Redox Regulation of Signaling Complex between Caveolin-1 and Neuronal Calcium Sensor Recoverin
Abstract
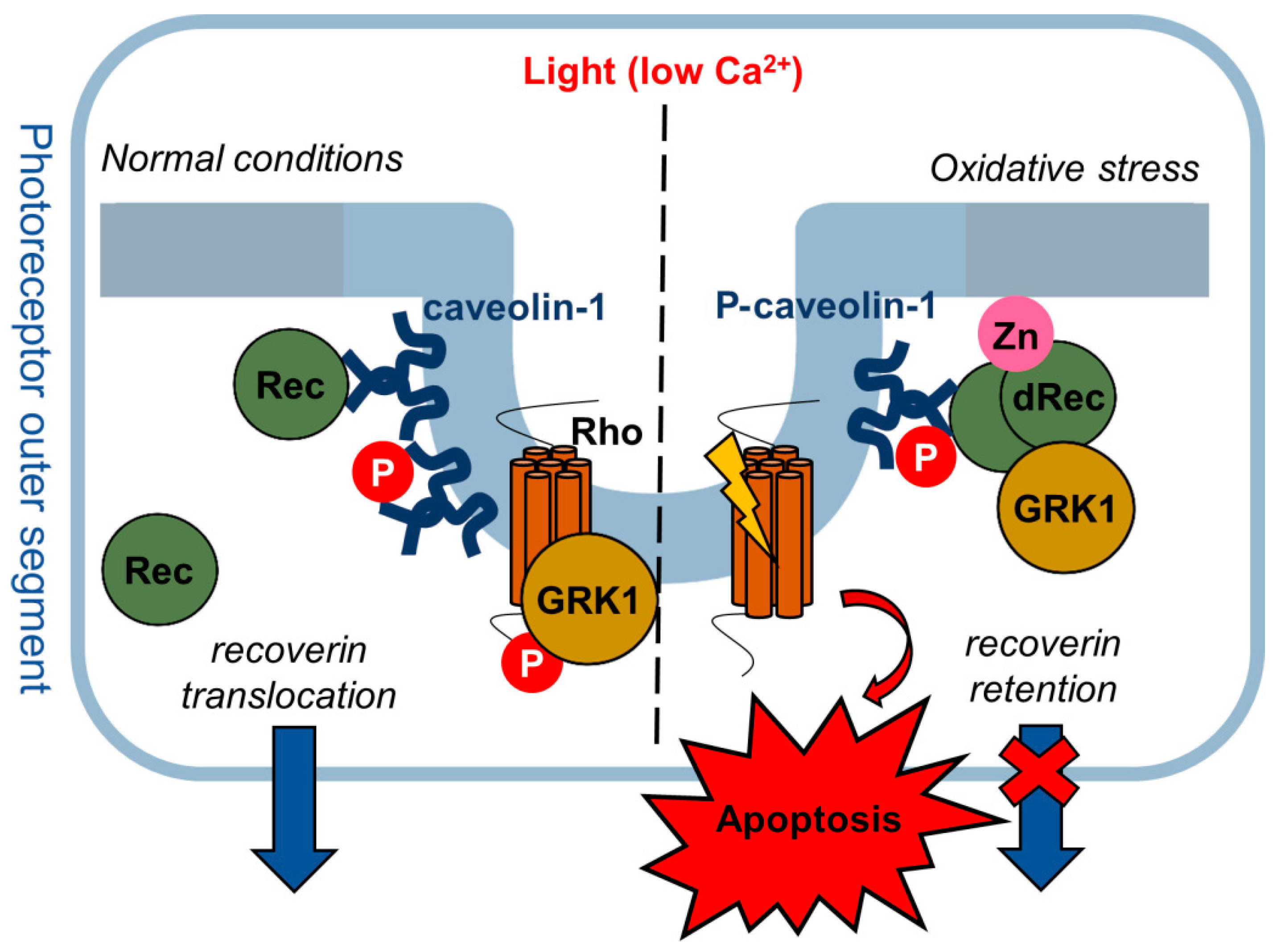
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Caveolin-1 Forms
2.3. Preparation of Recoverin Forms
2.4. Disulfide Dimerization of Recoverin In Vitro and Purification of dRec
2.5. Circular Dichroism Spectroscopy
2.6. Dynamic Light Scattering
2.7. Pull-Down Assay
2.8. Surface Plasmon Resonance Spectroscopy
2.9. Cell Culture
2.10. Western Blotting
2.11. Immunocytochemistry and Microscopy
2.12. Data Analysis
3. Results
3.1. Preparation and Characterization of Oxidative Stress-Related Forms of Recoverin and Caveolin-1
3.2. Effect of Oxidative Stress-Associated Modifications of Recoverin and Caveolin-1 on Their Interaction In Vitro
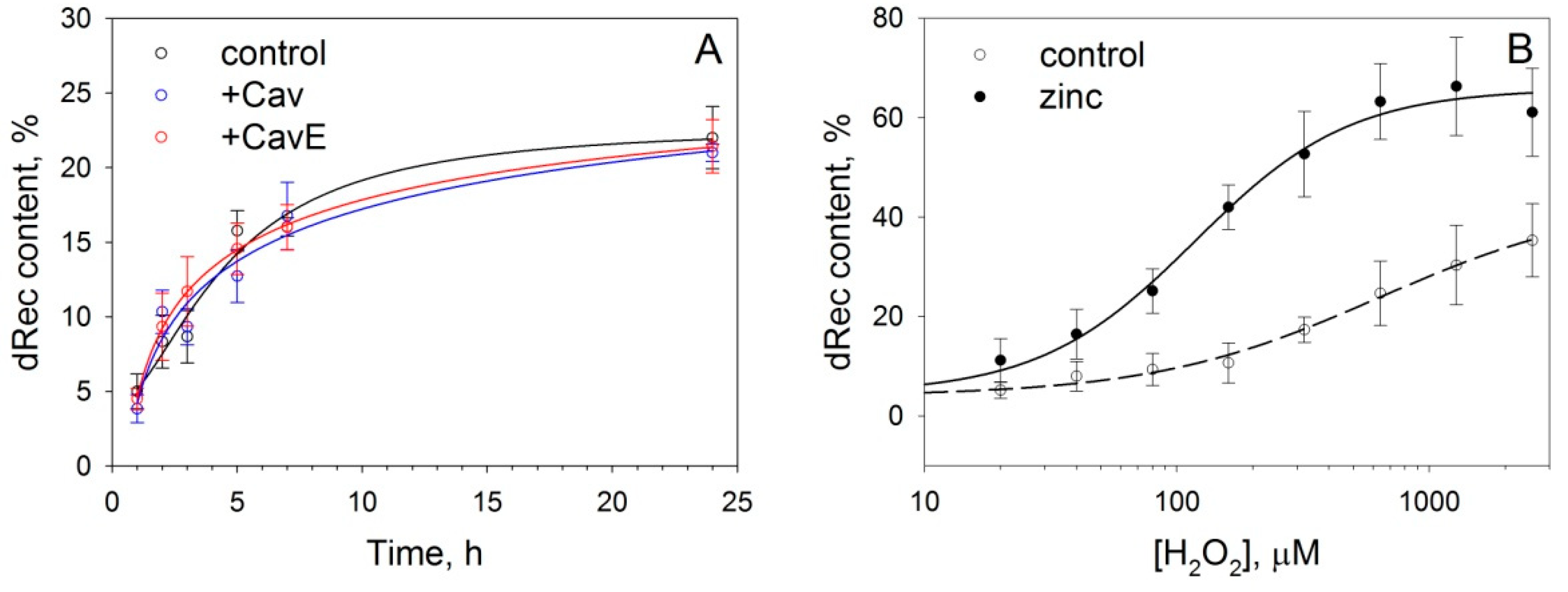
3.3. Effect of Caveolin-1 and Zinc on Disulfide Dimerization of Recoverin In Vitro
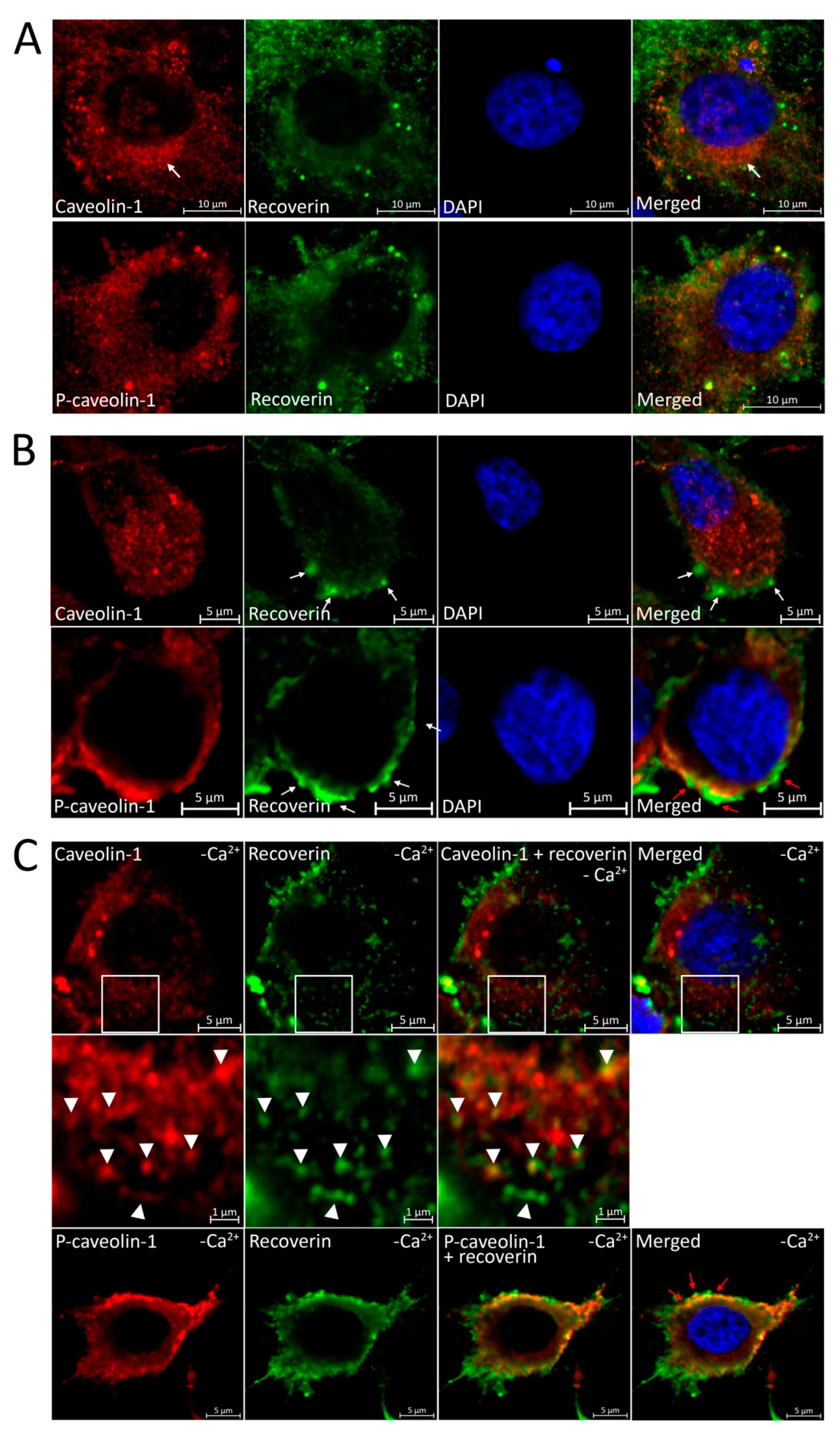
3.4. Disulfide Dimerization of Recoverin and Phosphorylation of Caveolin-1 in Living Cells during Oxidative Stress
3.5. Localization of Recoverin and Phosphorylated/Unphosphorylated Caveolin-1 during Oxidative Stress in Living Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, A.W.; Hnasko, R.; Schubert, W.; Lisanti, M.P. Role of caveolae and caveolins in health and disease. Physiol. Rev. 2004, 84, 1341–1379. [Google Scholar] [CrossRef] [PubMed]
- Boscher, C.; Nabi, I.R. Caveolin-1: Role in cell signaling. Adv. Exp. Med. Biol. 2012, 729, 29–50. [Google Scholar] [CrossRef]
