Design, Synthesis, Kinetic Analysis and Pharmacophore-Directed Discovery of 3-Ethylaniline Hybrid Imino-Thiazolidinone as Potential Inhibitor of Carbonic Anhydrase II: An Emerging Biological Target for Treatment of Cancer
Abstract
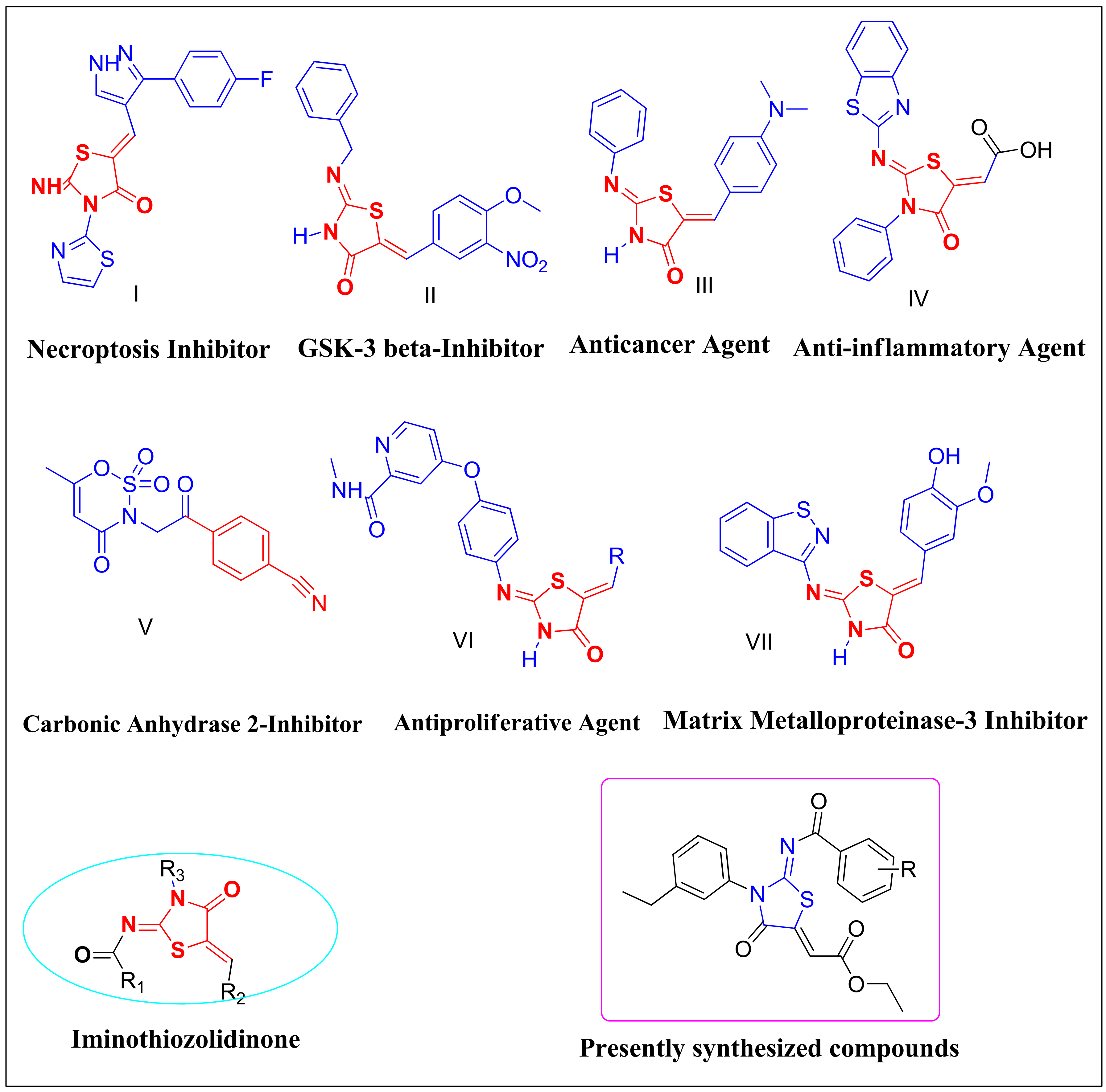
1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization
2.2. Carbonic Anhydrase Activity
2.3. Structure–Activity Relationship
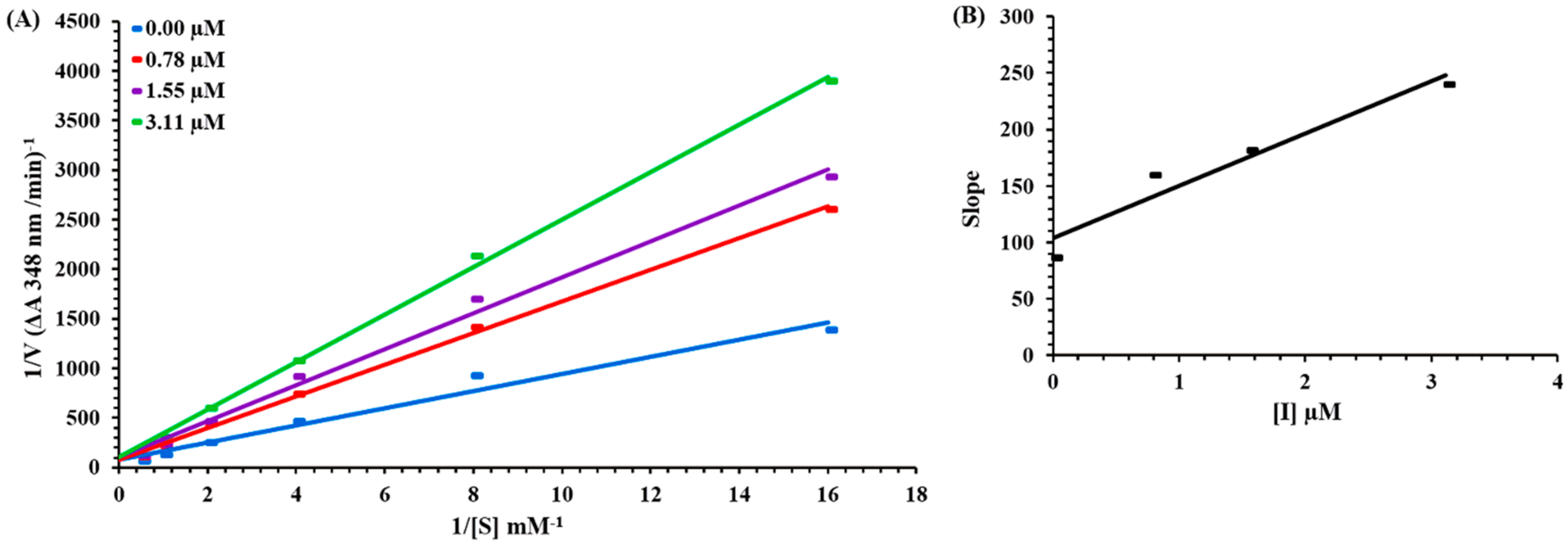
2.4. Kinetic Mechanism of Carbonic Anhydrase Inhibition
2.5. In Silico Studies
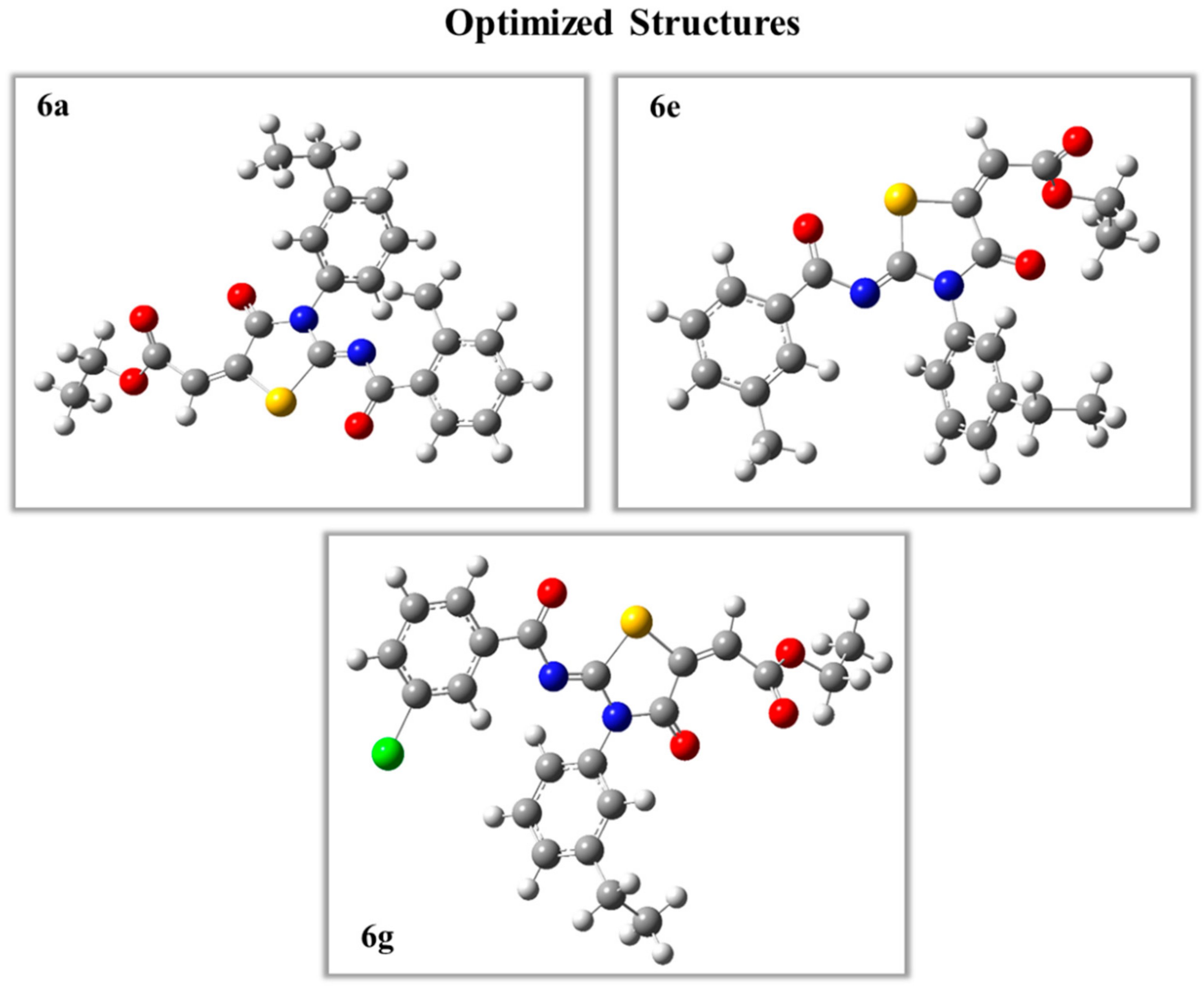
2.5.1. Density Functional Theory Studies
Optimized Structures
Frontier Molecular Orbital Analysis
Local and Global Reactivity Descriptors
2.5.2. E-Pharmacophore-Based Drug Discovery
2.5.3. Molecular Docking Studies
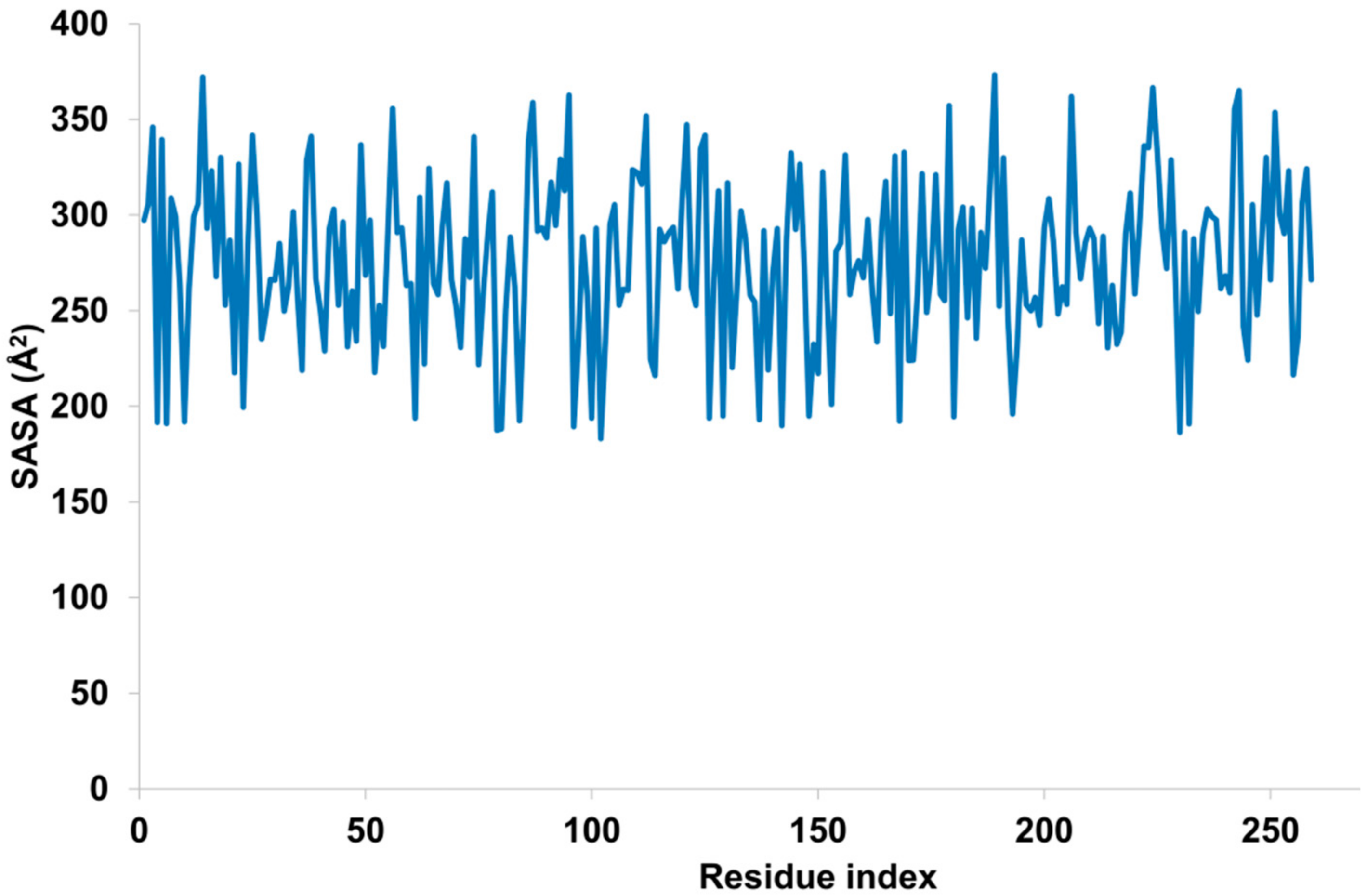
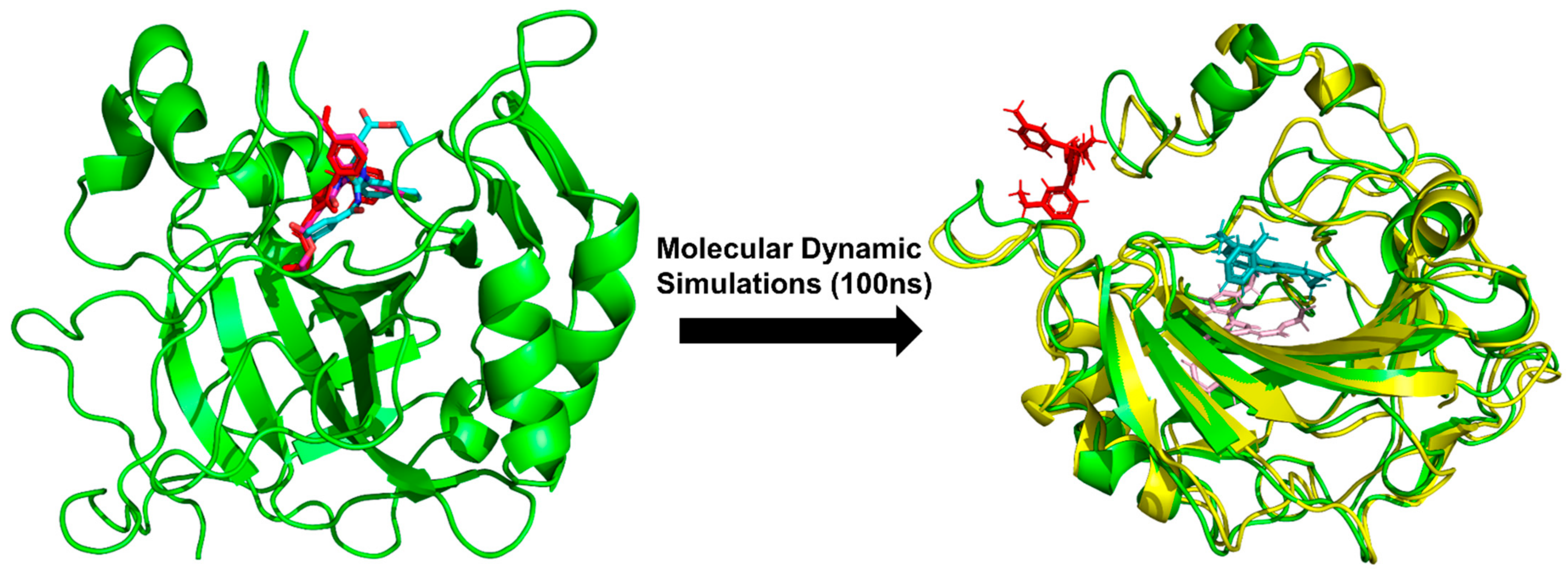
2.5.4. Molecular Dynamic Simulations
2.5.5. MM-GBSA Energy Calculations
2.5.6. In Silico ADMET Predictions
3. Materials and Methodology
3.1. General Information
3.2. Procedure for of Synthesis (6a–j)
3.3. Carbonic Anhydrase Assay
3.4. Kinetic Analysis
3.5. E-Pharmacophore Based Drug Discovery
3.6. Density Functional Theory (DFTs) Studies
3.7. Molecular Docking Studies
3.8. Molecular Dynamic Simulations
3.9. In Silico ADMET Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Viegas-Junior, C.; Danuello, A.; da Silva Bolzani, V.; Barreiro, E.J.; Fraga, C.A.M. Molecular Hybridization: A Useful Tool in the Design of New Drug Prototypes. Curr. Med. Chem 2007, 14, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Časar, Z. Synthesis of Heterocycles in Contemporary Medicinal Chemistry; Springer: Berlin/Heidelberg, Germany, 2016; Volume 44. [Google Scholar]
- Franklin, C.D.; Zavalij, P.Y.; Diamond, C.; Groth, A.J.; Lebron, D.J.; Holmes, S.G.; Anderson, D.R., Jr.; Paul, D.K.; Alvarez, J.C.; Sidorov, V. A Controlled Cyclization of Functionalized Thioureas and Unprecedented Termolecular Decyclization of Iminothiazolidinones. ChemistrySelect 2019, 4, 3567–3576. [Google Scholar] [CrossRef]
- Mahmoodi, N.O.; Mohammadi Zeydi, M.; Biazar, E.; Kazeminejad, Z. Synthesis of novel thiazolidine-4-one derivatives and their anticancer activity. Phosphorus Sulfur Silicon Relat. Elem. 2017, 192, 344–350. [Google Scholar] [CrossRef]
