Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains
Abstract
:1. Introduction
2. Material and Methods
2.1. Probiotic and Pathogenic Strains
2.2. Optimization of SCFAs Production and Conditions
2.3. Antimicrobial Activity of SCFAs against Pathogens
2.3.1. Agar Well Diffusion Method
2.3.2. Broth Microdilution Method
2.3.3. Checkerboard Assay
2.4. Effect of SCFAs on Growth Suppression of Caco-2 and HIEC-6 Cells
2.4.1. Cell Culture Preparation
2.4.2. Cell Suppression Assay
2.5. Effect of SCFAs on Antimicrobial Peptides and Anti-Inflammation
2.6. Effect of SCFAs on Suppression of Pro-Inflammatory Cytokines
The mRNA Expression by Real-Time PCR
2.7. Statistical Analysis
3. Results
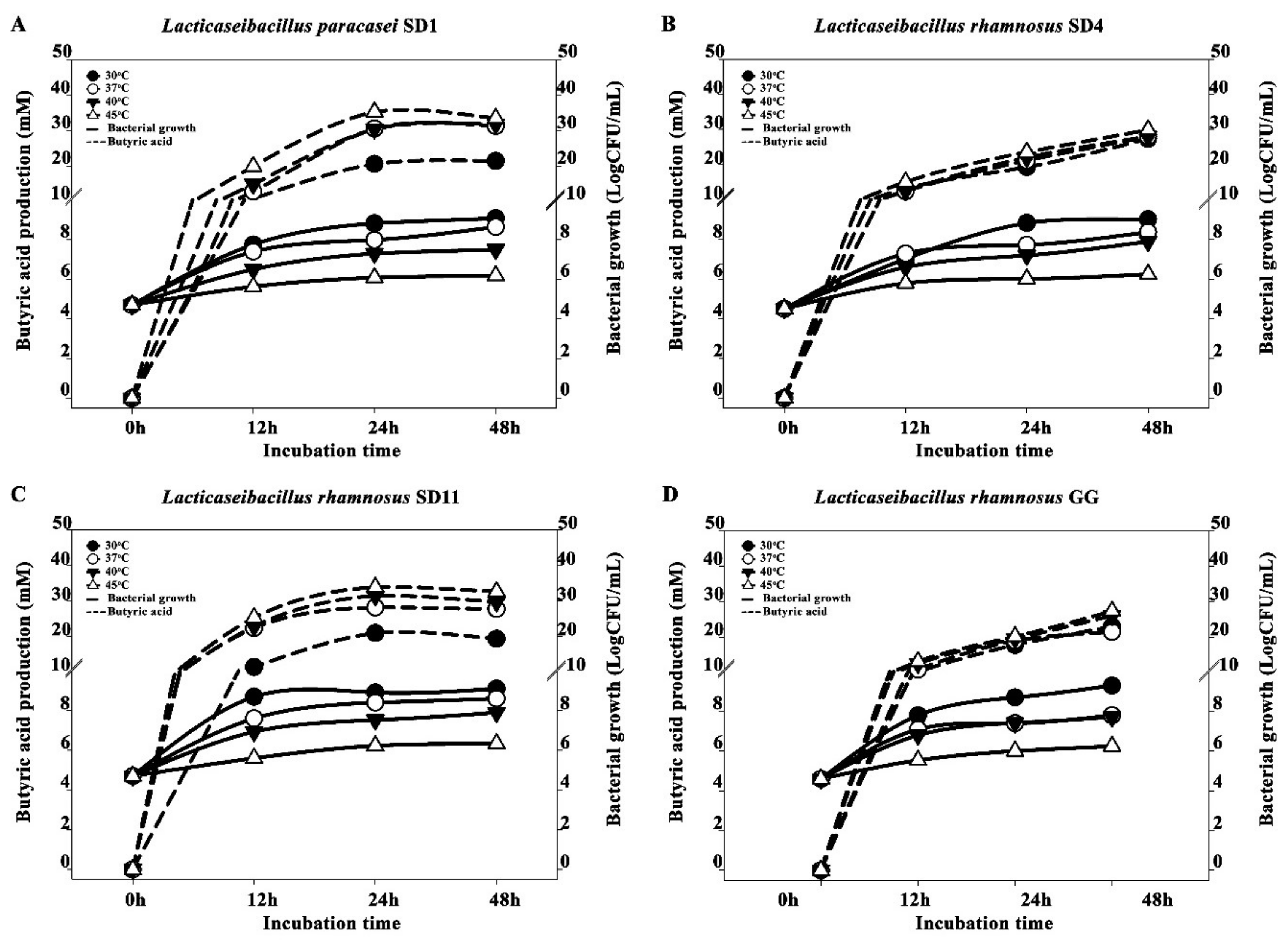
3.1. Optimization of SCFA Production and Conditions
3.2. Antimicrobial Activity of SCFAs Produced by Potential Probiotics against Pathogens
3.3. Effect of SCFAs on Growth Suppression of Caco-2 and HIEC-6 Cells
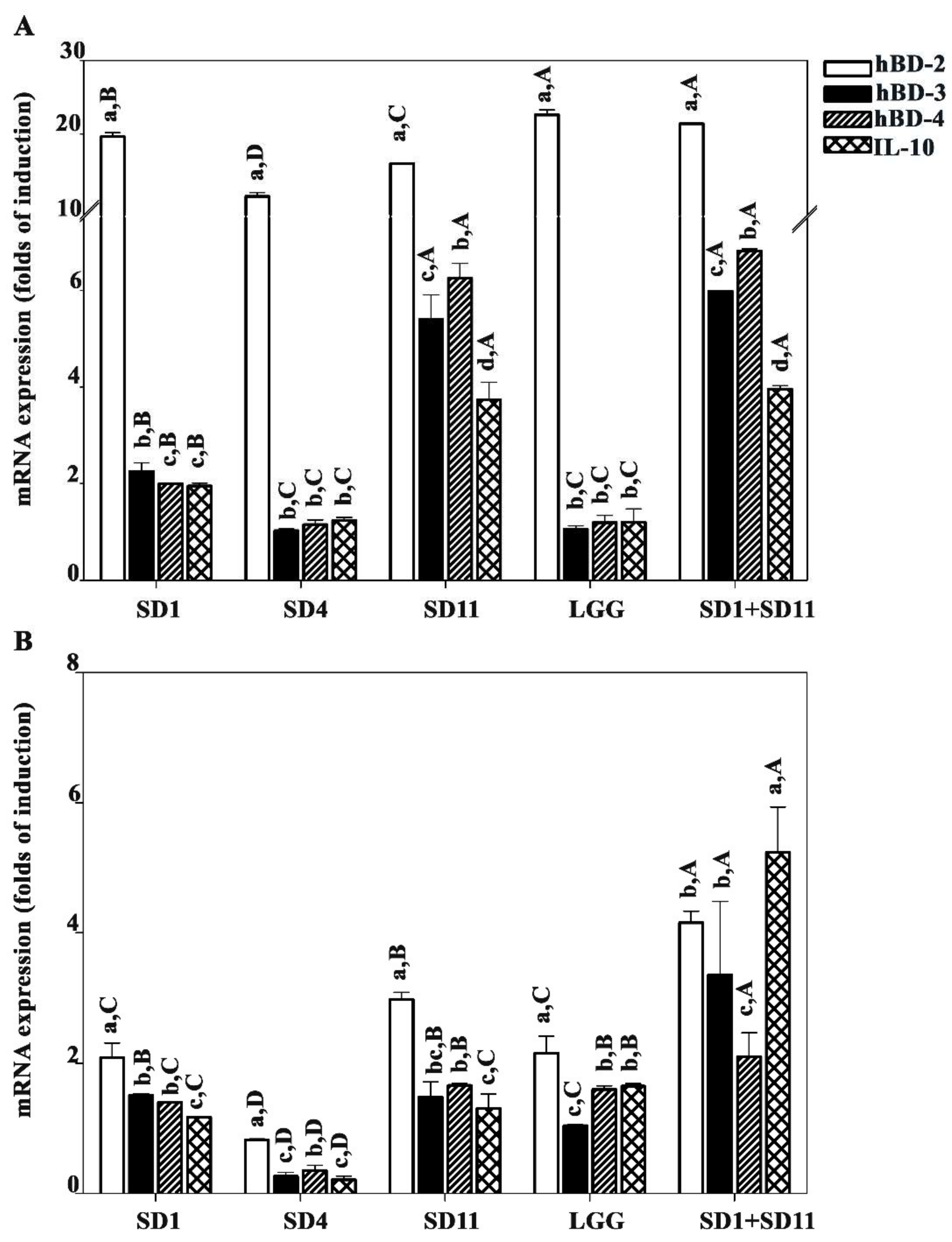
3.4. Effect of SCFAs Produced by Potential Probiotics on Antimicrobial Peptide and Anti-Inflammation IL10
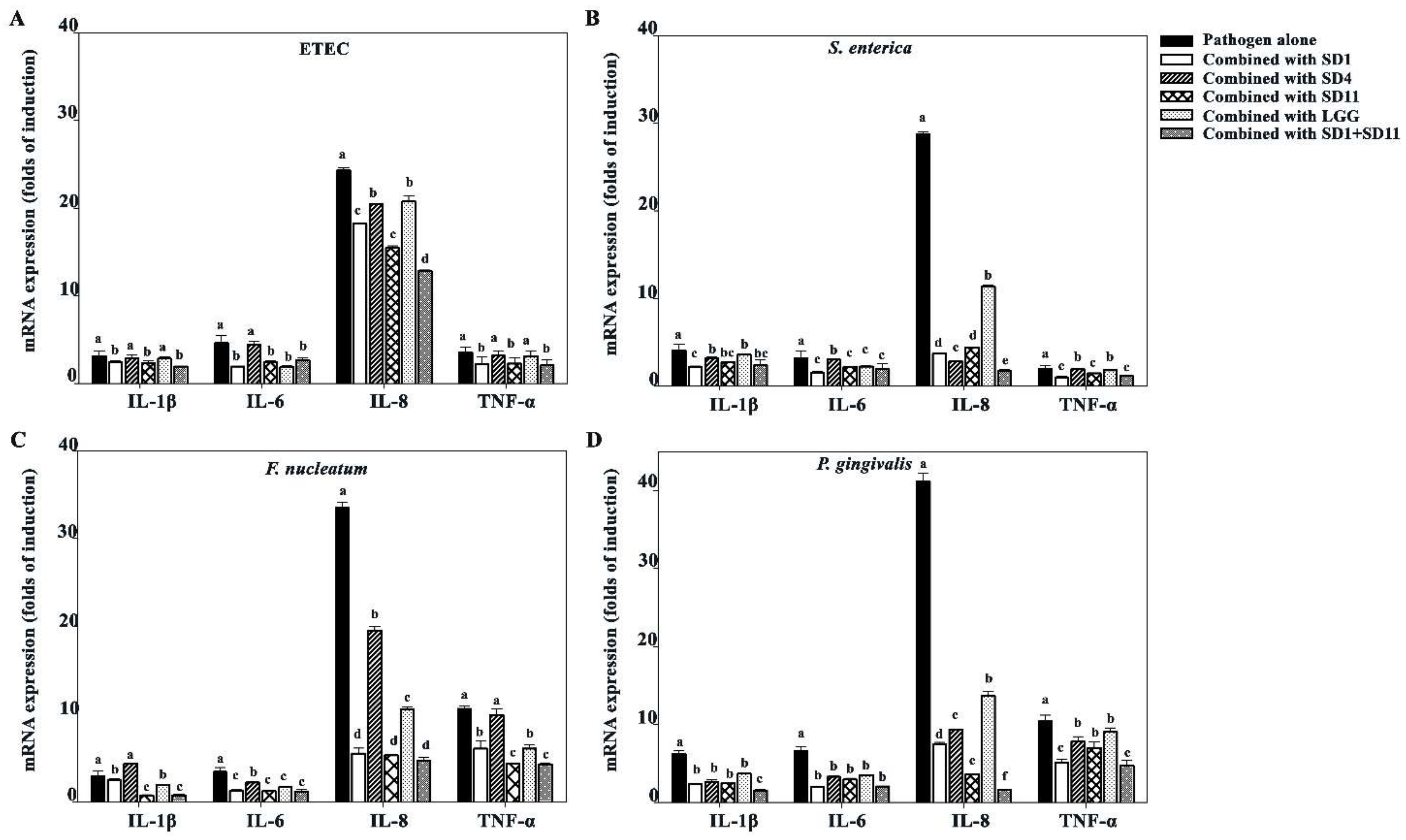
3.5. Effect of SCFAs on Suppression of Pro-Inflammatory Cytokines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alam, M.T.; Amos, G.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Mahallei, M.; Pormohammad, A.; Varshochi, M.; Ganbarov, K.; Zeinalzadeh, E.; Yousefi, B.; Bastami, M.; Tanomand, A.; Mahmood, S.S.; et al. Microbial balance in the intestinal microbiota and its association with diabetes, obesity and allergic disease. Microb. Pathog. 2018, 127, 48–55. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.-A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Lin, C.; Cai, X.; Zhang, J.; Wang, W.; Sheng, Q.; Hua, H.; Zhou, X. Role of Gut Microbiota in the Development and Treatment of Colorectal Cancer. Digestion 2018, 100, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2019, 469, 456–467. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef]
