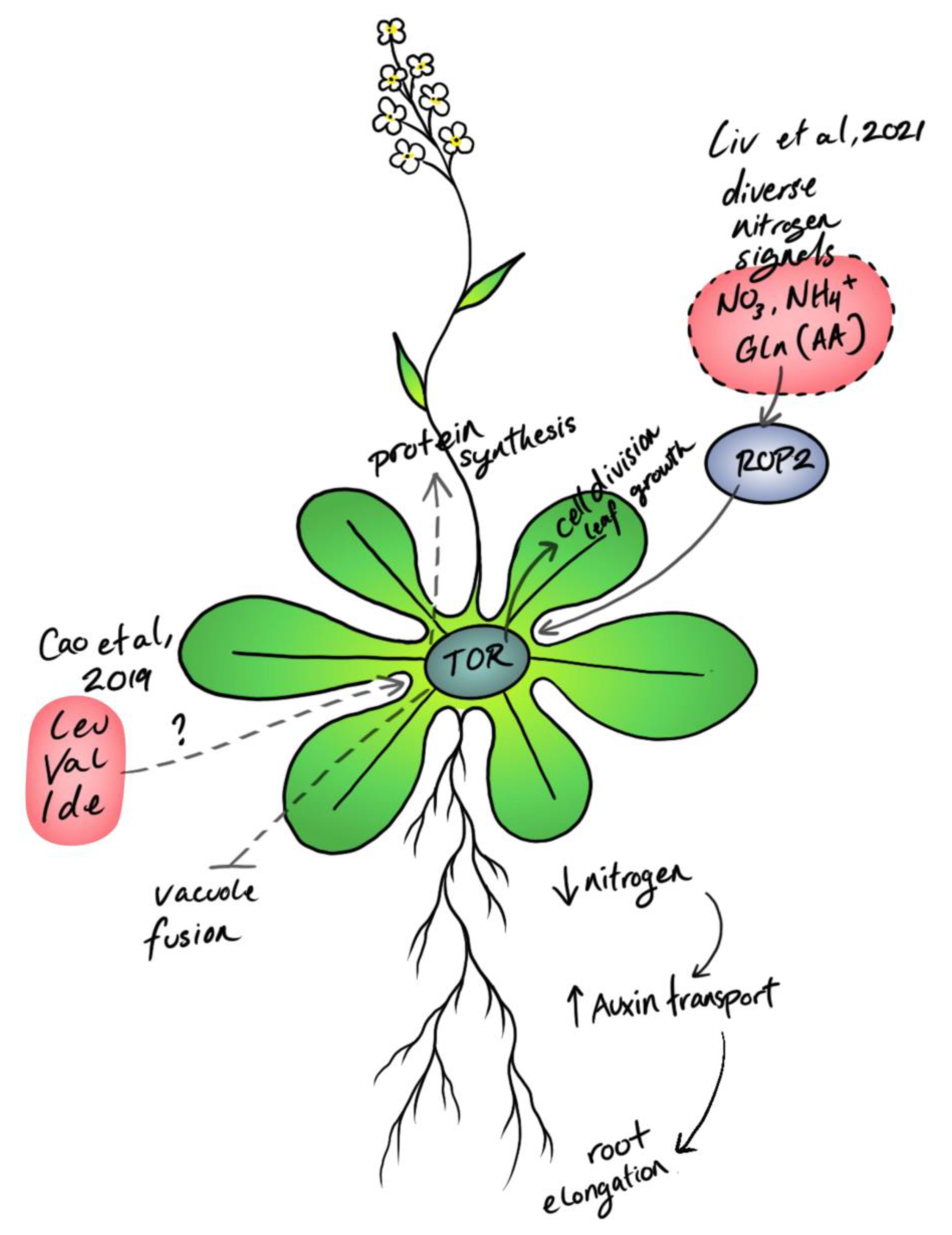
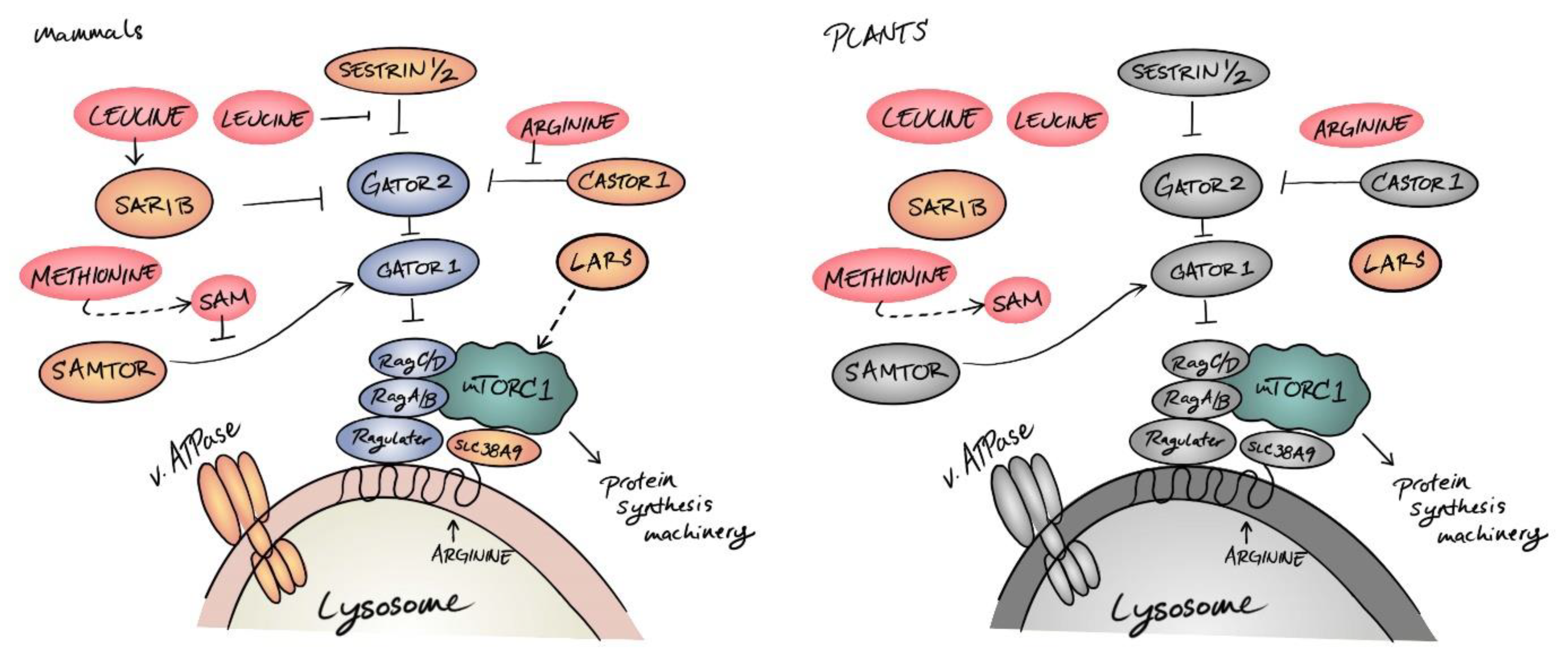
Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe
Abstract
1. Introduction
2. Sensors and Transducers in Metabolic Signaling
3. The Growing Chorus of Amino Acid Sensors and Transducers for Mammalian TOR
4. Plant Nutrient Sensing: From Inorganic Precursors to Organic Metabolites
5. The Future of Crops: Deploying TOR for a Sustainable Agricultural Future
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Valvezan, A.J.; Manning, B.D. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab. 2019, 1, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O. Exaptive Evolution of Target of Rapamycin Signaling in Multicellular Eukaryotes. Dev. Cell 2020, 54, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wu, Y.; Sheen, J. TOR signaling in plants: Conservation and innovation. Development 2018, 145, dev160887. [Google Scholar] [CrossRef] [PubMed]
- Burkart, G.M.; Brandizzi, F. A Tour of TOR Complex Signaling in Plants. Trends Biochem. Sci. 2021, 46, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Scarpin, M.R.; Leiboff, S.; Brunkard, J.O. Parallel global profiling of plant tor dynamics reveals a conserved role for larp1 in translation. Elife 2020, 9, e58795. [Google Scholar] [CrossRef]
- Thomas, G.; Hall, M.N. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 1997, 9, 782–787. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta—Gene Regul. Mech. 2015, 1849, 801–811. [Google Scholar] [CrossRef]
- Valvezan, A.J.; Turner, M.; Belaid, A.; Lam, H.C.; Miller, S.K.; McNamara, M.C.; Baglini, C.; Housden, B.E.; Perrimon, N.; Kwiatkowski, D.J.; et al. mTORC1 Couples Nucleotide Synthesis to Nucleotide Demand Resulting in a Targetable Metabolic Vulnerability. Cancer Cell 2017, 32, 624–638.e5. [Google Scholar] [CrossRef]
- Robitaille, A.M.; Christen, S.; Shimobayashi, M.; Cornu, M.; Fava, L.L.; Moes, S.; Prescianotto-Baschong, C.; Sauer, U.; Jenoe, P.; Hall, M.N. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 2013, 339, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sahra, I.; Howell, J.J.; Asara, J.M.; Manning, B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013, 339, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Busche, M.; Scarpin, M.R.; Hnasko, R.; Brunkard, J.O. TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell 2021, 33, 1615–1632. [Google Scholar] [CrossRef] [PubMed]
- Düvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Mol. Cell 2010, 39, 171–183. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. An Emerging Role of mTOR in Lipid Biosynthesis. Curr. Biol. 2009, 19, R1046–R1052. [Google Scholar] [CrossRef]
