Overexpressed c-Myc Sensitizes Cells to TH1579, a Mitotic Arrest and Oxidative DNA Damage Inducer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Culture Conditions
2.2. Drug Treatments
2.3. Transfection
2.4. Antibodies
2.5. DNA Fiber Analysis
2.6. EdU Incorporation
2.7. Immunofluorescence Microscopy
2.8. Flow Cytometry
2.9. qRT-PCR
2.10. Western Blotting
2.11. Resazurin Assay
2.12. Clonogenic Survival Assays
2.13. Statistical Analysis
3. Results
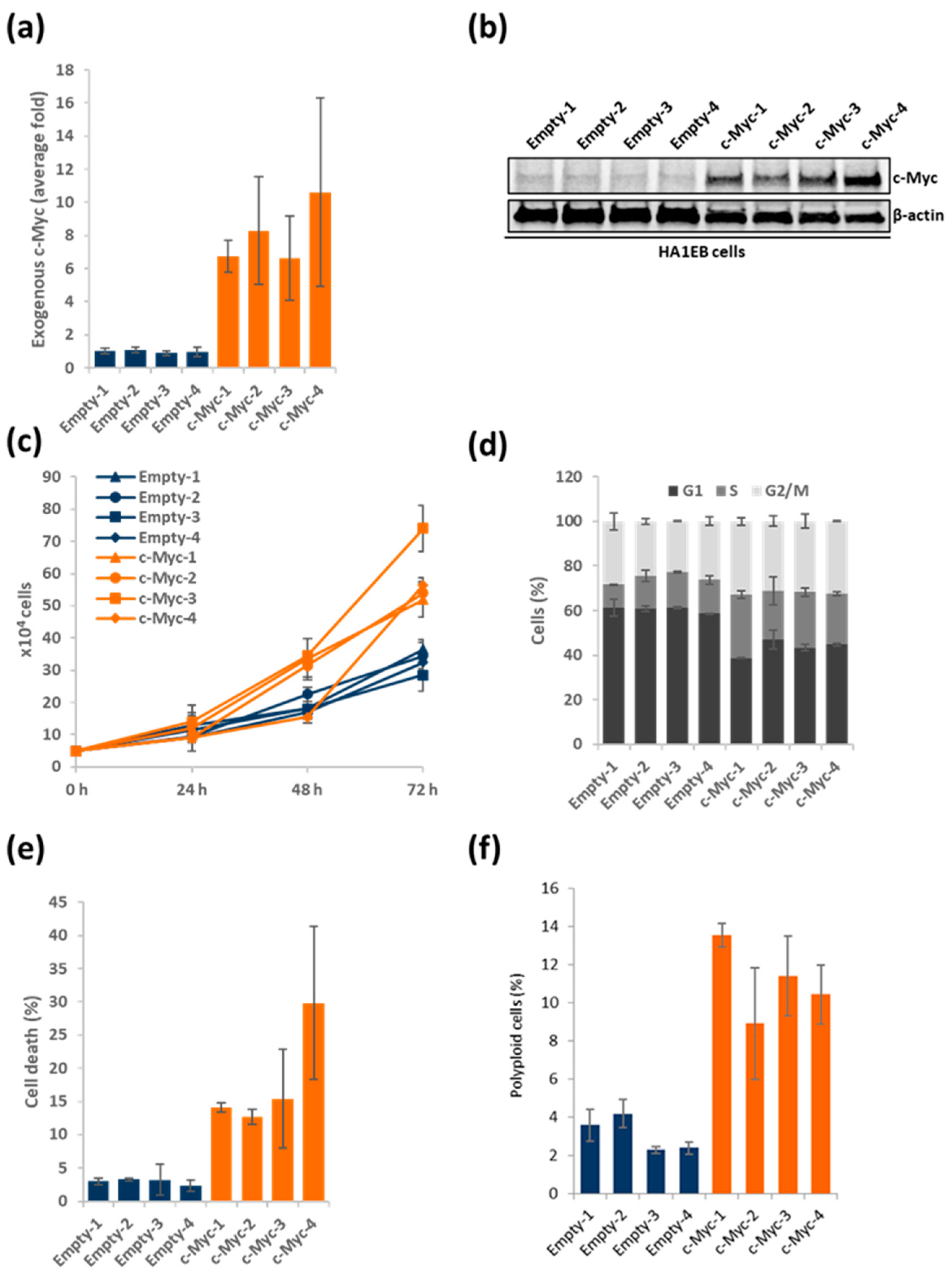
3.1. Generating and Characterizing c-Myc Overexpressing HA1EB Cells
3.2. c-Myc Overexpression Results in EdU and RPA32 Accumulation
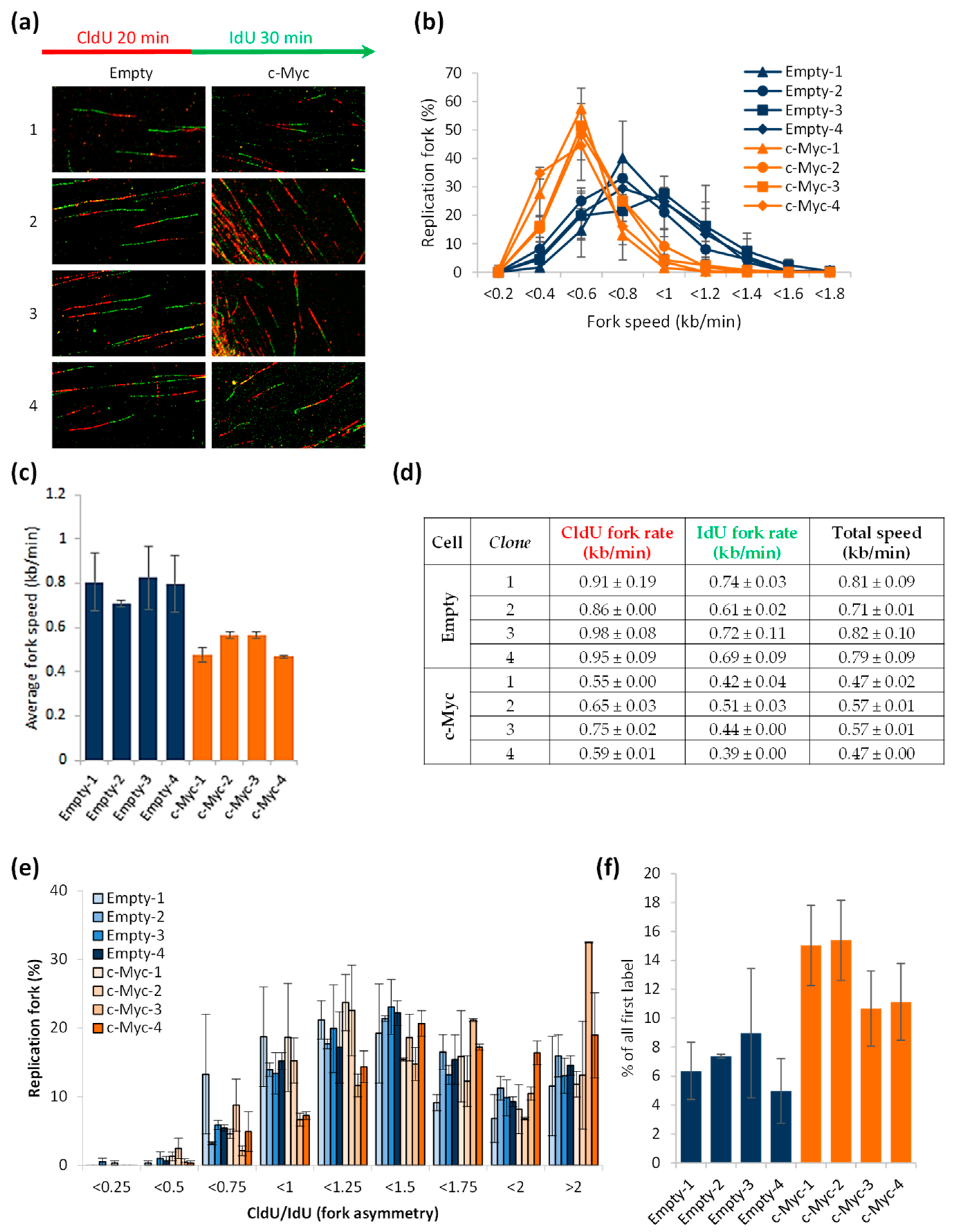
3.3. Excess of c-Myc Induces Replication Stress
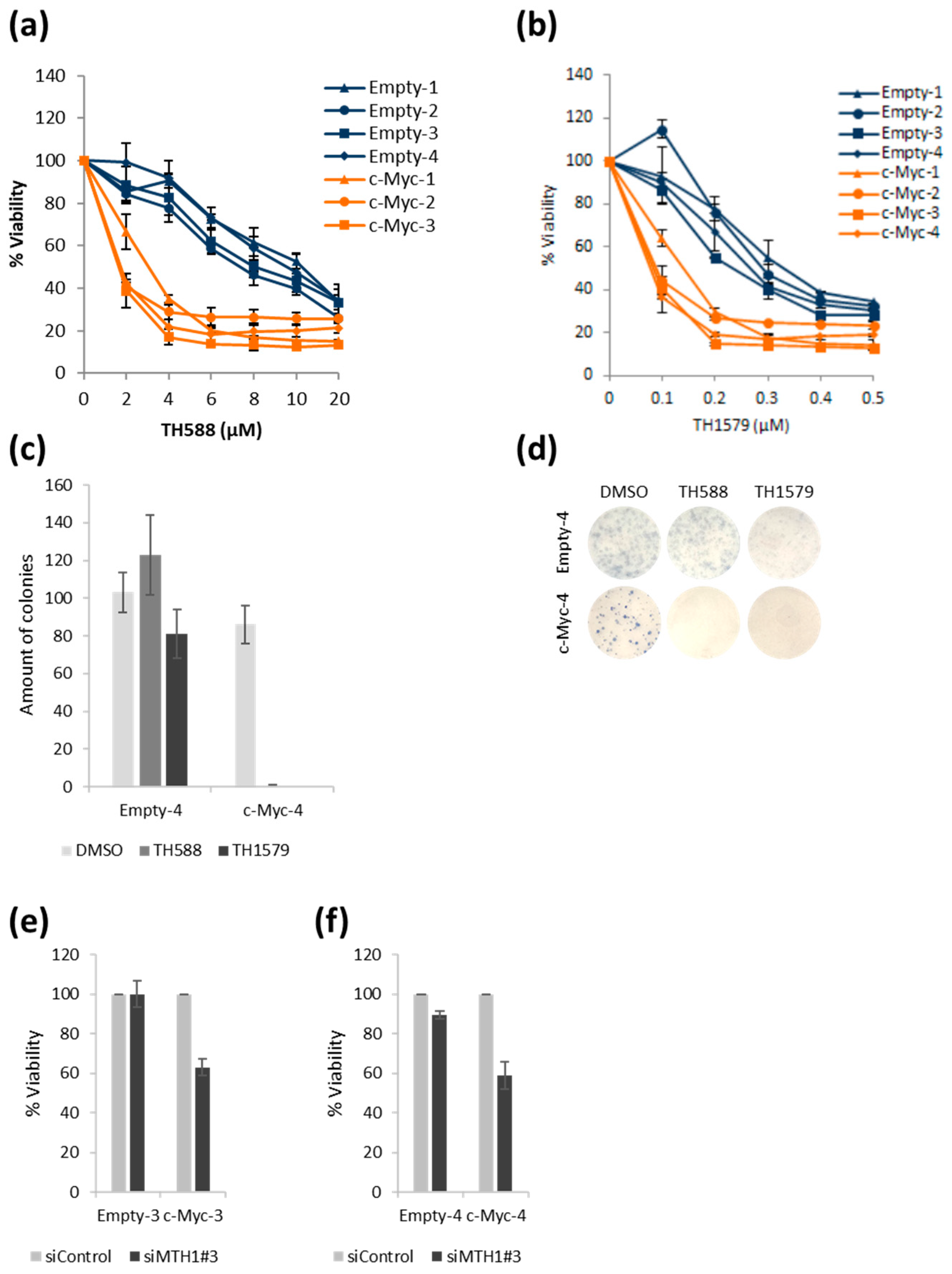
3.4. TH588 or TH1579 Treatment Decreases Viability of c-Myc Overexpressing Cells
3.5. Mitotic MTH1 Inhibitors TH588 and TH1579 Deplete c-Myc Protein and Cause Cell Death
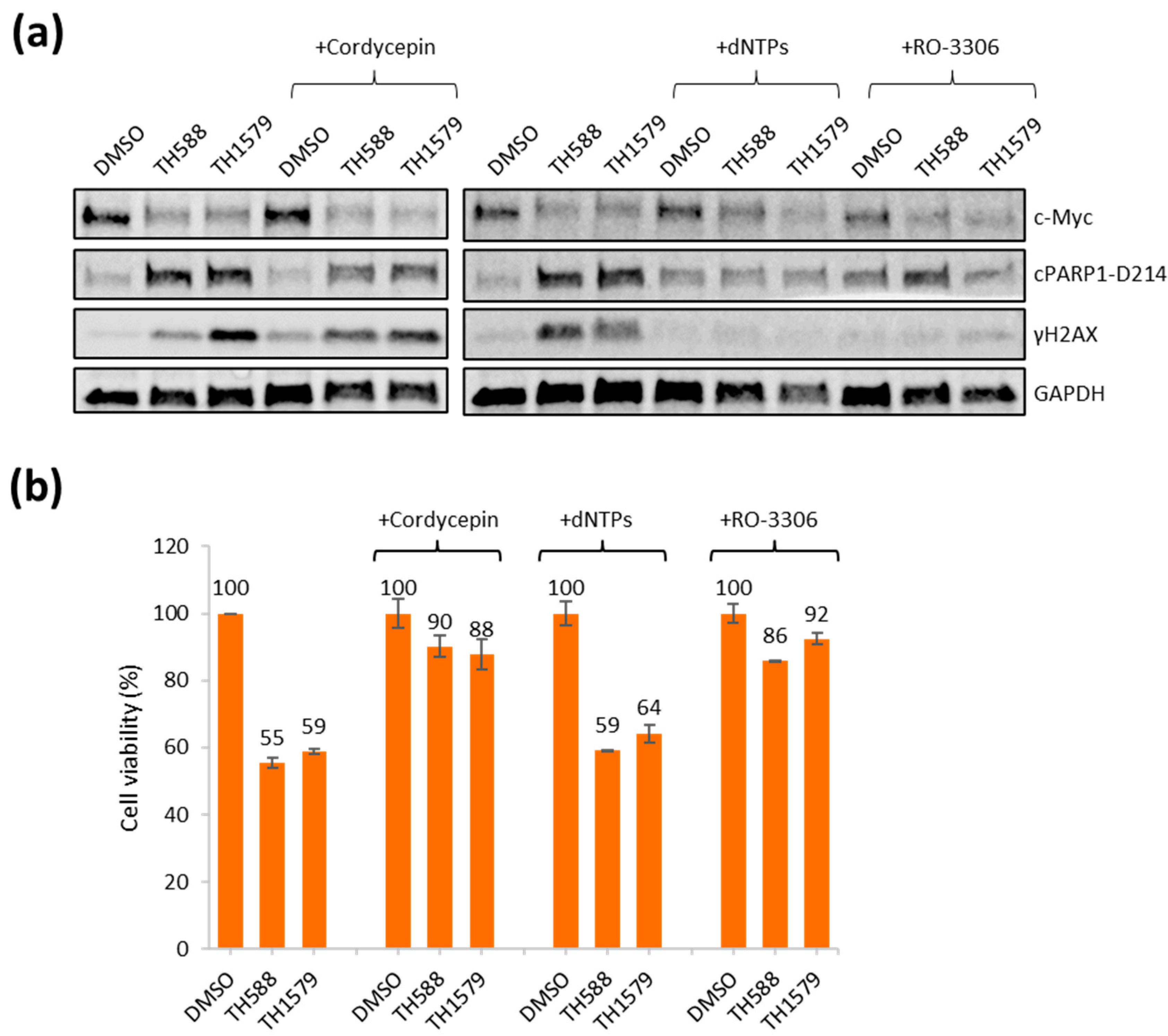
3.6. Transcription and CDK1 Inhibition Reverse Reduced Viability of Mitotic MTH1 Inhibitors TH588 and TH1579 in c-Myc Overexpressing Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gad, H.; Koolmeister, T.; Jemth, A.-S.; Eshtad, S.; Jacques, S.A.; Ström, C.E.; Svensson, L.M.; Schultz, N.; Lundbäck, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Berglund, U.W.; Sanjiv, K.; Gad, H.; Kalderén, C.; Koolmeister, T.; Pham, T.; Gokturk, C.; Jafari, R.; Maddalo, G.; Seashore-Ludlow, B.; et al. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. 