- Dong, G.; Liu, Y.; Wu, Y.; Tu, J.; Chen, S.; Liu, N.; Sheng, C. Novel non-peptidic small molecule inhibitors of secreted aspartic protease 2 (SAP2) for the treatment of resistant fungal infections. Chem. Commun. 2018, 54, 13535–13538. [Google Scholar] [CrossRef] [PubMed]
- El-Sharief, M.A.S.; Abbas, S.Y.; El-Sharief, A.M.S.; Sabry, N.M.; Moussa, Z.; El-Messery, S.M.; Elsheakh, A.R.; Hassan, G.S.; El Sayed, M.T. 5-Thioxoimidazolidine-2-one derivatives: Synthesis, anti-inflammatory activity, analgesic activity, COX inhibition assay and molecular modelling study. Bioorganic Chem. 2019, 87, 679–687. [Google Scholar] [CrossRef]
- Vicini, P.; Geronikaki, A.; Incerti, M.; Zani, F.; Dearden, J.; Hewitt, M. 2-Heteroarylimino-5-benzylidene-4-thiazolidinones analogues of 2-thiazolylimino-5-benzylidene-4-thiazolidinones with antimicrobial activity: Synthesis and structure–activity relationship. Bioorganic Med. Chem. 2008, 16, 3714–3724. [Google Scholar] [CrossRef]
- Saeed, A.; Tehseen, Y.; Rafique, H.; Furtmann, N.; Bajorath, J.; Flörke, U.; Iqbal, J. Benzothiazolyl substituted iminothiazolidinones and benzamido-oxothiazolidines as potent and partly selective aldose reductase inhibitors. MedChemComm 2014, 5, 1371–1380. [Google Scholar] [CrossRef]
- Ali, S.; Saeed, A.; Abbas, N.; Shahid, M.; Bolte, M.; Iqbal, J. Design, synthesis and molecular modelling of novel methyl [4-oxo-2-(aroylimino)-3-(substituted phenyl) thiazolidin-5-ylidene] acetates as potent and selective aldose reductase inhibitors. MedChemComm 2012, 3, 1428–1434. [Google Scholar] [CrossRef]
- Balzarini, J.; Orzeszko-Krzesińska, B.; Maurin, J.K.; Orzeszko, A. Synthesis and anti-HIV studies of 2-and 3-adamantyl-substituted thiazolidin-4-ones. Eur. J. Med. Chem. 2009, 44, 303–311. [Google Scholar] [CrossRef]
- Bell, M.; Foley, D.; Naylor, C.; Wood, G.; Robinson, C.; Riley, J.; Epemolu, O.; Ellis, L.; Scullion, P.; Shishikura, Y.; et al. Discovery of soft-drug topical tool modulators of Sphingosine-1-phosphate Receptor 1 (S1PR1). ACS Med. Chem. Lett. 2019, 10, 341–347. [Google Scholar] [CrossRef]
- Bell, M.; Foley, D.; Naylor, C.; Robinson, C.; Riley, J.; Epemolu, O.; Scullion, P.; Shishikura, Y.; Katz, E.; McLean, W.I.; et al. Discovery of super soft-drug modulators of sphingosine-1-phosphate receptor 1. Bioorganic Med. Chem. Lett. 2018, 28, 3255–3259. [Google Scholar] [CrossRef]