- Del Pozo, M.A.; Lolo, F.N.; Echarri, A. Caveolae: Mechanosensing and mechanotransduction devices linking membrane trafficking to mechanoadaptation. Curr. Opin. Cell Biol. 2021, 68, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Porta, J.C.; Han, B.; Gulsevin, A.; Chung, J.M.; Peskova, Y.; Connolly, S.; McHaourab, H.S.; Meiler, J.; Karakas, E.; Kenworthy, A.K.; et al. Molecular architecture of the human caveolin-1 complex. Sci. Adv. 2022, 8, eabn7232. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Reagan, A.M.; McClellan, M.E.; Elliott, M.H. Caveolins and caveolae in ocular physiology and pathophysiology. Prog. Retin. Eye Res. 2017, 56, 84–106. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, O.; Tillu, V.A.; Ariotti, N.; Parton, R.G.; Collins, B.M. Cavin family proteins and the assembly of caveolae. J. Cell Sci. 2015, 128, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Byrne, D.P.; Dart, C.; Rigden, D.J. Evaluating Caveolin Interactions: Do Proteins Interact with the Caveolin Scaffolding Domain through a Widespread Aromatic Residue-Rich Motif? PLoS ONE 2012, 7, e44879. [Google Scholar] [CrossRef]
- Root, K.T.; Plucinsky, S.M.; Glover, K.J. Recent progress in the topology, structure, and oligomerization of caveolin: A building block of caveolae. Curr. Top. Membr. 2015, 75, 305–336. [Google Scholar] [CrossRef]
- Wong, T.H.; Dickson, F.H.; Timmins, L.R.; Nabi, I.R. Tyrosine phosphorylation of tumor cell caveolin-1: Impact on cancer progression. Cancer Metastasis Rev. 2020, 39, 455–469. [Google Scholar] [CrossRef]
- Hoop, C.L.; Sivanandam, V.N.; Kodali, R.; Srnec, M.N.; van der Wel, P.C. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry 2012, 51, 90–99. [Google Scholar] [CrossRef]