- Wu, J.; Li, Q.; Fu, X. Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Transl. Oncol. 2019, 12, 846–851. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Cao, Y.; Fang, J.-Y.; Hong, J.; Chen, H. Fecal Fusobacterium nucleatum for the diagnosis of colorectal tumor: A systematic review and meta-analysis. Cancer Med. 2019, 8, 480–491. [Google Scholar] [CrossRef] [Green Version]
- Dos Reis, S.A.; da Conceição, L.L.; Siqueira, N.P.; Rosa, D.D.; da Silva, L.L.; Peluzio, M.D.C.G. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr. Res. 2017, 37, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Dong, S.; Wu, X.; Jin, R.; Chen, H. Probiotics in Cancer. Front. Oncol. 2021, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- Eslami, M.; Yousefi, B.; Kokhaei, P.; Hemati, M.; Nejad, Z.R.; Arabkari, V.; Namdar, A. Importance of probiotics in the prevention and treatment of colorectal cancer. J. Cell Physiol. 2019, 234, 17127–17143. [Google Scholar] [CrossRef]
- Górska, A.; Przystupski, D.; Niemczura, M.J.; Kulbacka, J. Probiotic Bacteria: A Promising Tool in Cancer Prevention and Therapy. Curr. Microbiol. 2019, 76, 939–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; Van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef]
- Schickel, R.; Boyerinas, B.; Park, S.-M.; Peter, M.E. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008, 27, 5959–5974. [Google Scholar] [CrossRef] [Green Version]
- Mahdavi, M.; Laforest-Lapointe, I.; Massé, E. Preventing Colorectal Cancer through Prebiotics. Microorganisms 2021, 9, 1325. [Google Scholar] [CrossRef]
- Javanmard, A.; Ashtari, S.; Sabet, B.; Davoodi, S.H.; Rostami-Nejad, M.; Esmaeil Akbari, M.; Niaz, A.; Mortazavian, A.M. Probiotics and their role in gastrointestinal cancers prevention and treatment; An overview. Gastroenterol. Hepatol. Bed Bench 2018, 11, 284–295. [Google Scholar]
- Pessione, A.; Bianco, G.L.; Mangiapane, E.; Cirrincione, S.; Pessione, E. Characterization of potentially probiotic lactic acid bacteria isolated from olives: Evaluation of short chain fatty acids production and analysis of the extracellular proteome. Food Res. Int. 2015, 67, 247–254. [Google Scholar] [CrossRef]