- Thelen, A.M.; Zoncu, R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol. 2017, 27, 833–850. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. TOR Is a Negative Regulator of Autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef]
- Kazibwe, Z.; Soto-Burgos, J.; MacIntosh, G.C.; Bassham, D.C. TOR mediates the autophagy response to altered nucleotide homeostasis in an RNase mutant. J. Exp. Bot. 2020, 71, 6907–6920. [Google Scholar] [CrossRef]
- Noda, T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules 2017, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Masvidal, L.; Hulea, L.; Furic, L.; Topisirovic, I.; Larsson, O. mTOR-sensitive translation: Cleared fog reveals more trees. RNA Biol. 2017, 14, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017, 26, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Binder, S. Branched-Chain Amino Acid Metabolism in Arabidopsis thaliana. Arab. B. 2010, 8, e0137. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant Nitrogen Assimilation and Use Efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Schaufelberger, M.; Galbier, F.; Herger, A.; De Brito Francisco, R.; Roffler, S.; Clement, G.; Diet, A.; Hörtensteiner, S.; Wicker, T.; Ringli, C. Mutations in the Arabidopsis ROL17/isopropylmalate synthase 1 locus alter amino acid content, modify the TOR network, and suppress the root hair cell development mutant lrx1. J. Exp. Bot. 2019, 70, 2313–2323. [Google Scholar] [CrossRef]
- Cao, P.; Kim, S.J.; Xing, A.; Schenck, C.A.; Liu, L.; Jiang, N.; Wang, J.; Last, R.L.; Brandizzi, F. Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. Elife 2019, 8, e50747. [Google Scholar] [CrossRef]
- O’Leary, B.M.; Oh, G.G.K.; Lee, C.P.; Millar, A.H. Metabolite Regulatory Interactions Control Plant Respiratory Metabolism via Target of Rapamycin (TOR) Kinase Activation. Plant Cell 2020, 32, 666–682. [Google Scholar] [CrossRef]
- Liu, Y.; Duan, X.; Zhao, X.; Ding, W.; Wang, Y.; Xiong, Y. Diverse nitrogen signals activate convergent ROP2-TOR signaling in Arabidopsis. Dev. Cell 2021, 56, 1283–1295.e5. [Google Scholar] [CrossRef]
- Vellai, T. How the amino acid leucine activates the key cell-growth regulator mTOR. Nature 2021, 596, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Ulijasz, A.T.; Cornilescu, G.; Cornilescu, C.C.; Zhang, J.; Rivera, M.; Markley, J.L.; Vierstra, R.D. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 2010, 463, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.A.R.; Kuchitsu, K.; Kwak, J.; Pei, Z.-M.; Iida, H. Sensors Make Sense of Signaling. Plant Cell Physiol. 2017, 58, 1121–1125. [Google Scholar] [CrossRef][Green Version]
