Polysorbate 21 Can Modulate the Antibacterial Potential of Two Pyrazol Derivatives
Abstract
:1. Introduction
2. Materials and Methods
2.1. MIC Evaluation
2.2. Time-Kill Assay
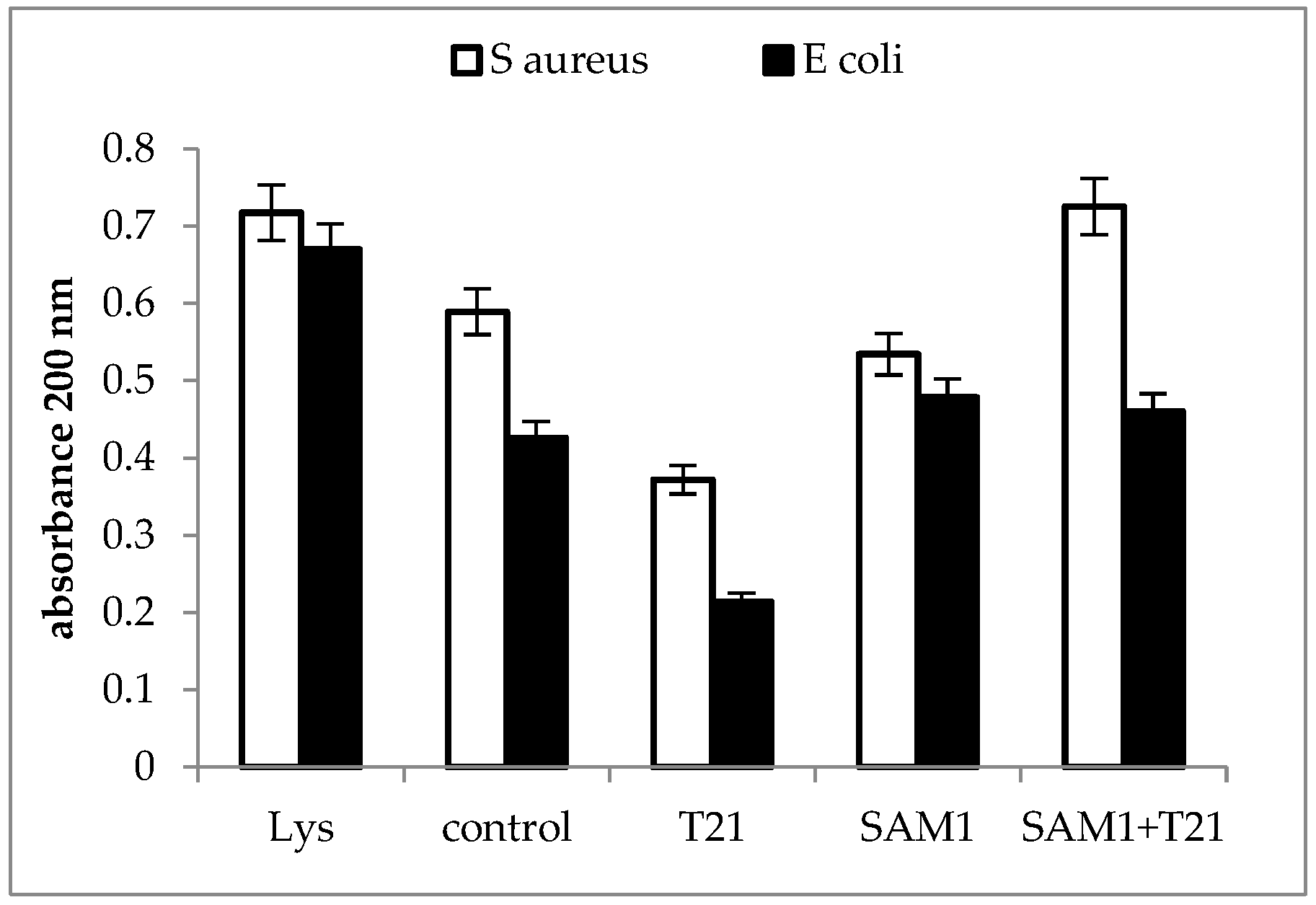
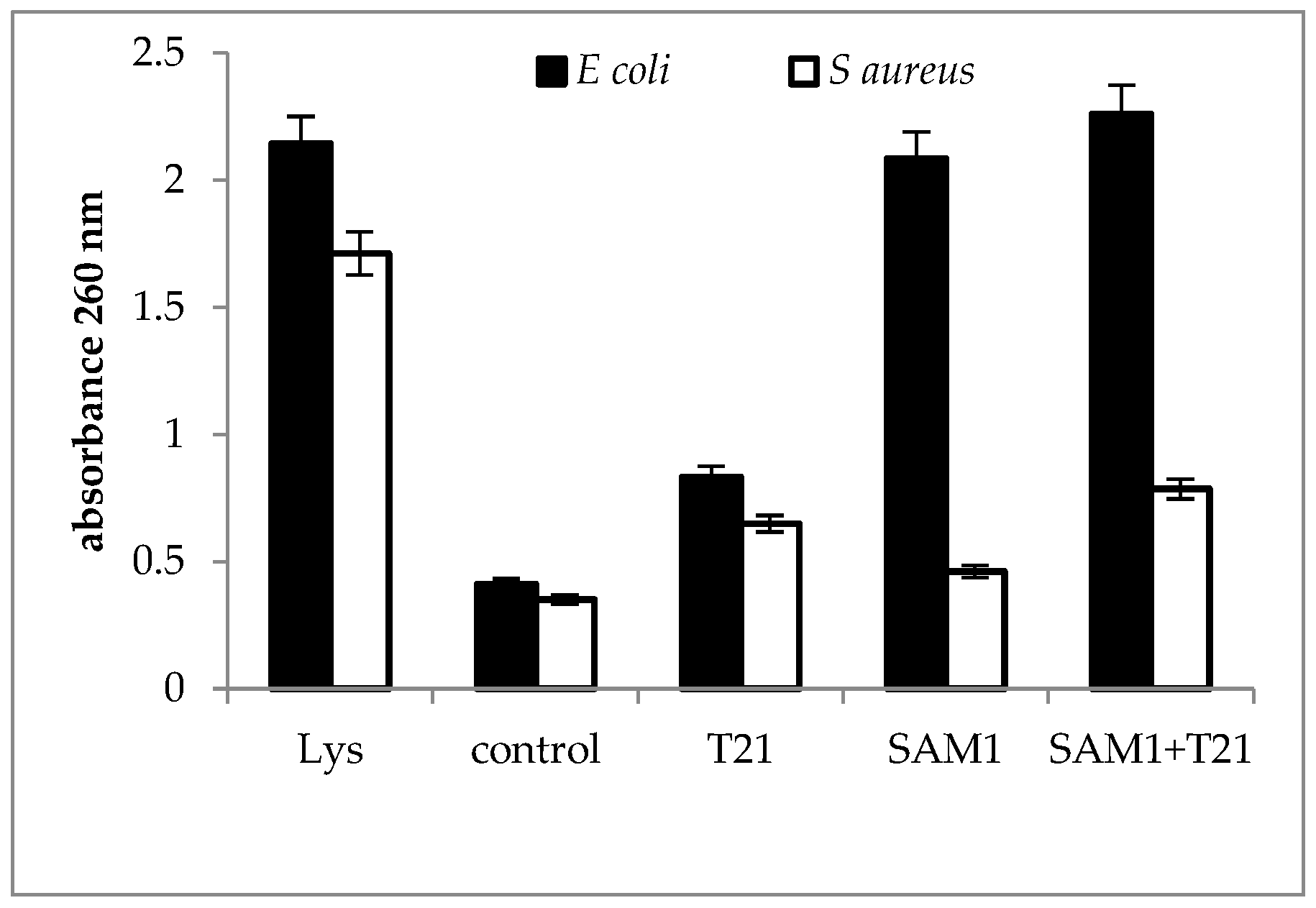
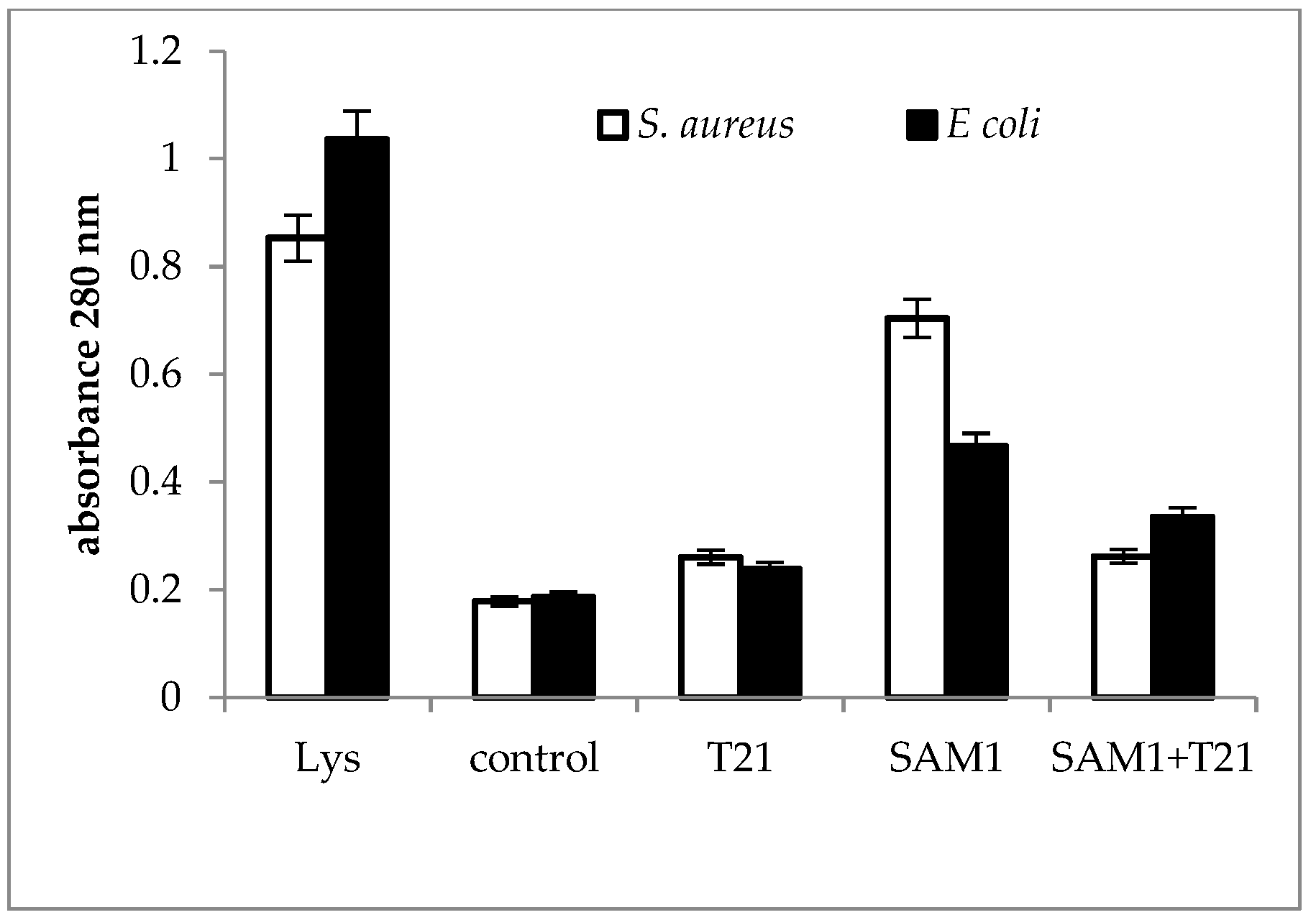
2.3. Cellular Material Release
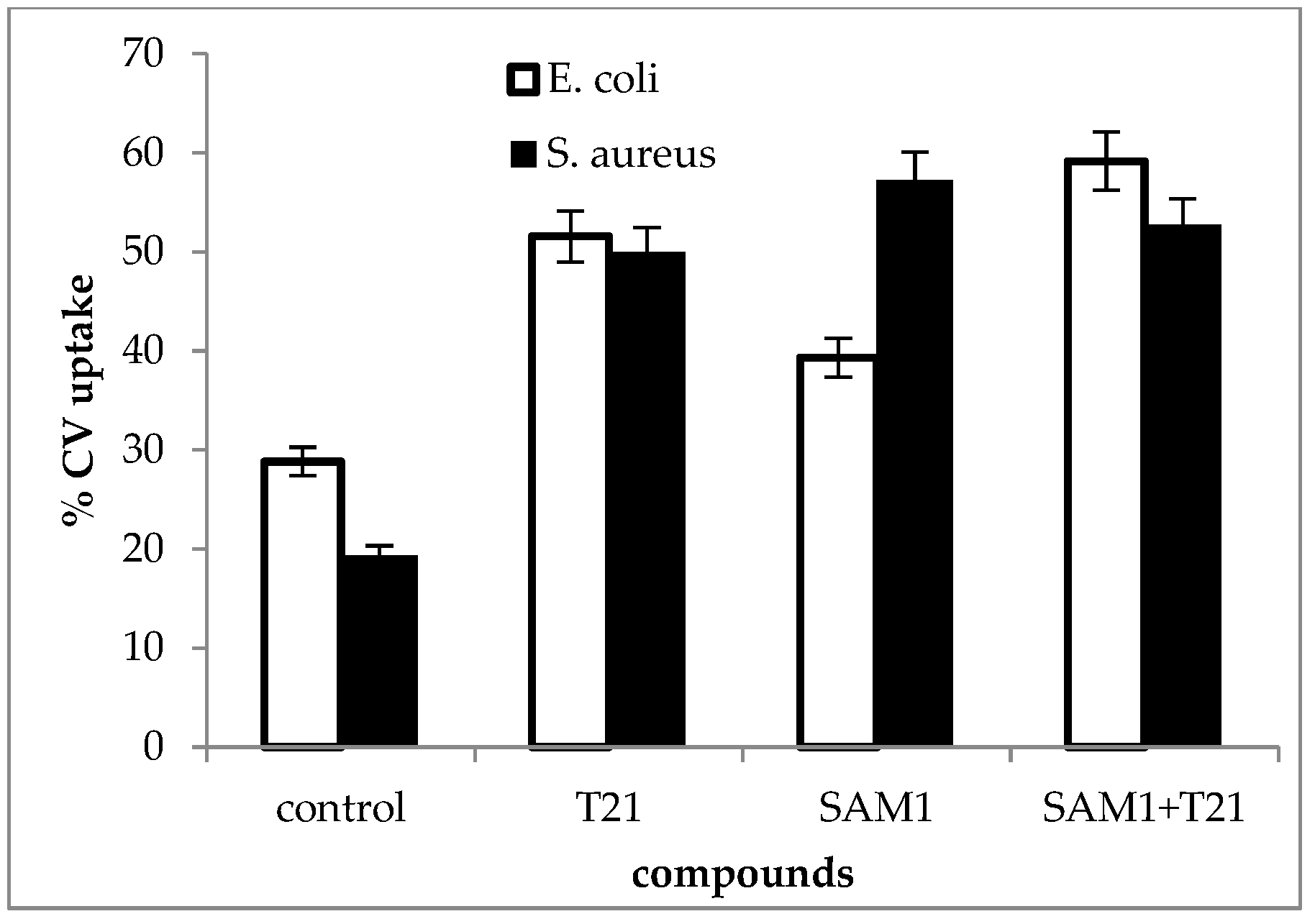
2.4. Uptake of Crystal Violet
3. Results
3.1. MIC Values of Pyrazoles and Their Combination with T21
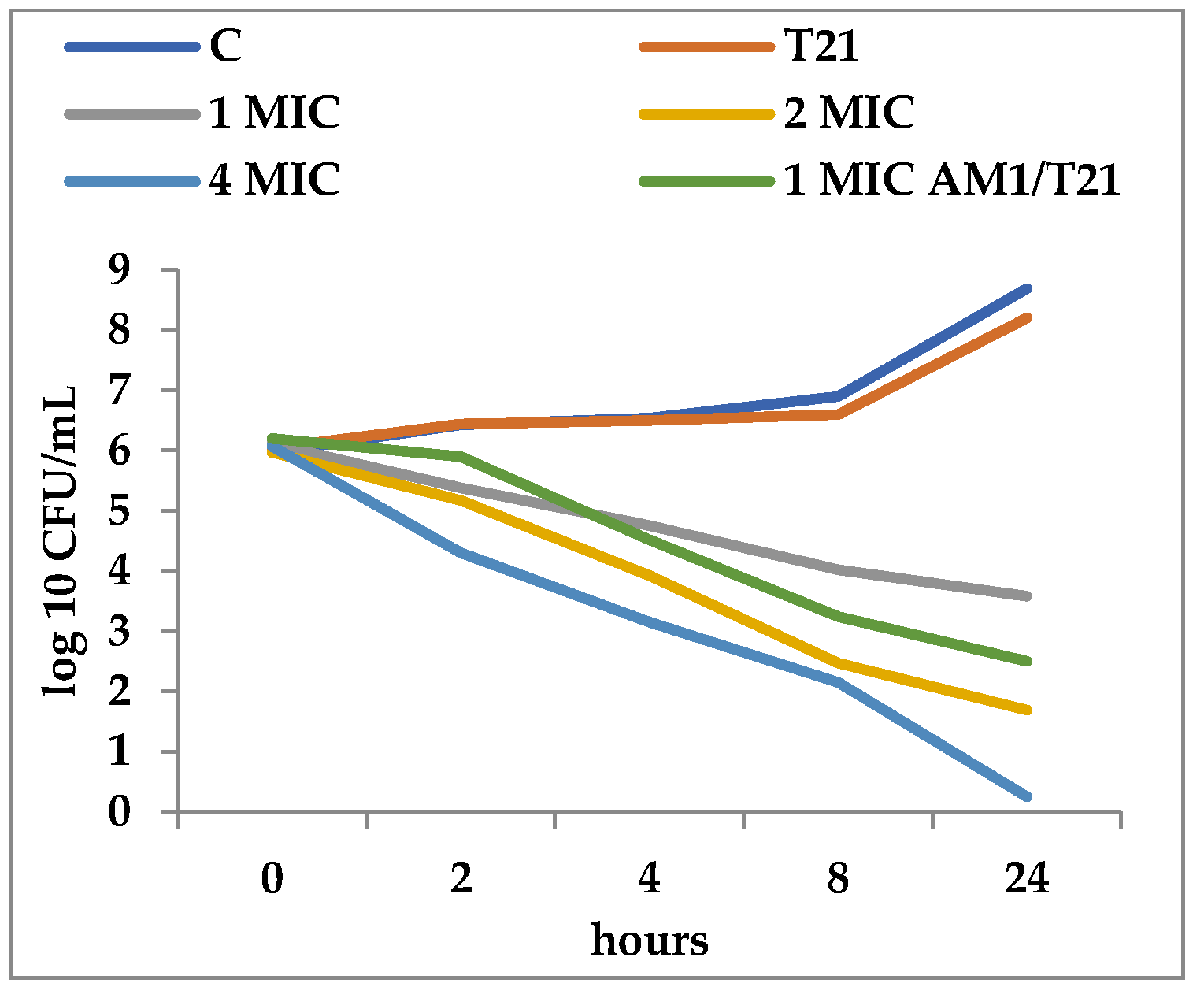
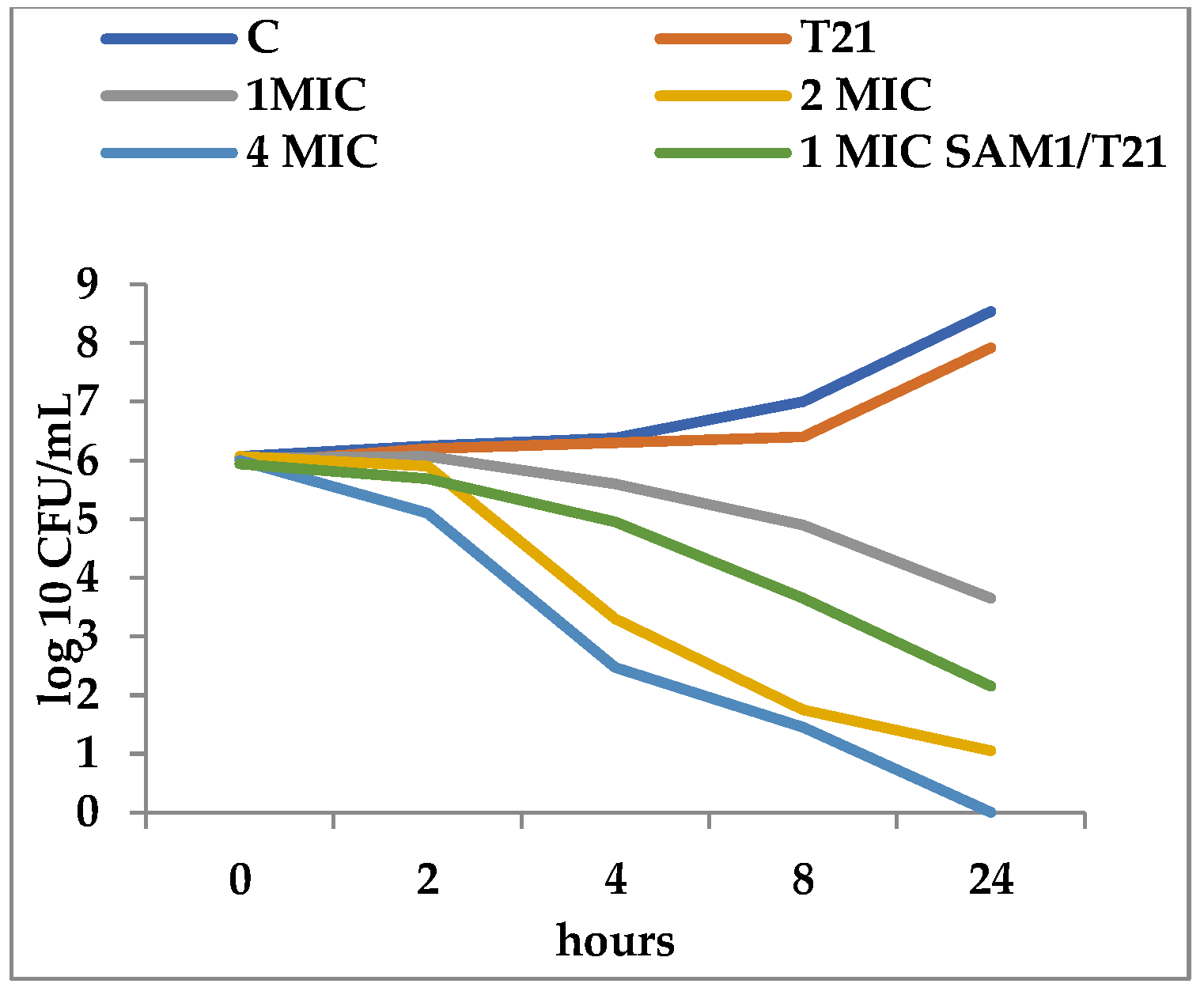
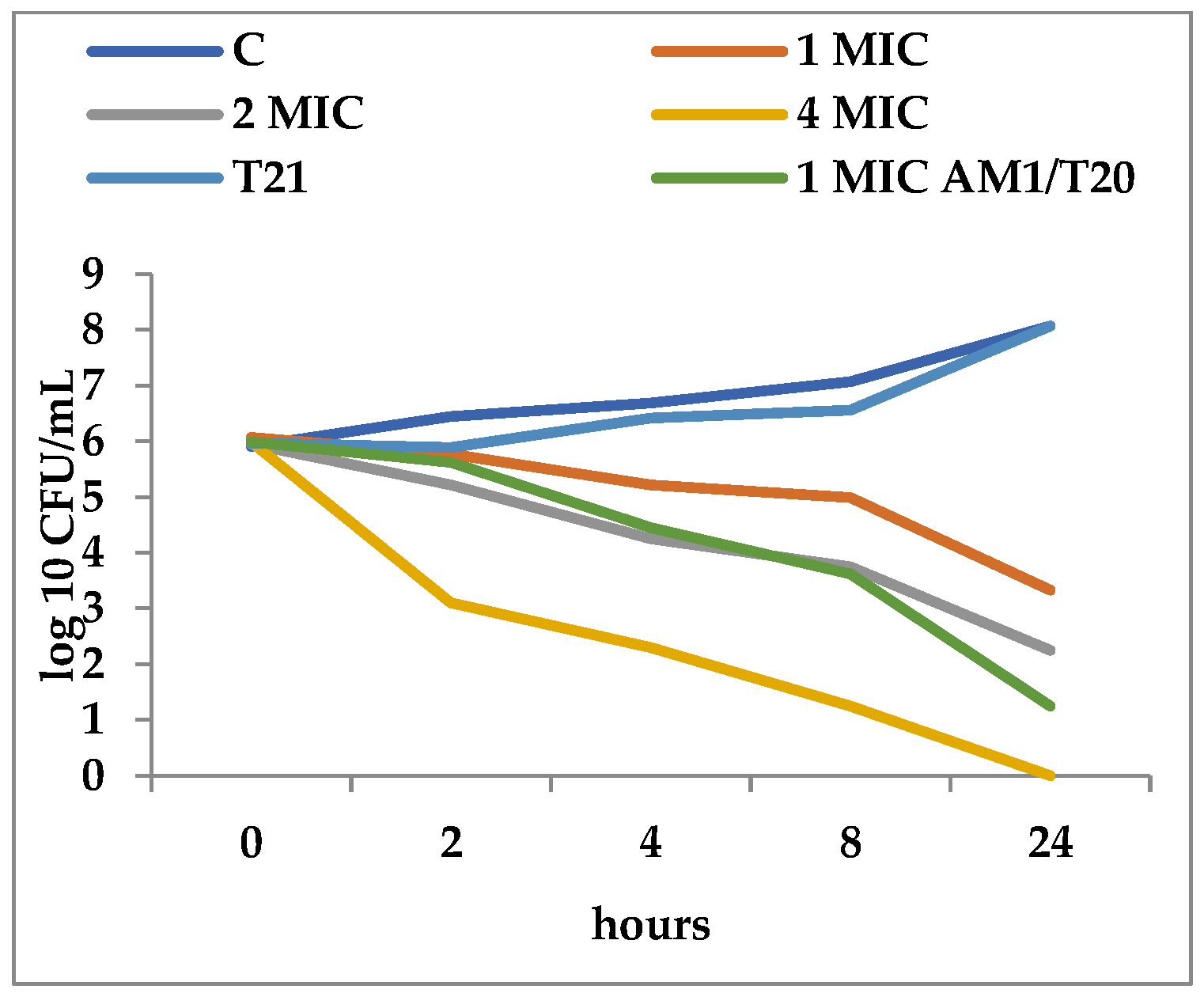
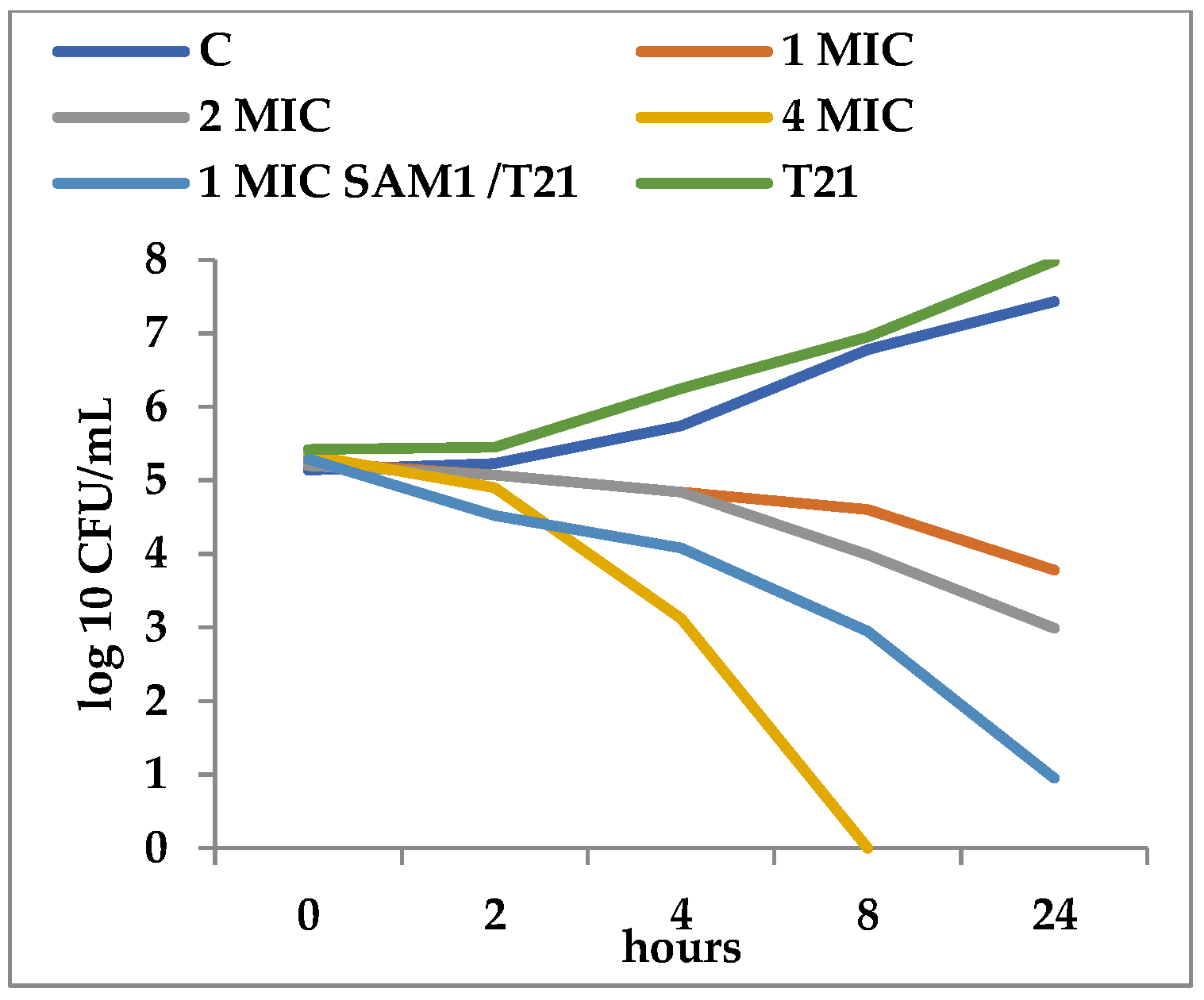
3.2. Time-Kill Dynamics
3.3. Cellular Material Release
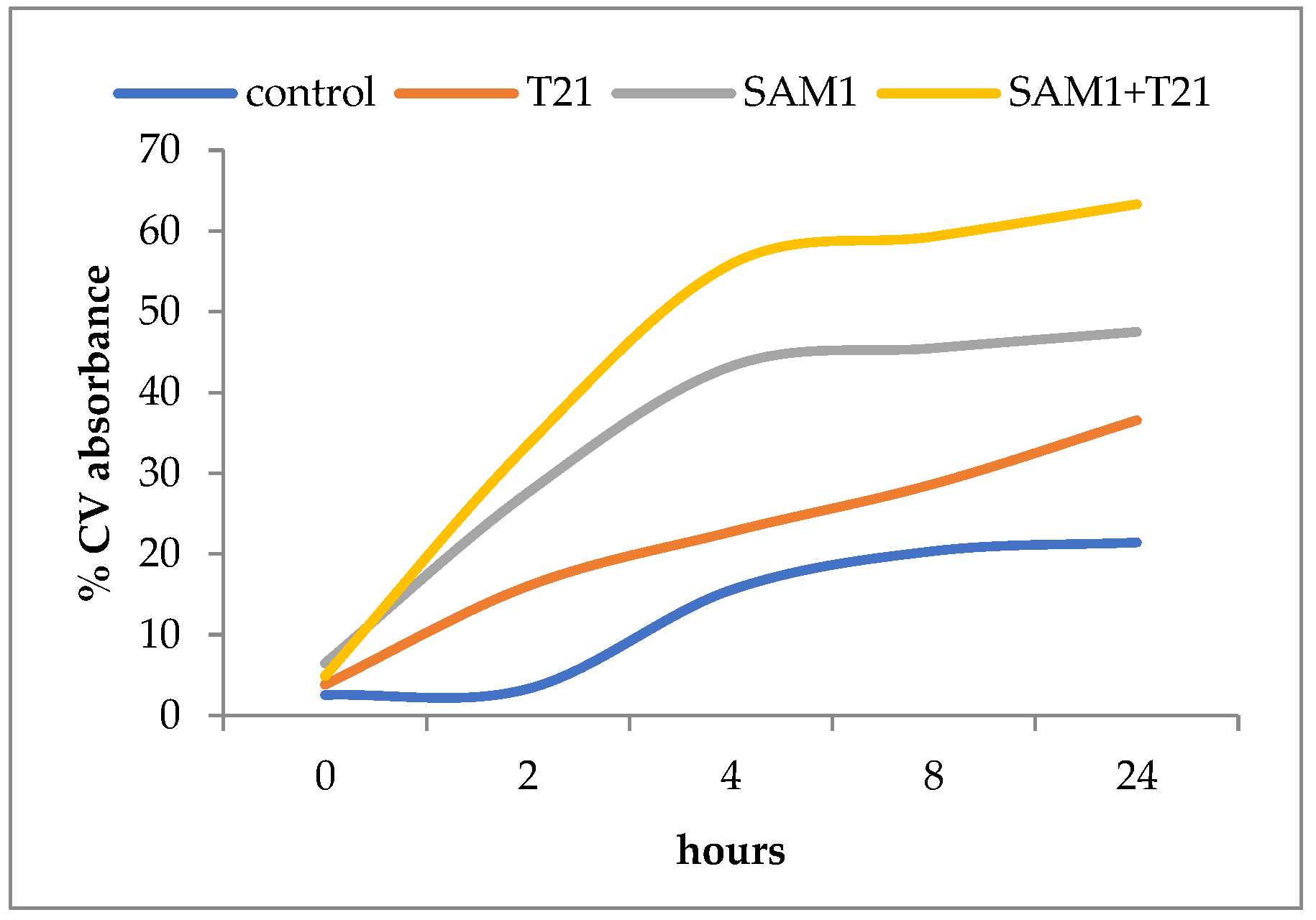
3.4. Crystal Violet Uptake
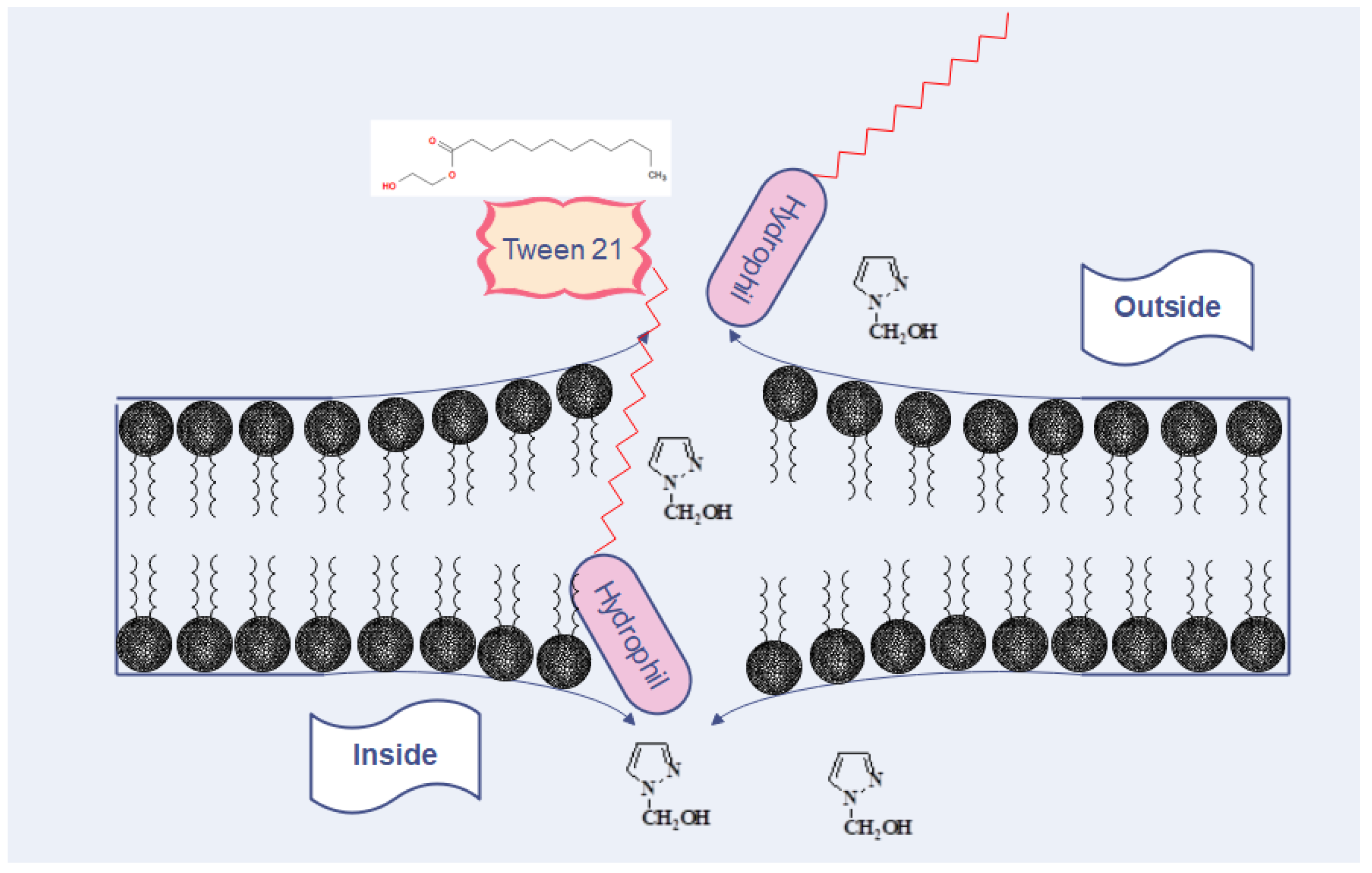
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Institute for Health Metrics and Evaluation (IHME), University of Oxford. Global Bacterial Antimicrobial Resistance Burden Estimates 2019; Institute for Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2022. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Naylor, N.R.; Atun, R.; Zhu, N.; Kulasabanathan, K.; Silva, S.; Chatterjee, A.; Knight, G.M.; Robotham, J.V. Estimating the burden of antimicrobial resistance: A systematic literature review. Antimicrob. Resist. Infect Control 2018, 7, 58. [Google Scholar] [CrossRef] [Green Version]
- Hasan, C.M.; Dutta, D.; Nguyen, A.N.T. Revisiting Antibiotic Resistance: Mechanistic Foundations to Evolutionary Outlook. Antibiotics 2021, 11, 40. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Kucukguzel, S.G.; Senkardes, S. Recent advances in bioactive pyrazoles. Eur. J. Med. Chem. 2014, 97, 786–815. [Google Scholar] [CrossRef]
- Raffa, D.; Maggio, B.; Raimondi, M.V.; Cascioferro, S.; Plescia, F.; Cancemi, G.; Daidone, G. Recent advanced in bioactive systems containing pyrazole fused with a five membered heterocycle. Eur. J. Med. Chem. 2014, 97, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Bekhit, A.A.; Ashour, H.M.A.; Abdel Ghany, Y.S.; Bekhit, A.E.; Baraka, A. Synthesis and biological evaluation of some thiazolyl and thiadiazolyl derivatives of 1H-pyrazole as anti-inflammatory antimicrobial agents. Eur. J. Med. Chem. 2008, 43, 456–463. [Google Scholar] [CrossRef]