- Plucinsky, S.M.; Glover, K.J. Secondary Structure Analysis of a Functional Construct of Caveolin-1 Reveals a Long C-Terminal Helix. Biophys. J. 2015, 109, 1686–1688. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A.; Arvan, P.; Lisanti, M.P. Caveolin-1 binding to endoplasmic reticulum membranes and entry into the regulated secretory pathway are regulated by serine phosphorylation. Protein sorting at the level of the endoplasmic reticulum. J. Biol. Chem. 2001, 276, 4398–4408. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; McClellan, M.E.; Tanito, M.; Garteiser, P.; Towner, R.; Bissig, D.; Berkowitz, B.A.; Fliesler, S.J.; Woodruff, M.L.; Fain, G.L.; et al. Loss of caveolin-1 impairs retinal function due to disturbance of subretinal microenvironment. J. Biol. Chem. 2012, 287, 16424–16434. [Google Scholar] [CrossRef]
- Vladimirov, V.I.; Zernii, E.Y.; Baksheeva, V.E.; Wimberg, H.; Kazakov, A.S.; Tikhomirova, N.K.; Nemashkalova, E.L.; Mitkevich, V.A.; Zamyatnin, A.A., Jr.; Lipkin, V.M.; et al. Photoreceptor calcium sensor proteins in detergent-resistant membrane rafts are regulated via binding to caveolin-1. Cell Calcium 2018, 73, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, Y.; Ohguro, H.; Odagiri, H.; Maruyama, I.; Maeda, T.; Maeda, A.; Sasaki, M.; Nakazawa, M. Aberrantly expressed recoverin is functionally associated with G-protein-coupled receptor kinases in cancer cell lines. Biochem. Biophys. Res. Commun. 2003, 300, 669–673. [Google Scholar] [CrossRef]