- Dever, T.E.; Feng, L.; Wek, R.C.; Cigan, A.M.; Donahue, T.F.; Hinnebusch, A.G. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 1992, 68, 585–596. [Google Scholar] [CrossRef]
- Lageix, S.; Lanet, E.; Pouch-Pélissier, M.N.; Espagnol, M.C.; Robaglia, C.; Deragon, J.M.; Pélissier, T. Arabidopsis eIF2α kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 2008, 8, 134. [Google Scholar] [CrossRef]
- Chikashige, Y.; Kato, H.; Thornton, M.; Pepper, W.; Hilgers, M.; Cecil, A.; Asano, I.; Yamada, H.; Mori, C.; Brunkow, C.; et al. Gcn2 eIF2α kinase mediates combinatorial translational regulation through nucleotide motifs and uORFs in target mRNAs. Nucleic Acids Res. 2020, 48, 8977–8992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Kanyuka, K.; Parry, M.A.J.; Powers, S.J.; Halford, N.G. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2α in Arabidopsis. J. Exp. Bot. 2008, 59, 3131–3141. [Google Scholar] [CrossRef]
- Tzamarias, D.; Thireos, G. Evidence that the GCN2 protein kinase regulates reinitiation by yeast ribosomes. EMBO J. 1988, 7, 3547–3551. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef]
- Li, M.W.; Auyeung, W.K.; Lam, H.M. The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biol. 2013, 15, 13–18. [Google Scholar] [CrossRef]
- Wek, S.A.; Zhu, S.; Wek, R.C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 1995, 15, 4497–4506. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Qiu, H.; Garcia-Barrio, M.; Anderson, J.; Hinnebusch, A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 2000, 6, 269–279. [Google Scholar] [CrossRef]
- Wek, R.C.; Jackson, B.M.; Hinnebusch, A.G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA 1989, 86, 4579–4583. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Ordonez, A.; Allen, F.; Parts, L.; Inglis, A.J.; Williams, R.L.; Ron, D. The ribosomal P-stalk couples amino acid starvation to GCN2 2 activation in mammalian cells. Elife 2019, 8, e50149. [Google Scholar] [CrossRef] [PubMed]
- Inglis, A.J.; Masson, G.R.; Shao, S.; Perisic, O.; McLaughlin, S.H.; Hegde, R.S.; Williams, R.L. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. USA 2019, 116, 4946–4954. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.-C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Pochopien, A.A.; Beckert, B.; Kasvandik, S.; Berninghausen, O.; Beckmann, R.; Tenson, T.; Wilson, D.N. Structure of Gcn1 bound to stalled and colliding 80S ribosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2022756118. [Google Scholar] [CrossRef]
- Mellado, W.; Willis, D.E. Stressing out translation. Science 2021, 373, 1089–1090. [Google Scholar] [CrossRef]