- Aonofriesei, F. Effect of methylpyrazoles and coumarin association on the growth of Gram-negative bacteria. Arch. Microbiol. 2022, 204, 160. [Google Scholar] [CrossRef]
- Vijesh, A.M.; Isloor, A.M.; Shetty, P.; Sundershan, S.; Fun, H.K. New pyrazole derivatives containing 1,2,4-triazoles and benzoxazoles as potent antimicrobial and analgesic agents. Eur. J. Med. Chem. 2013, 62, 410–415. [Google Scholar] [CrossRef]
- Cetin, A.; Bildirici, I. A study on synthesis and antimicrobial activity of 4-acyl-pyrazoles. J. Saudi Chem. Soc. 2018, 22, 279–296. [Google Scholar] [CrossRef] [Green Version]
- B’Bhatt, H.; Sharma, S. Synthesis and antimicrobial activity of pyrazole nucleus containing 2-thioxothiazolidin-4-one Derivatives. Arab. J. Chem. 2017, 10, S1590–S1596. [Google Scholar] [CrossRef] [Green Version]
- Rahimizadeh, M.; Pordel, M.; Bakavoli, M.; Rezaeian, S.; Sadeghian, A. Synthesis and antibacterial activity of some new derivatives of pyrazole. World J. Microbiol. Biotechnol. 2010, 26, 317–321. [Google Scholar] [CrossRef]
- Kumar, R.S.; Arif, I.A.; Ahamed, A.; Idhayadhulla, A. Anti-inflammatory and antimicrobial activities of novel pyrazole analogues. Saudi J. Biol. Sci. 2016, 23, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Nayak, N.; Ramprasad, J.; Dalimba, U. New INH–pyrazole analogs: Design, synthesis and evaluation of antitubercular and antibacterial activity. Bioorganic Med. Chem. Lett. 2015, 25, 5540–5545. [Google Scholar] [CrossRef]
- Pandit, U.; Dodiya, A. Synthesis and antitubercular activity of novel pyrazole–quinazolinone hybrid analogs. Med. Chem. Res. 2013, 22, 3364–3371. [Google Scholar] [CrossRef]
- Aragade, P.; Palkar, M.; Ronad, P.; Satyanarayana, D. Coumarinyl pyrazole derivatives of INH: Promising antimycobacterial agents. Med. Chem. Res. 2013, 22, 2279–2283. [Google Scholar] [CrossRef]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of Antibiotic Resistance in Important Gram-Positive and Gram-Negative Pathogens and Novel Antibiotic Solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Nielsen, K.M. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; López, C.A.; Gnanakaran, S. Permeability Barrier of Gram-Negative Cell Envelopes and Approaches to Bypass It. ACS Infect. Dis. 2016, 1, 512–522. [Google Scholar] [CrossRef] [Green Version]