2016, 27, 2275–2283. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Kawatani, M.; Muroi, M.; Kondoh, Y.; Futamura, Y.; Aono, H.; Tanaka, M.; Honda, K.; Osada, H. Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci. Rep. 2016, 6, 26521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gad, H.; Mortusewicz, O.; Rudd, S.G.; Stolz, A.; Amaral, N.; Brautigham, L. MTH1 promotes mitotic progression to avoid oxidative DNA damage in cancer cells. bioRxiv 2019, 575290. [Google Scholar] [CrossRef] [Green Version]
- Rudd, S.G.; Gad, H.; Sanjiv, K.; Amaral, N.; Hagenkort, A.; Groth, P.; Ström, C.E.; Mortusewicz, O.; Berglund, U.W.; Helleday, T. MTH1 Inhibitor TH588 Disturbs Mitotic Progression and Induces Mitosis-Dependent Accumulation of Genomic 8-oxodG. Cancer Res. 2020, 80, 3530–3541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuzuki, T.; Egashira, A.; Igarashi, H.; Iwakuma, T.; Nakatsuru, Y.; Tominaga, Y.; Kawate, H.; Nakao, K.; Nakamura, K.; Ide, F.; et al. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. USA 2001, 98, 11456–11461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garré, M.; Giovanni Nuciforo, P.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef]
- Bräutigam, L.; Pudelko, L.; Jemth, A.-S.; Gad, H.; Narwal, M.; Gustafsson, R.; Karsten, S.; Puigvert, J.C.; Homan, E.; Berndt, C.; et al. Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition. Cancer Res. 2016, 76, 2366–2375. [Google Scholar] [CrossRef] [Green Version]