- Zuko, A.; Mallik, M.; Thompson, R.; Spaulding, E.L.; Wienand, A.R.; Been, M.; Tadenev, A.L.D.; van Bakel, N.; Sijlmans, C.; Santos, L.A.; et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 2021, 373, 1161–1166. [Google Scholar] [CrossRef]
- Spaulding, E.L.; Hines, T.J.; Bais, P.; Tadenev, A.L.D.; Schneider, R.; Jewett, D.; Pattavina, B.; Pratt, S.L.; Morelli, K.H.; Stum, M.G.; et al. The integrated stress response contributes to tRNA synthetase–associated peripheral neuropathy. Science 2021, 373, 1156–1161. [Google Scholar] [CrossRef]
- Mendonsa, S.; von Kuegelgen, N.; Bujanic, L.; Chekulaeva, M. Charcot–Marie–Tooth mutation in glycyl-tRNA synthetase stalls ribosomes in a pre-accommodation state and activates integrated stress response. Nucleic Acids Res. 2021, 49, 10007–10017. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Baker, B. A two-headed monster to avert disaster: HBS1/SKI7 is alternatively spliced to build eukaryotic RNA surveillance complexes. Front. Plant Sci. 2018, 9, 1333. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Vrba, E.S. Exaptation—A Missing Term in the Science of Form. Paleobiology 1982, 1, 4–15. [Google Scholar] [CrossRef]
- Ban, H.; Shigemitsu, K.; Yamatsuji, T.; Haisa, M.; Nakajo, T.; Takaoka, M.; Nobuhisa, T.; Gunduz, M.; Tanaka, N.; Naomoto, Y. Arginine and Leucine regulate p70 S6 kinase and 4E-BP1 in intestinal epithelial cells. Int. J. Mol. Med. 2004, 13, 537–543. [Google Scholar] [CrossRef]
- Stipanuk, M.H. Leucine and Protein Synthesis: mTOR and Beyond. Nutr. Rev. 2008, 65, 122–129. [Google Scholar] [CrossRef]
- Bonfils, G.; Jaquenoud, M.; Bontron, S.; Ostrowicz, C.; Ungermann, C.; De Virgilio, C. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol. Cell 2012, 46, 105–110. [Google Scholar] [CrossRef]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef]
- Wang, S.; Tsun, Z.Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016, 351, 53–58. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 2021, 596, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Chantranupong, L.; Knockenhauer, K.E.; Schwartz, T.U.; Sabatini, D.M. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 2016, 536, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S -adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, U.S.; Karin, M. Sestrin regulation of TORC1: Is sestrin a leucine sensor? Sci. Signal. 2016, 9, re5. [Google Scholar] [CrossRef]
- Lang, E.J.; Cross, P.J.; Mittelstädt, G.; Jameson, G.B.; Parker, E.J. Allosteric ACTion: The varied ACT domains regulating enzymes of amino-acid metabolism. Curr. Opin. Struct. Biol. 2014, 29, 102–111. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef]