- Zheng, W.; Degterev, A.; Hsu, E.; Yuan, J.; Yuan, C. Structure–activity relationship study of a novel necroptosis inhibitor, necrostatin-7. Bioorganic Med. Chem. Lett. 2008, 18, 4932–4935. [Google Scholar] [CrossRef]
- Arfeen, M.; Bhagat, S.; Patel, R.; Prasad, S.; Roy, I.; Chakraborti, A.K.; Bharatam, P.V. Design, synthesis and biological evaluation of 5-benzylidene-2-iminothiazolidin-4-ones as selective GSK-3β inhibitors. Eur. J. Med. Chem. 2016, 121, 727–736. [Google Scholar] [CrossRef]
- Liu, Y.; Li, F.; Wu, L.; Wang, W.; Zhu, H.; Zhang, Q.; Zhou, H.; Yan, B. Improving both aqueous solubility and anti-cancer activity by assessing progressive lead optimization libraries. Bioorganic Med. Chem. Lett. 2015, 25, 1971–1975. [Google Scholar] [CrossRef]
- Sushilkumar, S.B.; Devanand, B.S. Synthesis and Anti-inflammatory Activity of [2-(Benzothiazol-2-ylimino)-4-oxo-3-phenylthiazolidin-5-yl]-acetic acid derivatives. J. Korean Chem. Soc. 2003, 47, 237–240. [Google Scholar]
- Zhai, X.; Li, W.; Chen, D.; Lai, R.; Liu, J.; Gong, P. Design and Synthesis of 2-Iminothiazolidin-4-one Moiety-Containing Compounds as Potent Antiproliferative Agents. Arch. Pharm. 2012, 345, 360–367. [Google Scholar] [CrossRef]
- Crascì, L.; Vicini, P.; Incerti, M.; Cardile, V.; Avondo, S.; Panico, A. 2-Benzisothiazolylimino-5-benzylidene-4-thiazolidinones as protective agents against cartilage destruction. Bioorganic Med. Chem. 2015, 23, 1551–1556. [Google Scholar] [CrossRef]
- Venta, P.J. Carbonic Anhydrases in Mammalian Cell Culture and Tumors. In The Carbonic Anhydrases; Springer: Berlin/Heidelberg, Germany, 1991; pp. 71–78. [Google Scholar]
- Thiry, A.; Dogne, J.-M.; Masereel, B.; Supuran, C.T. Targeting tumor-associated carbonic anhydrase IX in cancer therapy. Trends Pharmacol. Sci. 2006, 27, 566–573. [Google Scholar] [CrossRef]
- Thiry, A.; Supuran, C.T.; Masereel, B.; Dogné, J.-M. Recent developments of carbonic anhydrase inhibitors as potential anticancer drugs. J. Med. Chem. 2008, 51, 3051–3056. [Google Scholar] [CrossRef]
- Annan, D.A.; Maishi, N.; Soga, T.; Dawood, R.; Li, C.; Kikuchi, H.; Hojo, T.; Morimoto, M.; Kitamura, T.; Alam, M.T.; et al. Carbonic anhydrase 2 (CAII) supports tumor blood endothelial cell survival under lactic acidosis in the tumor microenvironment. Cell Commun. Signal. 2019, 17, 169. [Google Scholar] [CrossRef]