- Sophatha, B.; Piwat, S.; Teanpaisan, R. Adhesion, anti-adhesion and aggregation properties relating to surface charges of selected Lactobacillus strains: Study in Caco-2 and H357 cells. Arch. Microbiol. 2020, 202, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Wannun, P.; Piwat, S.; Teanpaisan, R. Purification and characterization of bacteriocin produced by oral Lactobacillus paracasei SD1. Anaerobe 2014, 27, 17–21. [Google Scholar] [CrossRef]
- Wannun, P.; Piwat, S.; Teanpaisan, R. Purification, Characterization, and Optimum Conditions of Fermencin SD11, a Bacteriocin Produced by Human Orally Lactobacillus fermentum SD11. Appl. Biochem. Biotechnol. 2016, 179, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Manmontri, C.; Nirunsittirat, A.; Piwat, S.; Wattanarat, O.; Pahumunto, N.; Makeudom, A.; Sastraruji, T.; Krisanaprakornkit, S.; Teanpaisan, R. Reduction of Streptococcus mutans by probiotic milk: A multicenter randomized controlled trial. Clin. Oral Investig. 2020, 24, 2363–2374. [Google Scholar] [CrossRef]
- Piwat, S.; Pahumunto, N.; Srisommai, P.; Mapaisansin, C.; Teanpaisan, R. Effect of probiotic delivery vehicles for probiotic Lactobacillus rhamnosus SD11 in caries prevention: A clinical study. J. Food Process. Preserv. 2019, 43, e14147. [Google Scholar] [CrossRef]
- Rungsri, P.; Akkarachaneeyakorn, N.; Wongsuwanlert, M.; Piwat, S.; Nantarakchaikul, P.; Teanpaisan, R. Effect of fermented milk containing Lactobacillus rhamnosus SD11 on oral microbiota of healthy volunteers: A randomized clinical trial. J. Dairy Sci. 2017, 100, 7780–7787. [Google Scholar] [CrossRef] [Green Version]
- Teanpaisan, R.; Piwat, S.; Tianviwat, S.; Sophatha, B.; Kampoo, T. Effect of Long-Term Consumption of Lactobacillus paracasei SD1 on Reducing Mutans streptococci and Caries Risk: A Randomized Placebo-Controlled Trial. Dent. J. 2015, 3, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Wattanarat, O.; Nirunsittirat, A.; Piwat, S.; Manmontri, C.; Teanpaisan, R.; Pahumunto, N.; Makeudom, A.; Sastraruji, T.; Krisanaprakornkit, S. Significant elevation of salivary human neutrophil peptides 1-3 levels by probiotic milk in preschool children with severe early childhood caries: A randomized controlled trial. Clin. Oral Investig. 