- Shen, K.; Huang, R.K.; Brignole, E.J.; Condon, K.J.; Valenstein, M.L.; Chantranupong, L.; Bomaliyamu, A.; Choe, A.; Hong, C.; Yu, Z.; et al. Architecture of the human GATOR1 and GATOR1–Rag GTPases complexes. Nature 2018, 556, 64–69. [Google Scholar] [CrossRef]
- Panchaud, N.; Péli-Gulli, M.-P.; De Virgilio, C. Amino Acid Deprivation Inhibits TORC1 Through a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr1. Sci. Signal. 2013, 6, ra42. [Google Scholar] [CrossRef]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; De Araújo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, G.G.; Papazotos, F.; Pawling, J.; Rajendran, D.; Knight, J.D.R.; Martinez, S.; Taipale, M.; Schramek, D.; Dennis, J.W.; Gingras, A.-C. The GATOR–Rag GTPase pathway inhibits mTORC1 activation by lysosome-derived amino acids. Science 2020, 370, 351–356. [Google Scholar] [CrossRef]
- Meng, D.; Yang, Q.; Wang, H.; Melick, C.H.; Navlani, R.; Frank, A.R.; Jewell, J.L. Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 2020, 295, 2890–2899. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.-X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.-L. Differential regulation of mTORC1 by leucine and glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Bodineau, C.; Tomé, M.; Courtois, S.; Costa, A.S.H.; Sciacovelli, M.; Rousseau, B.; Richard, E.; Vacher, P.; Parejo-Pérez, C.; Bessede, E.; et al. Two parallel pathways connect glutamine metabolism and mTORC1 activity to regulate glutamoptosis. Nat. Commun. 2021, 12, 4814. [Google Scholar] [CrossRef] [PubMed]
- Son, S.M.; Park, S.J.; Lee, H.; Siddiqi, F.; Lee, J.E.; Menzies, F.M.; Rubinsztein, D.C. Leucine Signals to mTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metab. 2019, 29, 192–201.e7. [Google Scholar] [CrossRef]
- Son, S.M.; Park, S.J.; Stamatakou, E.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Leucine regulates autophagy via acetylation of the mTORC1 component raptor. Nat. Commun. 2020, 11, 3148. [Google Scholar] [CrossRef]
- Mariño, G.; Pietrocola, F.; Eisenberg, T.; Kong, Y.; Malik, S.A.; Andryushkova, A.; Schroeder, S.; Pendl, T.; Harger, A.; Niso-Santano, M.; et al. Regulation of Autophagy by Cytosolic Acetyl-Coenzyme A. Mol. Cell 2014, 53, 710–725. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Liu, K.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Liu, K.-H.; Diener, A.; Lin, Z.; Liu, C.; Sheen, J. Primary nitrate responses mediated by calcium signalling and diverse protein phosphorylation. J. Exp. Bot. 2020, 71, 4428–4441. [Google Scholar] [CrossRef]
- Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.-H.; Liu, Y.-X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis Glucose Sensor HXK1 in Nutrient, Light, and Hormonal Signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Frommer, W.B.; Schulze, W.X.; Lalonde, S. Hexokinase, Jack-of-All-Trades. Science 2003, 300, 261–263. [Google Scholar] [CrossRef]