- Carradori, S.; Secci, D.; De Monte, C.; Mollica, A.; Ceruso, M.; Akdemir, A.; Sobolev, A.P.; Codispoti, R.; De Cosmi, F.; Guglielmi, P.; et al. A novel library of saccharin and acesulfame derivatives as potent and selective inhibitors of carbonic anhydrase IX and XII isoforms. Bioorganic Med. Chem. 2016, 24, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Secci, D.; Carradori, S.; Bizzarri, B.; Chimenti, P.; De Monte, C.; Mollica, A.; Rivanera, D.; Zicari, A.; Mari, E.; Zengin, G.; et al. Novel 1, 3-thiazolidin-4-one derivatives as promising anti-Candida agents endowed with anti-oxidant and chelating properties. Eur. J. Med. Chem. 2016, 117, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Saeed, A.; Ejaz, S.A.; Aziz, M.; Hashmi, M.Z.; Channar, P.A.; Abbas, Q.; Raza, H.; Shafiq, Z.; El-Seedi, H.R. Novel adamantyl clubbed iminothiazolidinones as promising elastase inhibitors: Design, synthesis, molecular docking, ADMET and DFT studies. RSC Adv. 2022, 12, 11974–11991. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, B.L.A.; Silva, M.V.A.S.; de Oliveira, E.B.; de Assis Soares, W.R.; Góes-Neto, A.; Santos, G.; Andrade, B.S. Virtual screening and molecular docking for arylalkylamine-N-acetyltransferase (aaNAT) inhibitors, a key enzyme of Aedes (Stegomyia) aegypti (L.) metabolism. Comput. Mol. Biosci. 2015, 5, 35–44. [Google Scholar] [CrossRef][Green Version]
- Yu, H.S.; Li, S.L.; Truhlar, D.G. Perspective: Kohn-Sham density functional theory descending a staircase. J. Chem. Phys. 2016, 145, 130901. [Google Scholar] [CrossRef]
- Thanikaivelan, P.; Subramanian, V.; Rao, J.R.; Nair, B.U. Application of quantum chemical descriptor in quantitative structure activity and structure property relationship. Chem. Phys. Lett. 2000, 323, 59–70. [Google Scholar] [CrossRef]
- Vijya, P.; Patel, D.; Desai, S.; Meshram, D. Development and validation of derivative spectrophotometric method for simultaneous estimation of brimonidine tartrate and brinzolamide in combined dosage form. Indo Am. J. Pharm. Res. 2014, 4, 1478. [Google Scholar]
- Silver, L.H.; The Brinzolamide Primary Therapy Study Group. Clinical efficacy and safety of brinzolamide (Azopt™), a new topical carbonic anhydrase inhibitor for primary open-angle glaucoma and ocular hypertension. Am. J. Ophthalmol. 1998, 126, 400–408. [Google Scholar] [CrossRef]
- MOE (Molecular Operating Environment) Version 2016.01; Chemical Computing Group: Montreal, QC, Canada, 2016.