2020, 25, 2891–2903. [Google Scholar] [CrossRef]
- Piwat, S.; Teanpaisan, R.; Thitasomakul, S.; Thearmontree, A.; Dahlén, G. Lactobacillus species and genotypes associated with dental caries in Thai preschool children. Mol. Oral Microbiol. 2010, 25, 157–164. [Google Scholar] [CrossRef]
- Teanpaisan, R.; Dahlen, G. Use of polymerase chain reaction techniques and sodium dodecyl sulfate-polyacrylamide gel electrophoresis for differentiation of oral Lactobacillus species. Oral Microbiol. Immunol. 2006, 21, 79–83. [Google Scholar] [CrossRef]
- Piwat, S.; Teanpaisan, R. 16S rRNA PCR-Denaturing Gradient Gel Electrophoresis of Oral Lactobacillus casei Group and Their Phenotypic Appearances. ISRN Microbiol. 2013, 2013, 342082. [Google Scholar] [CrossRef]
- Pahumunto, N.; Chotjumlong, P.; Makeudom, A.; Krisanaprakornkit, S.; Dahlen, G.; Teanpaisan, R. Pro-inflammatory cytokine responses in human gingival epithelial cells after stimulation with cell wall extract of Aggregatibacter actinomycetemcomitans subtypes. Anaerobe 2017, 48, 103–109. [Google Scholar] [CrossRef]
- Fung, K.Y.C.; Cosgrove, L.; Lockett, T.; Head, R.; Topping, D.L. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br. J. Nutr. 2012, 108, 820–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Śliżewska, K.; Chlebicz-Wójcik, A. Growth Kinetics of Probiotic Lactobacillus Strains in the Alternative, Cost-Efficient Semi-Solid Fermentation Medium. Biology 2020, 9, 423. [Google Scholar] [CrossRef]
- Monteiro, S.S.; Da Silva, W.P.; Monteiro, S.S.; Gomes, J.P.; Pereira, E.M.; Queiroz, A.J.D.M.; De Figueirêdo, R.M.F.; Rocha, A.P.T.; Silva, H.A.; De Almeida, L.R.B.; et al. Optimization of the Effects of Different Temperatures and Compositions of Filmogenic Solution on Lactobacillus Salivarius Using Predictive Mathematical Models. Foods 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Tomás, M.S.J.; Ocaña, V.S.; Wiese, B.; Nader-Macías, M.E. Growth and lactic acid production by vaginal Lactobacillus acidophilus CRL 1259, and inhibition of uropathogenic Escherichia coli. J. Med. Microbiol. 