- Zernii, E.Y.; Zinchenko, D.V.; Vladimirov, V.I.; Grigoriev, I.I.; Skorikova, E.E.; Baksheeva, V.E.; Lipkin, V.M.; Philippov, P.P.; Senin, I.I. Ca2+-dependent regulatory activity of recoverin in photoreceptor raft structures: The role of caveolin-1. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 2014, 8, 44–49. [Google Scholar] [CrossRef]
- Philippov, P.P.; Zernii, E.Y. Recoverin. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 4556–4563. [Google Scholar] [CrossRef]
- Zang, J.; Neuhauss, S.C.F. The Binding Properties and Physiological Functions of Recoverin. Front. Mol. Neurosci. 2018, 11, 473. [Google Scholar] [CrossRef]
- Strissel, K.J.; Lishko, P.V.; Trieu, L.H.; Kennedy, M.J.; Hurley, J.B.; Arshavsky, V.Y. Recoverin undergoes light-dependent intracellular translocation in rod photoreceptors. J. Biol. Chem. 2005, 280, 29250–29255. [Google Scholar] [CrossRef]
- Makino, C.L.; Dodd, R.L.; Chen, J.; Burns, M.E.; Roca, A.; Simon, M.I.; Baylor, D.A. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 2004, 123, 729–741. [Google Scholar] [CrossRef]
- Baksheeva, V.E.; Tiulina, V.V.; Tikhomirova, N.K.; Gancharova, O.S.; Komarov, S.V.; Philippov, P.P.; Zamyatnin, A.A., Jr.; Senin, I.I.; Zernii, E.Y. Suppression of Light-Induced Oxidative Stress in the Retina by Mitochondria-Targeted Antioxidant. Antioxidants 2018, 8, 3. [Google Scholar] [CrossRef]
- Organisciak, D.T.; Vaughan, D.K. Retinal light damage: Mechanisms and protection. Prog. Retin. Eye Res. 2010, 29, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Fleckenstein, M.; Keenan, T.D.L.; Guymer, R.H.; Chakravarthy, U.; Schmitz-Valckenberg, S.; Klaver, C.C.; Wong, W.T.; Chew, E.Y. Age-related macular degeneration. Nat. Rev. Dis. Prim. 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Grahn, B.H.; Paterson, P.G.; Gottschall-Pass, K.T.; Zhang, Z. Zinc and the eye. J. Am. Coll. Nutr. 2001, 20, 106–118. [Google Scholar] [CrossRef]