- Yamaguchi, H.; Kidachi, Y.; Kamiie, K.; Noshita, T.; Umetsu, H. Structural insight into the ligand-receptor interaction between glycyrrhetinic acid (GA) and the high-mobility group protein B1 (HMGB1)-DNA complex. Bioinformation 2012, 8, 1147. [Google Scholar] [CrossRef]
- Aziz, M.; Ejaz, S.A.; Rehman, H.M.; Alsubaie, A.S.; Mahmoud, K.; Siddique, F.; Al-Buriahi, M.; Alrowaili, Z. Identification of NEK7 inhibitors: Structure based virtual screening, molecular docking, density functional theory calculations and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Makeneni, S.; Thieker, D.F.; Woods, R.J. Applying pose clustering and MD simulations to eliminate false positives in molecular docking. J. Chem. Inf. Model. 2018, 58, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Fu, T.; Khan, D.; Abid, A.; Abdalla, A.; Abid, A.; Glass, L.M.; Zitnik, M.; Xiao, C.; Sun, J. Moldesigner: Interactive design of efficacious drugs with deep learning. arXiv 2020, arXiv:2010.03951. [Google Scholar]
- Khan, F.M.; Abbasi, M.A.; Siddiqui, S.Z.; Sadiq Butt, A.R.; Raza, H.; Zafar, A.; Ali Shah, S.A.; Shahid, M.; Seo, S.Y. Convergent synthesis of carbonic anhydrase inhibiting bi-heterocyclic benzamides: Structure–activity relationship and mechanistic explorations through enzyme inhibition, kinetics, and computational studies. J. Heterocycl. Chem. 2021, 58, 1089–1103. [Google Scholar] [CrossRef]
- Ahmed, A.; Channar, P.A.; Saeed, A.; Kalesse, M.; Kazi, M.A.; Larik, F.A.; Abbas, Q.; Hassan, M.; Raza, H.; Seo, S.-Y. Synthesis of sulfonamide, amide and amine hybrid pharmacophore, an entry of new class of carbonic anhydrase II inhibitors and evaluation of chemo-informatics and binding analysis. Bioorganic Chem. 2019, 86, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Appling, D.R. Prism 4 GraphPad Software, Inc. San Diego, CA. See Web Site for Pricing Information. ACS Publications. 2003. Available online: www.graphpad.com (accessed on 8 August 2022).
- Wermuth, C.; Ganellin, C.; Lindberg, P.; Mitscher, L. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998). Pure Appl. Chem. 1998, 70, 1129–1143. [Google Scholar] [CrossRef]
- Seidel, T.; Wolber, G.; Murgueitio, M. Pharmacophore Perception and Applications. In Applied Chemoinformatics; Wiley: Weinheim, Germany, 2018. [Google Scholar]
- Wilson, G.L.; Lill, M.A. Integrating structure-based and ligand-based approaches for computational drug design. Future Med. Chem. 2011, 3, 735–750. [Google Scholar] [CrossRef]