- Bueno, J. Antimicrobial Adjuvants Drug Discovery, the Challenge of Avoid the Resistance and Recover the Susceptibility of Multidrug-Resistant Strains. J. Microb. Biochem. Technol. 2016, 8, 169–176. [Google Scholar] [CrossRef]
- Pedersen, J.N.; Lyngsø, J.; Zinn, T.; Otzen, D.E.; Pedersen, J.S. A complete picture of protein unfolding and refolding in surfactants. Chem. Sci. 2020, 11, 699–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otzen, D.E. Biosurfactants and surfactants interacting with membranes and proteins: Same but different? Biochim. Biophys. Acta-Biomembr. 2017, 1859, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Anestopoulos, I.; Kiousi, D.E.; Klavaris, A.; Galanis, A.; Salek, K.; Euston, S.R.; Pappa, A.; Panayiotidis, M.I. Surface Active Agents and Their Health-Promoting–Properties: Molecules of Multifunctional Significance. Pharmaceutics 2020, 12, 688. [Google Scholar] [CrossRef] [PubMed]
- Lupsor, S.; Aonofriesei, F.; Iovu, M. Antibacterial activity of aminals and hemiaminals of pyrazole and imidazole. Med. Chem. Res. 2012, 21, 3035–3042. [Google Scholar] [CrossRef]
- Lupsor, S.; Tarcomnicu, I.; Aonofriesei, F.; Iovu, M. Microwave-Assisted Synthesis of 1-Hydroxymethylazoles. Rev. Chim. 2011, 62, 493–498. [Google Scholar]
- Falk, N.A. Surfactants as Antimicrobials: A Brief Overview of Microbial Interfacial Chemistry and Surfactant Antimicrobial Activity. J. Surfact. Deterg. 2019, 22, 1119–1127. [Google Scholar] [CrossRef]
- Hindler, J.F.; Munro, S. Antimicrobial Susceptibility Testing. In Microbiology Procedures Handbook, Section 5; Isenberg, H.D., Garcia, L.S., Eds.; ASM Press American Society for Microbiology: Washington, DC, USA, 2007; pp. 2036–2904. [Google Scholar]