- Doménech, E.; Maestre, C.; Esteban-Martínez, L.; Partida, D.; Pascual, R.; Fernández-Miranda, G.; Seco, E.; Campos-Olivas, R.; Pérez, M.; Megias, D.; et al. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat. Cell Biol. 2015, 17, 1304–1316. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Lobrich, M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.-V.F.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.-C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartkova, J.; Hamerlik, P.; Stockhausen, M.-T.; Ehrmann, J.; Hlobilkova, A.; Laursen, H.; Kalita, O.; Kolar, Z.; Poulsen, H.S.; Broholm, H.; et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene 2010, 29, 5095–5102. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Murga, M.; Campaner, S.; Lopez-Contreras, A.J.; Toledo, L.I.; Soria, R.; Montaña, M.F.; Artista, L.D.; Schleker, T.; Guerra, C.; Garcia, E.O.; et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011, 18, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and Cancer Metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef] [Green Version]
- Maya-Mendoza, A.; Ostrakova, J.; Kosar, M.; Hall, A.; Duskova, P.; Mistrik, M.; Merchut-Maya, J.M.; Hodny, Z.; Bartkova, J.; Christensen, C.; et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 2015, 9, 601–616. [Google Scholar] [CrossRef]
- Bouchard, C.; Thieke, K.; Maier, A.; Saffrich, R.; Hanley-Hyde, J.; Ansorge, W.; Reed, S.; Sicinski, P.; Bartek, J.; Eilers, M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999, 18, 5321–5333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohban, S.; Campaner, S. Myc induced replicative stress response: How to cope with it and exploit it. Biochim. Biophys. Acta 2015, 1849, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Le, H.-V.; Massague, J. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 2002, 419, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Mannava, S.; Grachtchouk, V.; Wheeler, L.J.; Im, M.; Zhuang, D.; Slavina, E.G.; Mathews, C.K.; Shewach, D.S.; Nikiforov, M.A. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 2008, 7, 2392–2400. [Google Scholar] [CrossRef] [PubMed]
- Kotsantis, P.; Petermann, E.; Boulton, S.J. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov. 2018, 8, 537–555. [Google Scholar] [CrossRef] [Green Version]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef] [Green Version]
- Sanjiv, K.; Hagenkort, A.; Calderón-Montaño, J.M.; Koolmeister, T.; Reaper, P.M.; Mortusewicz, O.; Jacques, S.A.; Kuiper, R.V.; Schultz, N.; Scobie, M.; et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016, 14, 298–309. [Google Scholar] [CrossRef]
- Petermann, E.; Helleday, T.; Caldecott, K.W. Claspin Promotes Normal Replication Fork Rates in Human Cells. Mol. Biol. Cell 2008, 19, 2373–2378. [Google Scholar] [CrossRef] [Green Version]
- Pipas, J.M.; Levine, A.J. Role of T antigen interactions with p53 in tumorigenesis. Semin. Cancer Biol. 2001, 11, 23–30. [Google Scholar] [CrossRef]