- Beck, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Shahab, S.; Sheikhi, M.; Filippovich, L.; Anatol’evich, D.E.; Yahyaei, H. Quantum chemical modeling of new derivatives of (E, E)-azomethines: Synthesis, spectroscopic (FT-IR, UV/Vis, polarization) and thermophysical investigations. J. Mol. Struct. 2017, 1137, 335–348. [Google Scholar] [CrossRef]
- Frisch, M.J.E.A.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.; et al. Gaussian 09, Revision A; Gaussian Inc.: Wallingford, CT, USA, 2009; Volume 121, pp. 150–166. [Google Scholar]
- Dennington, R.; Keith, T.; Millam, J.; Eppinnett, K.; Hovell, W.; Gilliland, R. GaussView, Version 5.0.9; Visualizer and Builder; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Morris, G.M.; Lim-Wilby, M. Molecular Docking. In Molecular Modeling of Proteins; Springer: Berlin/Heidelberg, Germany, 2008; pp. 365–382. [Google Scholar]
- Prieto-Martínez, F.D.; Arciniega, M.; Medina-Franco, J.L. Molecular docking: Current advances and challenges. TIP Rev. Espec. Cienc. Químic. Biol. 2018, 21, 65–87. [Google Scholar]
- Houston, D.R.; Walkinshaw, M.D. Consensus docking: Improving the reliability of docking in a virtual screening context. J. Chem. Inf. Model. 2013, 53, 384–390. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L., III; Lee, H.S.; Im, W. CHARMM-GUI Ligand Reader and Modeler for CHARMM Force Field Generation of Small Molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Rasheed, S.; Aziz, M.; Saeed, A.; Ejaz, S.A.; Channar, P.A.; Zargar, S.; Abbas, Q.; Alanazi, H.; Hussain, M.; Alharbi, M.; et al. Analysis of 1-Aroyl-3-[3-chloro-2-methylphenyl] Thiourea Hybrids as Potent Urease Inhibitors: Synthesis, Biochemical Evaluation and Computational Approach. Int. J. Mol. Sci. 2022, 23, 11646. [Google Scholar] [CrossRef]
- Barclay, P.L.; Zhang, D.Z. Periodic boundary conditions for arbitrary deformations in molecular dynamics simulations. J. Comput. Phys. 2021, 435, 110238. [Google Scholar] [CrossRef]
- Campos, D.; Ji, H. IMG2SMI: Translating Molecular Structure Images to Simplified Molecular-input Line-entry System. arXiv 2021, arXiv:2109.04202. [Google Scholar]
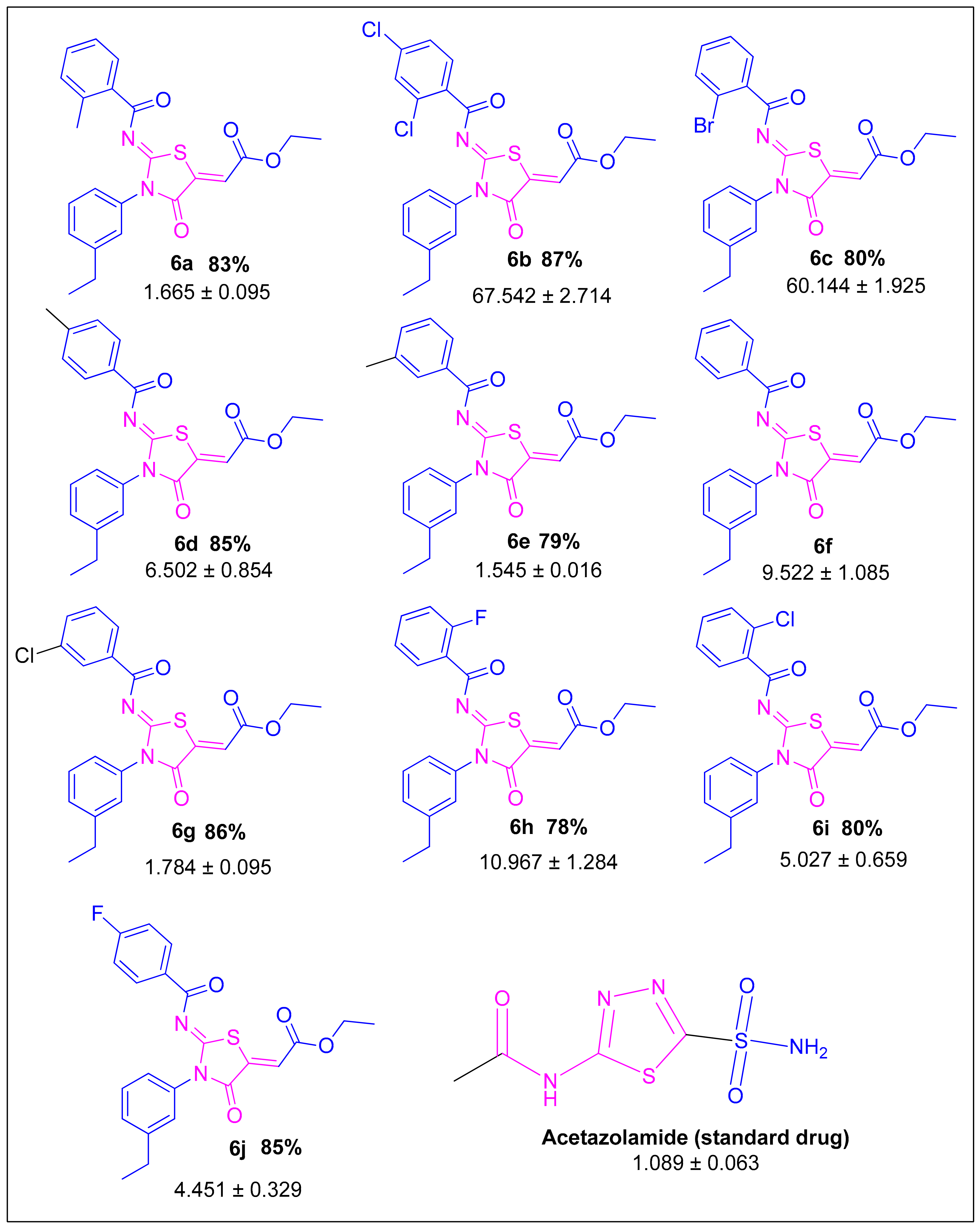


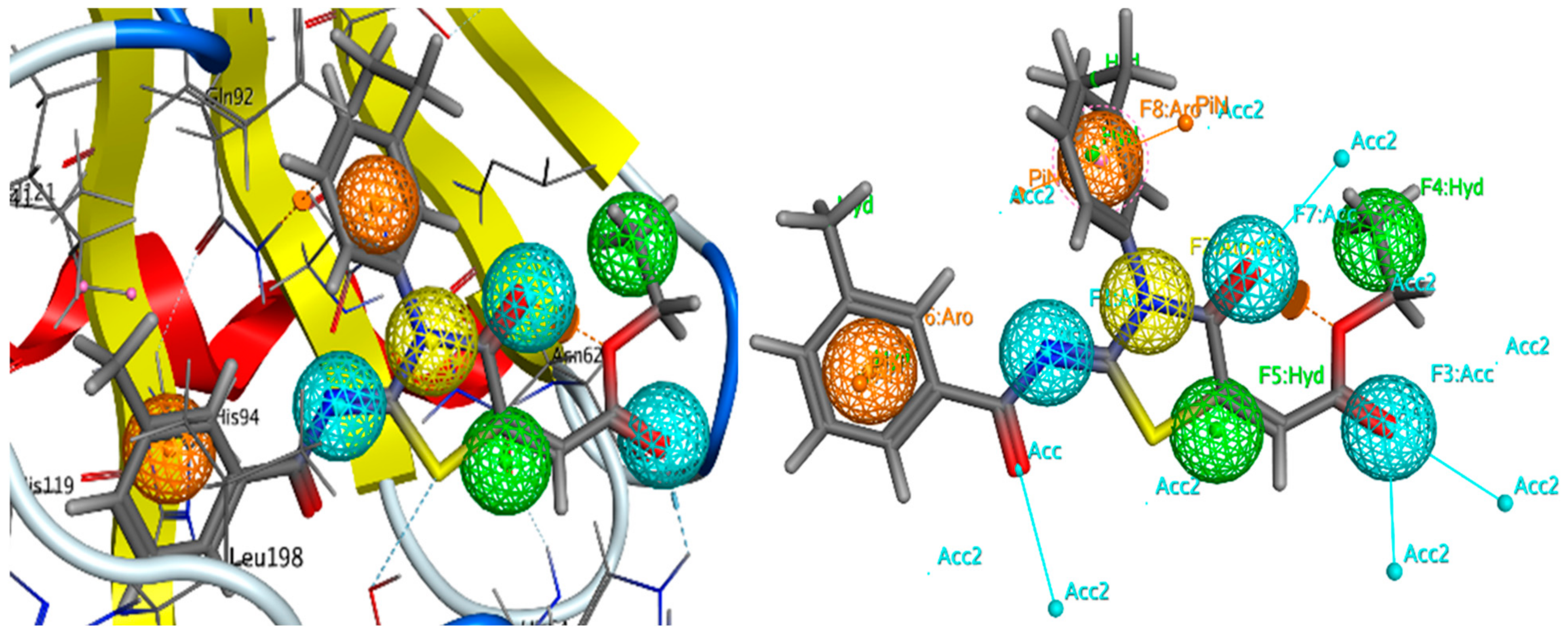
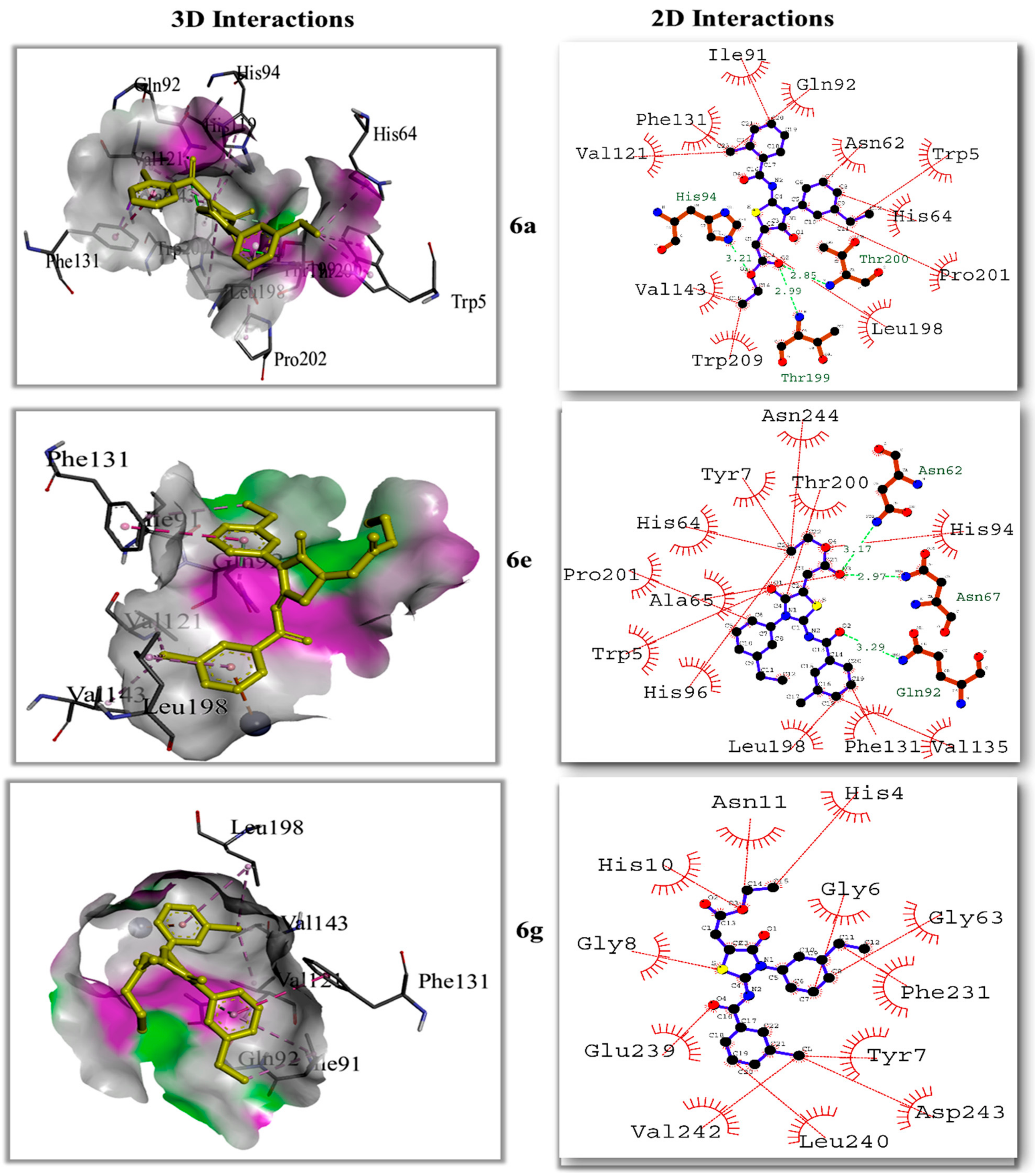
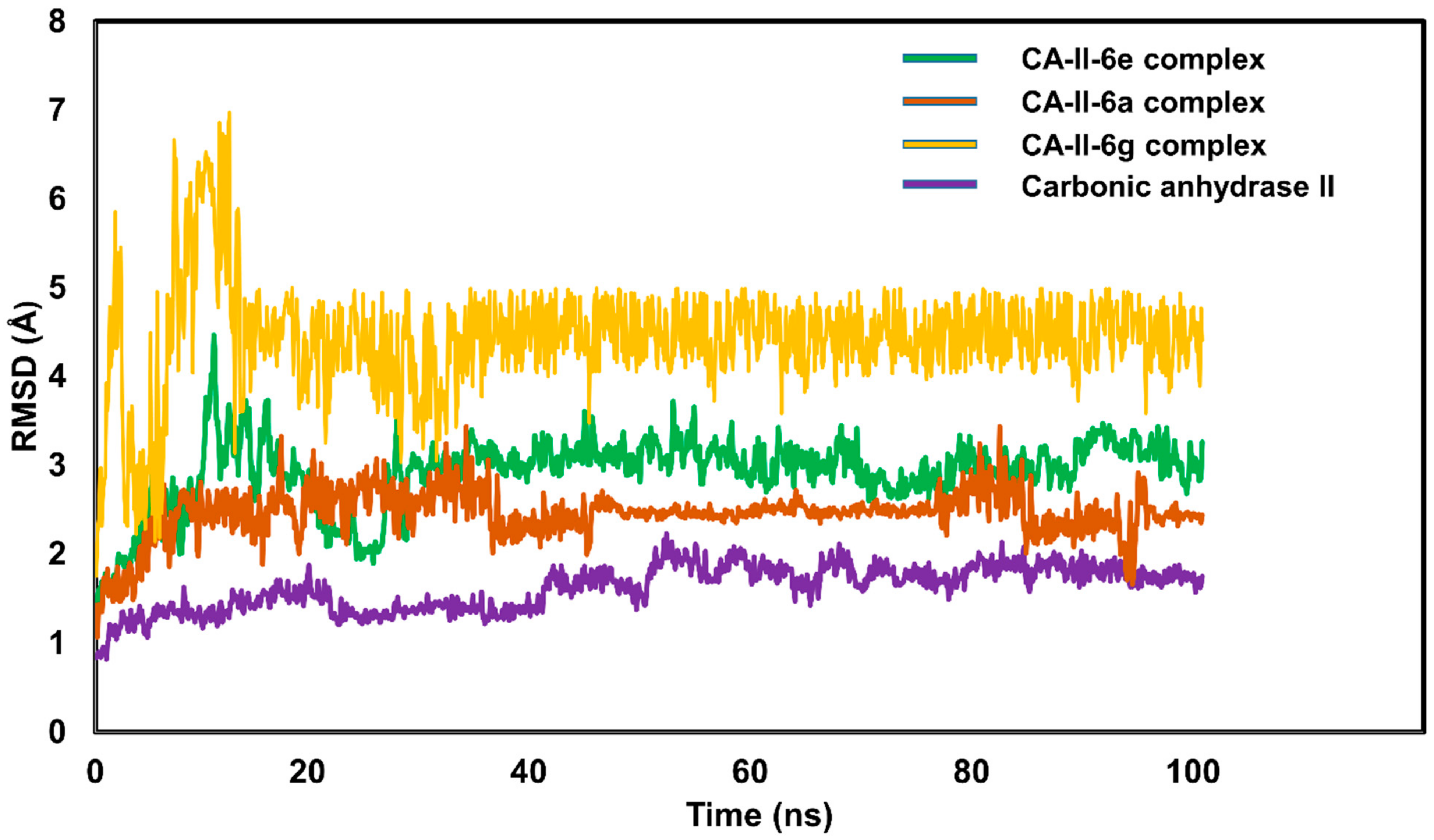
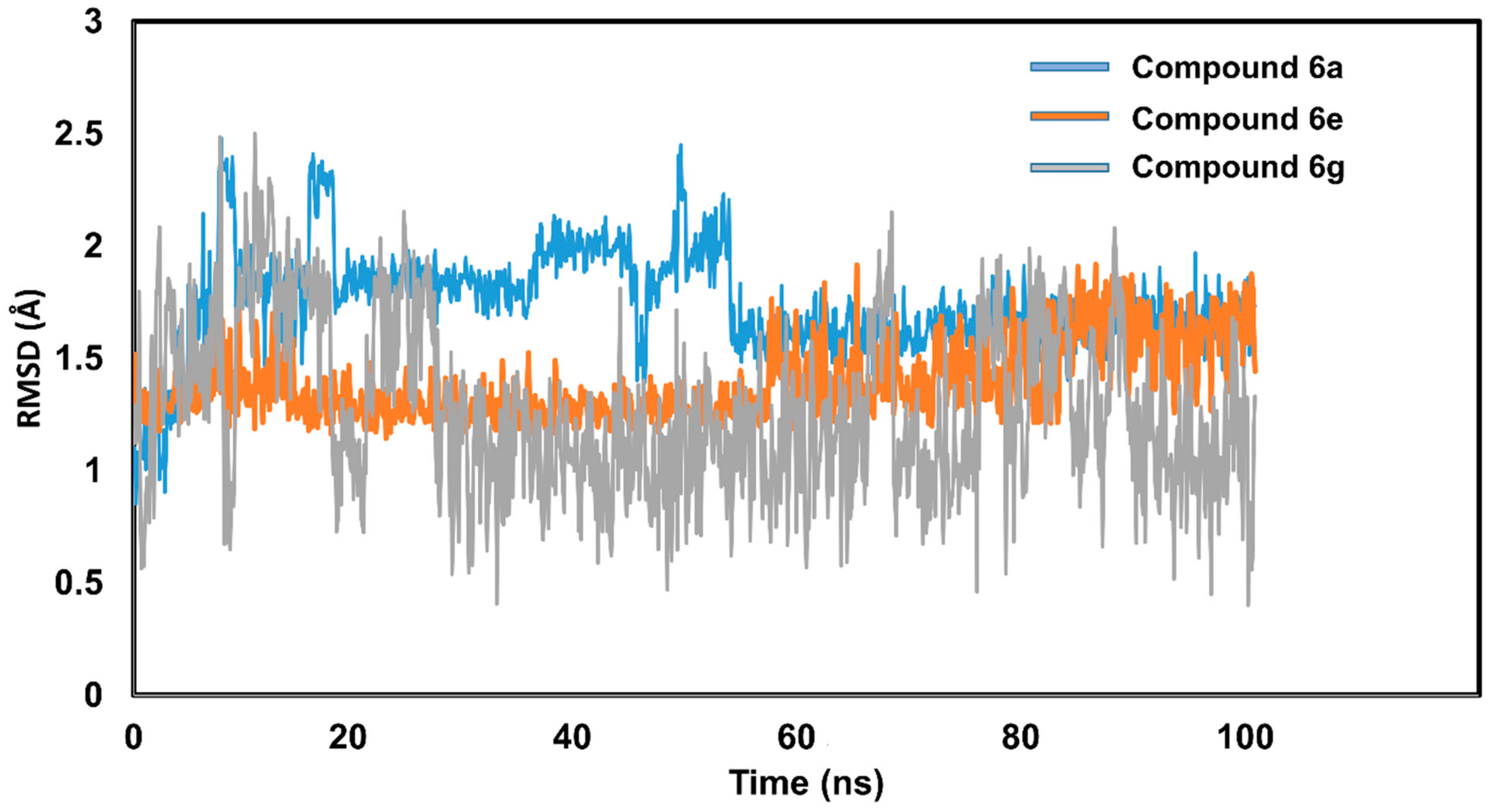