- Zhang, J.; Zhang, X.; Pan, D.-D.; Sun, Y.-Y.; Cao, J.-X. Antibacterial Activity and Mechanism of Action of Black Pepper Essential Oil on Meat-Borne Escherichia coli. Front. Microbiol. 2017, 7, 2094. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Bahuguna, A.; Kumar, P.; Bajpai, V.K.; Kang, S.C. Antimicrobial Potential of Carvacrol against Uropathogenic Escherichia coli via Membrane Disruption, Depolarization, and Reactive Oxygen Species Generation. Front. Microbiol. 2017, 8, 2421. [Google Scholar] [CrossRef] [Green Version]
- Desai, N.; Rajpara, C.K.M.; Joshi, V.V. Synthesis of pyrazole encompassing 2-pyridone derivatives as antibacterial agents. Bioorganic Med. Chem. Lett. 2013, 23, 2714–2717. [Google Scholar] [CrossRef]
- Mohi El-Deen, E.M.; Nossier, E.S.; Karam, E.A. New Quinazolin-4(3H)-one Derivatives Incorporating Hydrazone and Pyrazole Scaffolds as Antimicrobial Agents Targeting DNA Gyraze Enzyme. Sci. Pharm. 2022, 90, 52. [Google Scholar] [CrossRef]
- Addoum, B.; El Khalfi, B.; Derdak, R.; Sakoui, S.; Elmakssoudi, A.; Soukri, A. Synthesis, in vitro Antimicrobial Activity, and Docking Studies of some Pyrano[2,3-c] Pyrazole Derivatives. Biointerface Res. Appl. Chem. 2022, 12, 4705–4730. [Google Scholar] [CrossRef]
- Mohamed, H.A.; Ammar, Y.A.; Gameel, A.M.; Elhagali, G.A.M.; Eyada, H.A.; Aboul-Magd, D.S.; Ragab, A. In Vitro Antimicrobial Evaluation, Single-Point Resistance Study, and Radiosterilization of Novel Pyrazole Incorporating Thiazol-4-one/Thiophene Derivatives as Dual DNA Gyrase and DHFR Inhibitors against MDR Pathogens. ACS Omega 2022, 7, 4970–4990. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Askar, A.A.; Naglah, A.M.; Almehizia, A.A.; Ragab, A. Discovery of New Schiff Bases Tethered 36. Pyrazole Moiety: Design, Synthesis, Biological Evaluation, and Molecular Docking Study as Dual Targeting DHFR/DNA Gyrase Inhibitors with Immunomodulatory Activity. Molecules 2020, 25, 2593. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.M.; Mondragón, A. Basic residues at the C-gate of DNA gyrase are involved in DNA supercoiling. J. Biol. Chem. 2021, 297, 101000. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making Use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, V.; Goveas, L.C.; Narayan, B.; Halami, P.M. Comparison of lipase production by Enterococcus faecium MTCC, 5695 and Pediococcus acidilactici MTCC, 11361 using fish waste as substrate: Optimization of culture conditions by response surface methodology. Int. Sch. Res. Not. Biotechnol. 2013, 2013, 980562. [Google Scholar]