- Hahn, W.C.; Counter, C.M.; Lundberg, A.S.; Beijersbergen, R.L.; Brooks, M.W.; Weinberg, R.A. Creation of human tumour cells with defined genetic elements. Nature 1999, 400, 464–468. [Google Scholar] [CrossRef]
- Zimonjic, D.; Brooks, M.W.; Popescu, N.; Weinberg, R.A.; Hahn, W.C. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 2001, 61, 8838–8844. [Google Scholar] [PubMed]
- Grignani, F.; Lombardi, L.; Inghirami, G.; Sternas, L.; Cechova, K.; Dalla-Favera, R. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. EMBO J. 1990, 9, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, M.V.; Adams, C.M.; Kushinsky, S.; Eischen, C.M. Smarcal1 and Zranb3 Protect Replication Forks from Myc-Induced DNA Replication Stress. Cancer Res. 2019, 79, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Campaner, S.; Amati, B. Two sides of the Myc-induced DNA damage response: From tumor suppression to tumor maintenance. Cell Div. 2012, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef] [Green Version]
- Kettle, J.G.; Alwan, H.; Bista, M.; Breed, J.; Davies, N.L.; Eckersley, K.; Fillery, S.; Foote, K.M.; Goodwin, L.; Jones, D.R.; et al. Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival. J. Med. Chem. 2016, 59, 2346–2361. [Google Scholar] [CrossRef]
- Frenzel, A.; Zirath, H.; Vita, M.; Albihn, A.; Henriksson, M.A. Identification of Cytotoxic Drugs That Selectively Target Tumor Cells with MYC Overexpression. PLoS ONE 2011, 6, e27988. [Google Scholar] [CrossRef]