| Compound | Carbonic Anhydrase IC50 ± SEM (µM) | Compound | Carbonic Anhydrase IC50 ± SEM (µM) |
|---|---|---|---|
| 6a | 1.665 ± 0.095 | 6f | 9.522 ± 1.085 |
| 6b | 67.542 ± 2.714 | 6g | 1.784 ± 0.095 |
| 6c | 60.144 ± 1.925 | 6h | 10.967 ± 1.284 |
| 6d | 6.502 ± 0.854 | 6i | 5.027 ± 0.659 |
| 6e | 1.545 ± 0.016 | 6j | 4.451 ± 0.329 |
| Acetazolamide | 1.089 ± 0.063 | ||
| Compound | ∆Egap (eV) | Hardness (η) | Softness (S) | Electronegativity (X) | Electrophilicity Index (Δω±) |
|---|---|---|---|---|---|
| 6a | 0.14278 | 0.071 | 7.004 | 0.111 | 0.087 |
| 6b | 0.13858 | 0.069 | 7.216 | 0.112 | 0.090 |
| 6c | 0.14053 | 0.070 | 7.116 | 0.112 | 0.089 |
| 6d | 0.15224 | 0.076 | 6.569 | 0.122 | 0.098 |
| 6e | 0.14917 | 0.075 | 6.707 | 0.121 | 0.098 |
| 6f | 0.14457 | 0.072 | 6.917 | 0.122 | 0.102 |
| 6g | 0.14241 | 0.071 | 7.022 | 0.121 | 0.103 |
| 6h | 0.14086 | 0.070 | 7.099 | 0.121 | 0.102 |
| 6i | 0.13998 | 0.070 | 7.144 | 0.118 | 0.100 |
| 6j | 0.14344 | 0.072 | 6.972 | 0.117 | 0.095 |
| Sr # | Ionization Energy (eV) | Electron Affinity (eV) | Electron Donating Power (ω−) | Electron Accepting Power (ω+) |
|---|---|---|---|---|
| 6a | 0.2452 | 0.1024 | 0.307 | 0.134 |
| 6b | 0.2488 | 0.1102 | 0.331 | 0.151 |
| 6c | 0.2454 | 0.1048 | 0.315 | 0.139 |
| 6d | 0.2378 | 0.0856 | 0.262 | 0.100 |
| 6e | 0.2478 | 0.0987 | 0.297 | 0.124 |
| 6f | 0.2473 | 0.1028 | 0.309 | 0.134 |
| 6g | 0.2500 | 0.1076 | 0.323 | 0.144 |
| 6h | 0.2431 | 0.1022 | 0.307 | 0.134 |
| 6i | 0.2450 | 0.1051 | 0.315 | 0.140 |
| 6j | 0.2491 | 0.1057 | 0.317 | 0.140 |
| Compounds | Docking Scores (kcal/mol) | Interaction Type | Amino Acid Residues |
|---|---|---|---|
| 6a | −6.12 | Hydrogen bonding | THR200, THR199, HIS94 |
| Hydrophobic interactions | LEU198, VAL143, HIS119, HIS64, TRP209, VAL121, HIS64, TRP5, TRP206, ILE91, GLN92 | ||
| 6e | −6.99 | Hydrogen bonding | GLN92, ASN62, ASN67 |
| Hydrophobic interactions | ASN244, THR200, TYR7, HIS64, PRO201, ALA65, TRP5, HIS96, LEU198, PHE131, VAL135 | ||
| 6g | −6.76 | Hydrogen bonding | - |
| Hydrophobic interactions | GLY6, GLY63, PHE231, TRY7, ASP243, LEU240, VAL242, GLU239, GLY8, HIS10, ASN11, HIS4 |
| Protein–Ligand Complex | Gibbs Free Energy ΔEBind (kcal/mol) | ΔEvdw (kcal/mol) | ΔEH-bond (kcal/mol) | ΔEcoloumb (kcal/mol) |
|---|---|---|---|---|
| CA-II-6e complex | −55.932 | −30.211 | −1.058 | 115.843 |
| CA-II-6a complex | −47.343 | −23.342 | −0.221 | 110.221 |