- Thomas, S.M.; Kavitha, S. Isolation and screening of lipase producing microorganism from soil and comparative study of enzyme activity with different substrates. Int. J. Sci. Eng. Technol. Res. 2015, 4, 5778–5781. [Google Scholar]
- Longshaw, C.M.; Farrell, A.M.; Wright, J.D.; Holland, K.T. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: Comparison of sequence with those of other staphylococcal lipases. Microbiology 2000, 146, 1419–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakinc, T.; Woznowski, M.; Ebsen, E.; Gatermann, S.G. The Surface-Associated Protein of Staphylococcus saprophyticus is a Lipase. Infect. Immun. 2005, 73, 6419–6428. [Google Scholar] [CrossRef] [Green Version]
- Toutain-Kidd, C.M.; Kadivar, S.C.; Bramante, C.T.; Bobin, S.A.; Zegans, M.E. Polysorbate 80 Inhibition of Pseudomonas aeruginosa Biofilm Formation and Its Cleavage by the Secreted Lipase LipA. Antimicrob. Agents Chemother. 2009, 53, 136–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Lim, J.Y.; Fuller, G.G.; Cegelski, L. Disruption of E. coli amyloid-integrated biofilm formation at the air-liquid interface by a polysorbate surfactant. Langmuir 2013, 22, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Taoka, Y.; Nagano, N.; Okita, Y.; Izumida, H.; Sugimoto, S.; Hayashi, M. Effect of Tween 80 on the growth, lipid accumulation and fatty acid composition of Thraustochytrium aureum ATCC34304. J. Biosci. Bioen. 2011, 111, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Bouke, K.; Boekema, H.L.; Beselin, A.; Breuer, M.; Hauer, B.; Koster, M.; Rosenau, F.; Jaeger, K.-E.; Tommassen, J. Hexadecane and Tween 80 Stimulate Lipase Production in Burkholderia glumae by Different Mechanisms. Appl. Environ. Microbiol. 2007, 73, 3838–3844. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, C.K.; Kjems, J.; Mygind, T.; Snabe, T.; Meyer, R.L. Effects of Tween 80 on Growth and Biofilm Formation in Laboratory Media. Front. Microbiol. 2016, 7, 1878. [Google Scholar] [CrossRef]