- Petermann, E.; Woodcock, M.; Helleday, T. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. USA 2010, 107, 16090–16095. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.M.; Mortusewicz, O.; Afzal, I.; Lorvellec, M.; García, P.; Helleday, T.; Petermann, E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 2012, 32, 3744–3753. [Google Scholar] [CrossRef] [Green Version]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide Deficiency Promotes Genomic Instability in Early Stages of Cancer Development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henriksson, S.; Calderón-Montaño, J.M.; Solvie, D.; Warpman Berglund, U.; Helleday, T. Overexpressed c-Myc Sensitizes Cells to TH1579, a Mitotic Arrest and Oxidative DNA Damage Inducer. Biomolecules 2022, 12, 1777. https://doi.org/10.3390/biom12121777
Henriksson S, Calderón-Montaño JM, Solvie D, Warpman Berglund U, Helleday T. Overexpressed c-Myc Sensitizes Cells to TH1579, a Mitotic Arrest and Oxidative DNA Damage Inducer. Biomolecules. 2022; 12(12):1777. https://doi.org/10.3390/biom12121777
Chicago/Turabian StyleHenriksson, Sofia, José Manuel Calderón-Montaño, Daniel Solvie, Ulrika Warpman Berglund, and Thomas Helleday. 2022. "Overexpressed c-Myc Sensitizes Cells to TH1579, a Mitotic Arrest and Oxidative DNA Damage Inducer" Biomolecules 12, no. 12: 1777. https://doi.org/10.3390/biom12121777
APA StyleHenriksson, S., Calderón-Montaño, J. M., Solvie, D., Warpman Berglund, U., & Helleday, T. (2022). Overexpressed c-Myc Sensitizes Cells to TH1579, a Mitotic Arrest and Oxidative DNA Damage Inducer. Biomolecules, 12(12), 1777. https://doi.org/10.3390/biom12121777