2003, 52, 1117–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, J.M.W.; de Souza, R.; Kendall, C.W.C.; Emam, A.; Jenkins, D.J.A. Colonic Health: Fermentation and Short Chain Fatty Acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor−κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866e24. [Google Scholar] [CrossRef] [Green Version]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. The Role of Probiotics in Colorectal Cancer Management. Evidence-Based Complement. Altern. Med. 2020, 2020, 3535982. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 3100. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, K.-N.; Vitetta, L. Effects of Intestinal Microbial–Elaborated Butyrate on Oncogenic Signaling Pathways. Nutrients 2019, 11, 1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.-J.; Hong, Y.-H. In situ delivery of biobutyrate by probiotic Escherichia coli for cancer therapy. Sci. Rep. 2021, 11, 18172. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.-J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [Green Version]
- Davie, J.R. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef] [Green Version]
- Otte, J.-M.; Werner, I.; Brand, S.; Chromik, A.M.; Schmitz, F.; Kleine, M.; Schmidt, W.E. Human beta defensin 2 promotes intestinal wound healing in vitro. J. Cell. Biochem. 2008, 104, 2286–2297. [Google Scholar] [CrossRef]
- O’Neil, D.A.; Porter, E.M.; Elewaut, D.; Anderson, G.M.; Eckmann, L.; Ganz, T.; Kagnoff, M.F. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 1999, 163, 6718–6724. [Google Scholar]