- Prasad, S.; Mandal, I.; Singh, S.; Paul, A.; Mandal, B.; Venkatramani, R.; Swaminathan, R. Near UV-Visible electronic absorption originating from charged amino acids in a monomeric protein. Chem. Sci. 2017, 8, 5416. [Google Scholar] [CrossRef] [Green Version]
- Saraiva, M.A. Interpretation of α-synuclein UV absorption spectra in the peptide bond and the aromatic regions. J. Photochem. Photobiology. B Biol. 2020, 212, 112022. [Google Scholar] [CrossRef]
- Perdomo, R.; Montero, V. Purification of E. coli 055:B5 lipopolysaccharides by size exclusion chromatography. Biotecnol. Appl. 2006, 23, 124–129. [Google Scholar]
- McHowat, J.; Jones, J.H.; Creer, M.H. Quantitation of individual phospholipid molecular species by UV absorption measurements. J. Lipid Res. 1996, 371, 2450–2460. [Google Scholar] [CrossRef]
- Olson, N.D.; Morrow, J.B. DNA extract characterization process for microbial detection methods development and validation. BMC Res. Notes 2012, 5, 668. [Google Scholar] [CrossRef] [Green Version]
- Wilfinger, W.W.; Mackey, K.; Chomczynski, P. Assessing the quantity, purity and integrity of RNA and DNA following nucleic acid purification. In DNA Sequencing. II Optimizing Preparation and Cleanup; Sudbury, K.J., Ed.; Jones and Bartlett Publishers: Burlington, MA, USA, 2006; pp. 291–312. [Google Scholar]
- Schmid, F.-X. Biological Macromolecules: UV-visible Spectrophotometry. In Encyclopedia of Life Sciences; Nature Publishing Group: London, UK, 2001; pp. 1–4. Available online: www.els.net (accessed on 25 June 2021). [CrossRef]
- Sana, S.; Datta, S.; Biswas, D.; Sengupta, D. Assessment of synergistic antibacterial activity of combined biosurfactants revealed by bacterial cell envelop damage. BBA–Biomembr. 2018, 1860, 579–585. [Google Scholar] [CrossRef]
- Paul, S.; Dubey, R.C.; Maheswari, D.K.; Kang, S.H. Trachyspermum ammi (L.) fruit essential oil influencing on membrane permeability and surface characteristics in inhibiting food-borne pathogens. Food Control 2011, 22, 725–731. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of Action of Melaleuca alternifolia (Tea Tree) Oil on Staphylococcus aureus Determined by Time-Kill, Lysis, Leakage, and Salt Tolerance Assays and Electron Microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Davidson, P.M.; Critzer, F.; Zhong, Q. Antimicrobial activities of lauric arginate and cinnamon oil combination against foodborne pathogens: Improvement by ethylenediaminetetraacetate and possible mechanisms. LWT-Food Sci. Technol. 2016, 72, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Luo, M.; Fu, Y.; Zu, Y.; Wang, W.; Zhang, L.; Yao, L.; Zhao, C.; Sun, Y. Effect of Corilagin on Membrane Permeability of Escherichia coli, Staphylococcus aureus and Candida albicans. Phytother. Res. 2013, 27, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.P.; Sakthivel, R.; Arif Nisha, S.; Suganthy, N.; Karutha Pandia, S. Eugenol alters the integrity of cell membrane and acts against the nosocomial pathogen Proteus mirabilis. Arch. Pharm. Res. 2013, 36, 282–292. [Google Scholar] [CrossRef]
- Guo, N.; Gai, Q.-Y.; Jiao, J.; Wang, W.; Zu, Y.G.; Fu, Y.J. Antibacterial Activity of Fructus forsythia Essential Oil and the Application of EO-Loaded Nanoparticles to Food-Borne Pathogens. Foods 2016, 5, 73. [Google Scholar] [CrossRef] [Green Version]
- Thorgeirsdottir, T.O.; Thormar, H.; Kristmundsdottir, T. Effects of polysorbates on antiviral and antibacterial activity of monoglyceride in pharmaceutical formulations. Pharmazie 2003, 58, 286–287. [Google Scholar]
- Churchward, C.P.; Kinani, A.A.; Abdelkader, H.; Swinden, J.; Siwoku, O.; Varnakulasingam, T.; Alany, R.G.; Snyder, L.A.S. Monocaprin eye drop formulation to combat antibiotic resistant gonococcal blindness. Sci. Rep. 2020, 10, 12010. [Google Scholar] [CrossRef]
| Crt. No. | Strain | Phenotypic Characteristics | Abbreviation |
|---|---|---|---|
| 1 | Proteus mirabilis | Clinical strain isolated from urinary tract infection, resistant to ampicillin, aztreonam cefuroxime, imipenem, nitrofurantoin, and tetracycline | PM1 |
| 2 | Escherichia coli | Clinical strain isolated from urinary tract infection, resistant to nalidixic acid, amoxicillin, cotrimoxazole, and cefalotin | EC1 |