- Li, L.; Liu, K.; Sheen, J. Dynamic Nutrient Signaling Networks in Plants. Annu. Rev. Cell Dev. Biol. 2021, 37, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.C.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-Related Protein Kinase1 Activity and Regulation of Metabolic Pathways by Trehalose-6-Phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-Phosphate Positively Regulates Fatty Acid Synthesis by Stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef]
- Baena-González, E.; Lunn, J.E. SnRK1 and trehalose 6-phosphate—Two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 2020, 55, 52–59. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Riegler, S.; Servi, L.; Scarpin, M.R.; Godoy Herz, M.A.; Kubaczka, M.G.; Venhuizen, P.; Meyer, C.; Brunkard, J.O.; Kalyna, M.; Barta, A.; et al. Light regulates alternative splicing outcomes via the TOR kinase pathway. Cell Rep. 2021, 36, 109676. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.M.; Krawczyk, P.A.; Scaria, S.M.; Cangelosi, A.L.; Chan, S.H.; Kunchok, T.; Lewis, C.A.; Sabatini, D.M. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat. Metab. 2020, 2, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Xu, M.; Regina Scarpin, M.; Chatterjee, S.; Shemyakina, E.A.; Goodman, H.M.; Zambryski, P. TOR dynamically regulates plant cell-cell transport. Proc. Natl. Acad. Sci. USA 2020, 117, 5049–5058. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Xu, M.; Shemyakina, E.A.; Brunkard, J.O. Pontin/Reptin-associated complexes differentially impact plant development and viral pathology. bioRxiv 2021. [Google Scholar] [CrossRef]
- Takeuchi, T.; Benning, C. Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas. Biotechnol. Biofuels 2019, 12, 292. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Kawase, Y.; Kobayashi, I.; Sone, T.; Era, A.; Miyagishima, S.; Shimojima, M.; Ohta, H.; Tanaka, K. Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 2015, 89, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Kawase, Y.; Kobayashi, I.; Shimojima, M.; Ohta, H.; Tanaka, K. TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal. Behav. 2016, 11, e1149285. [Google Scholar] [CrossRef]
- Caldana, C.; Li, Y.; Leisse, A.; Zhang, Y.; Bartholomaeus, L.; Fernie, A.R.; Willmitzer, L.; Giavalisco, P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013, 73, 897–909. [Google Scholar] [CrossRef]
- Mukaida, S.; Ogawa, T.; Ohishi, K.; Tanizawa, Y.; Ohta, D.; Arita, M. The effect of rapamycin on biodiesel-producing protist Euglena gracilis. Biosci. Biotechnol. Biochem. 2016, 80, 1223–1229. [Google Scholar] [CrossRef]