- Wehkamp, J.; Harder, J.; Wehkamp, K.; Meissner, B.W.-V.; Schlee, M.; Enders, C.; Sonnenborn, U.; Nuding, S.; Bengmark, S.; Fellermann, K.; et al. NF-κB- and AP-1-Mediated Induction of Human Beta Defensin-2 in Intestinal Epithelial Cells by Escherichia coli Nissle 1917: A Novel Effect of a Probiotic Bacterium. Infect. Immun. 2004, 72, 5750–5758. [Google Scholar] [CrossRef] [Green Version]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008, 23, 1298–1303. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2011, 6, 320–329. [Google Scholar] [CrossRef]
- Monleón, D.; Morales, J.M.; Barrasa, A.; López, J.A.; Vázquez, C.; Celda, B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2008, 22, 342–348. [Google Scholar] [CrossRef]
- Chen, H.-M.; Yu, Y.-N.; Wang, J.-L.; Lin, Y.-W.; Kong, X.; Yang, C.-Q.; Yang, L.; Liu, Z.-J.; Yuan, Y.-Z.; Liu, F.; et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013, 97, 1044–1052. [Google Scholar] [CrossRef] [Green Version]
- Simeoli, R.; Raso, G.M.; Pirozzi, C.; Lama, A.; Santoro, A.; Russo, R.; Montero-Melendez, T.; Canani, R.B.; Calignano, A.; Perretti, M.; et al. An orally administered butyrate-releasing derivative reduces neutrophil recruitment and inflammation in dextran sulphate sodium-induced murine colitis. Br. J. Pharmacol. 2016, 174, 1484–1496. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFarland, L.V. Efficacy of single-strain probiotics versus multi-strain Mixtures: Systematic review of strain and disease specificity. Dig. Dis. Sci. 2021, 66, 694–704. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, Y.; Fang, K.; Mao, K.; Dou, J.; Fan, H.; Zhou, C.; Wang, H. Lactobacillus acidophilus and Clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef. Microbes 2018, 9, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Pahumunto, N.; Teanpaisan, R. Anti-cancer properties of potential probiotics and their cell-free supernatants for the prevention of colorectal cancer: An in vitro study. Probiotics Antimicrob. Prot. 2022. [CrossRef] [PubMed]
- Flach, J.; van der Waal, M.; Nieuwboer, M.V.D.; Claassen, H.; Larsen, O.F.A. The underexposed role of food matrices in probiotic products: Reviewing the relationship between carrier matrices and product parameters. Crit. Rev. Food Sci. Nutr. 2017, 58, 2570–2584. [Google Scholar] [CrossRef] [PubMed]