| 3 | Pseudomonas aeruginosa | Clinical strain isolated from ear infection, pyocianin production, resistant to aztreonam ciprofloxacin, cefpirome, gentamicin, levofloxacin, minocycline ticarcilin, netilmicin, tobramycin, and pefloxacin | PA1 |
| 4 | Staphylococcus aureus | Clinical strain isolated from skin infection, methicillin-resistant (MRSA), coagulase-positive, haemolytic, resistant to cefoxitin, penicillin, penicillin, ciprofloxacin, erythromycin, clindamycin, and gentamycin | SA1 |
| 5 | Staphylococcus aureus | Clinical strain isolated from skin infection, (MRSA), coagulase-positive, haemolytic, resistant to azithromycin, cloxacillin, ciprofloxacin, co-trimoxazole, chloramphenicol, clindamycin, tetracycline, and gentamycin | SA2 |
| 6 | Staphylococcus aureus | Clinical strain isolated from skin infection, (MRSA), coagulase-positive, haemolytic, resistant to ampicillin ciprofloxacin cefoxitin, clindamycin penicillin, erythromycin gentamycin, and tetracycline | SA3 |
| 7 | Escherichia coli ATCC 11229 | Reference strains | ECATTC |
| 8 | Pseudomonas aeruginosa ATCC 27853 | Reference strains | PAATTC |
| 9 | Staphylococcus aureus ATCC 33952 | Reference strains | SAATTC |
| Crt. No. | Strain | Compounds | |||
|---|---|---|---|---|---|
| SAM1 | AM1 | SAM1 + T21 | AM1 + T21 | ||
| 1 | PM 1 | 200 | 400 | 100 | 100 |
| 2 | PAATCC | 200 | 400 | 50 | 200 |
| 3 | PA 1 | 400 | 400 | 50 | 200 |
| 4 | EC1 | 200 | 400 | 50 | 200 |
| 5 | ECATTC | 200 | 400 | 50 | 200 |
| 6 | SAATTC | 100 | 200 | 50 | 100 |
| 7 | SA 1 | 100 | 200 | 50 | 100 |
| 8 | SA 2 | 100 | 200 | 50 | 100 |
| 9 | SA 3 | 100 | 200 | 50 | 100 |
| Mean | 177.77 | 311.11 | 55.55 | 144.44 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aonofriesei, F. Polysorbate 21 Can Modulate the Antibacterial Potential of Two Pyrazol Derivatives. Biomolecules 2022, 12, 1819. https://doi.org/10.3390/biom12121819
Aonofriesei F. Polysorbate 21 Can Modulate the Antibacterial Potential of Two Pyrazol Derivatives. Biomolecules. 2022; 12(12):1819. https://doi.org/10.3390/biom12121819
Chicago/Turabian StyleAonofriesei, Florin. 2022. "Polysorbate 21 Can Modulate the Antibacterial Potential of Two Pyrazol Derivatives" Biomolecules 12, no. 12: 1819. https://doi.org/10.3390/biom12121819