| CA-II-6g complex | −46.564 | −19.212 | −0.005 | 90.212 |
| Property | Prediction |
|---|---|
| Solubility | −6.69 log mol/L |
| Lipophilicity | 2.82 (log-ratio) |
| (Absorption) Caco-2 | −5.08 cm/s |
| (Absorption) HIA | 92.64% |
| (Absorption) Pgp | 21.46% |
| (Absorption) Bioavailability F20 | 75.78% |
| (Distribution) BBB | 40.55% |
| (Distribution) PPBR | 92.21% |
| (Metabolism) CYP2C19 | 91.68% |
| (Metabolism) CYP2D6 | 7.16% |
| (Metabolism) CYP3A4 | 47.23% |
| (Metabolism) CYP1A2 | 56.16% |
| (Metabolism) CYP2C9 | 62.06% |
| (Execretion) Half life | 8.27 h |
| (Execretion) Clearance | 7.80 mL/min/kg |
| Clinical toxicity | 14.24% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, A.; Aziz, M.; Ejaz, S.A.; Channar, P.A.; Saeed, A.; Zargar, S.; Wani, T.A.; Hamad, A.; Abbas, Q.; Raza, H.; et al. Design, Synthesis, Kinetic Analysis and Pharmacophore-Directed Discovery of 3-Ethylaniline Hybrid Imino-Thiazolidinone as Potential Inhibitor of Carbonic Anhydrase II: An Emerging Biological Target for Treatment of Cancer. Biomolecules 2022, 12, 1696. https://doi.org/10.3390/biom12111696
Ahmed A, Aziz M, Ejaz SA, Channar PA, Saeed A, Zargar S, Wani TA, Hamad A, Abbas Q, Raza H, et al. Design, Synthesis, Kinetic Analysis and Pharmacophore-Directed Discovery of 3-Ethylaniline Hybrid Imino-Thiazolidinone as Potential Inhibitor of Carbonic Anhydrase II: An Emerging Biological Target for Treatment of Cancer. Biomolecules. 2022; 12(11):1696. https://doi.org/10.3390/biom12111696
Chicago/Turabian StyleAhmed, Atteeque, Mubashir Aziz, Syeda Abida Ejaz, Pervaiz Ali Channar, Aamer Saeed, Seema Zargar, Tanveer A. Wani, Asad Hamad, Qamar Abbas, Hussain Raza, and et al. 2022. "Design, Synthesis, Kinetic Analysis and Pharmacophore-Directed Discovery of 3-Ethylaniline Hybrid Imino-Thiazolidinone as Potential Inhibitor of Carbonic Anhydrase II: An Emerging Biological Target for Treatment of Cancer" Biomolecules 12, no. 11: 1696. https://doi.org/10.3390/biom12111696
APA StyleAhmed, A., Aziz, M., Ejaz, S. A., Channar, P. A., Saeed, A., Zargar, S., Wani, T. A., Hamad, A., Abbas, Q., Raza, H., & Kim, S. J. (2022). Design, Synthesis, Kinetic Analysis and Pharmacophore-Directed Discovery of 3-Ethylaniline Hybrid Imino-Thiazolidinone as Potential Inhibitor of Carbonic Anhydrase II: An Emerging Biological Target for Treatment of Cancer. Biomolecules, 12(11), 1696. https://doi.org/10.3390/biom12111696