| Probiotics | S. enterica | ETEC | F. nucleatum | P. gingivalis |
|---|---|---|---|---|
| Antimicrobial activity by agar diffusion method (mean ± SD in mm) | ||||
| SD1 | 12.5 ± 0.0 c,B | 12.5 ± 0.4 c,A | 15.0 ± 0.4 b,A | 17.0 ± 0.4 a,A |
| SD4 | 9.0 ± 0.0 c,C | 10.0 ± 1.1 c,B | 12.0 ± 0.4 b,C | 14.0 ± 0.0 a,C |
| SD11 | 12.3 ± 0.0 c,A | 12.0 ± 0.4 c,A | 15.0 ± 0.7 b,A | 17.0 ± 0.4 a,A |
| LGG | 10.6 ± 1.1 c,B | 10.0 ± 0.0 c,B | 13.0 ± 0.4 b,B | 15.5 ± 0.4 a,B |
| MIC by broth microdilution method (mean ± SD in µg/mL) | ||||
| SD1 | 517.3 ± 0.0 a,B | 517.3 ± 0.0 a,B | 358.6 ± 0.0 b,B | 358.6 ± 0.0 b,B |
| SD4 | 537.6 ± 0.0 a,A | 537.6 ± 0.0 a,A | 368.8 ± 0.0 b,A | 368.8 ± 0.0 b,A |
| SD11 | 517.3 ± 0.0 a,B | 517.3 ± 0.0 a,B | 358.6 ± 0.0 b,B | 358.6 ± 0.0 b,B |
| FIC index | ||||
| L. paracasei SD1 + L. rhamnosus SD4 | 1.0 (258.7/268.8) | 1.0 (258.7/268.8) | 0.5 (89.7/92.2) | 0.5 (89.7/92.2) |
| L. paracasei SD1+ L. rhamnosus SD11 | 1.0 (258.7/258.7) | 1.0 (258.7/258.7) | 0.5 (89.7/89.7) | 0.5 (89.7/89.7) |
| L. rhamnosus SD4+ L. rhamnosus SD11 | 1.0 (268.8/258.7) | 1.0 (268.8/258.7) | 0.5 (92.2/89.7) | 0.5 (92.2/89.7) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thananimit, S.; Pahumunto, N.; Teanpaisan, R. Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains. Biomolecules 2022, 12, 1829. https://doi.org/10.3390/biom12121829
Thananimit S, Pahumunto N, Teanpaisan R. Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains. Biomolecules. 2022; 12(12):1829. https://doi.org/10.3390/biom12121829
Chicago/Turabian StyleThananimit, Suchera, Nuntiya Pahumunto, and Rawee Teanpaisan. 2022. "Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains" Biomolecules 12, no. 12: 1829. https://doi.org/10.3390/biom12121829
APA StyleThananimit, S., Pahumunto, N., & Teanpaisan, R. (2022). Characterization of Short Chain Fatty Acids Produced by Selected Potential Probiotic Lactobacillus Strains. Biomolecules, 12(12), 1829. https://doi.org/10.3390/biom12121829