- Prioretti, L.; Avilan, L.; Carrière, F.; Montané, M.-H.; Field, B.; Grégori, G.; Menand, B.; Gontero, B. The inhibition of TOR in the model diatom Phaeodactylum tricornutum promotes a get-fat growth regime. Algal Res. 2017, 26, 265–274. [Google Scholar] [CrossRef]
- Jüppner, J.; Mubeen, U.; Leisse, A.; Caldana, C.; Wiszniewski, A.; Steinhauser, D.; Giavalisco, P. The target of rapamycin kinase affects biomass accumulation and cell cycle progression by altering carbon/nitrogen balance in synchronized Chlamydomonas reinhardtii cells. Plant J. 2018, 93, 355–376. [Google Scholar] [CrossRef]
- Mubeen, U.; Jüppner, J.; Alpers, J.; Hincha, D.K.; Giavalisco, P. Target of Rapamycin Inhibition in Chlamydomonas reinhardtii Triggers de Novo Amino Acid Synthesis by Enhancing Nitrogen Assimilation. Plant Cell 2018, 30, 2240–2254. [Google Scholar] [CrossRef]
- Upadhyaya, S.; Agrawal, S.; Gorakshakar, A.; Rao, B.J. TOR kinase activity in Chlamydomonas reinhardtii is modulated by cellular metabolic states. FEBS Lett. 2020, 594, 3122–3141. [Google Scholar] [CrossRef] [PubMed]
- Stewart, W.M.; Dibb, D.W.; Johnston, A.E.; Smyth, T.J. The contribution of commercial fertilizer nutrients to food production. Agron. J. 2005, 97, 1–6. [Google Scholar] [CrossRef]
- Tilman, D. The greening of the green revolution. Nature 1998, 396, 211–212. [Google Scholar] [CrossRef]
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.R.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012, 195, 306–320. [Google Scholar] [CrossRef]
- Cordell, D.; White, S. Peak phosphorus: Clarifying the key issues of a vigorous debate about long-term phosphorus security. Sustainability 2011, 3, 2027–2049. [Google Scholar] [CrossRef]
- Springmann, M.; Clark, M.; Mason-D’Croz, D.; Wiebe, K.; Bodirsky, B.L.; Lassaletta, L.; de Vries, W.; Vermeulen, S.J.; Herrero, M.; Carlson, K.M.; et al. Options for keeping the food system within environmental limits. Nature 2018, 562, 519–525. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lutt, N.; Brunkard, J.O. Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe. Biomolecules 2022, 12, 387. https://doi.org/10.3390/biom12030387
Lutt N, Brunkard JO. Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe. Biomolecules. 2022; 12(3):387. https://doi.org/10.3390/biom12030387
Chicago/Turabian StyleLutt, Nanticha, and Jacob O. Brunkard. 2022. "Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe" Biomolecules 12, no. 3: 387. https://doi.org/10.3390/biom12030387
APA StyleLutt, N., & Brunkard, J. O. (2022). Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe. Biomolecules, 12(3), 387. https://doi.org/10.3390/biom12030387