- Ugarte, M.; Osborne, N.N. Recent advances in the understanding of the role of zinc in ocular tissues. Metallomics 2014, 6, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Sheline, C.T.; Zhou, Y.; Bai, S. Light-induced photoreceptor and RPE degeneration involve zinc toxicity and are attenuated by pyruvate, nicotinamide, or cyclic light. Mol. Vis. 2010, 16, 2639–2652. [Google Scholar]
- Organisciak, D.; Wong, P.; Rapp, C.; Darrow, R.; Ziesel, A.; Rangarajan, R.; Lang, J. Light-induced retinal degeneration is prevented by zinc, a component in the age-related eye disease study formulation. Photochem. Photobiol. 2012, 88, 1396–1407. [Google Scholar] [CrossRef]
- Sanguinetti, A.R.; Mastick, C.C. c-Abl is required for oxidative stress-induced phosphorylation of caveolin-1 on tyrosine 14. Cell. Signal. 2003, 15, 289–298. [Google Scholar] [CrossRef]
- Chen, D.B.; Li, S.M.; Qian, X.X.; Moon, C.; Zheng, J. Tyrosine phosphorylation of caveolin 1 by oxidative stress is reversible and dependent on the c-src tyrosine kinase but not mitogen-activated protein kinase pathways in placental artery endothelial cells. Biol. Reprod. 2005, 73, 761–772. [Google Scholar] [CrossRef]
- Wehinger, S.; Ortiz, R.; Diaz, M.I.; Aguirre, A.; Valenzuela, M.; Llanos, P.; Mc Master, C.; Leyton, L.; Quest, A.F. Phosphorylation of caveolin-1 on tyrosine-14 induced by ROS enhances palmitate-induced death of beta-pancreatic cells. Biochim. Biophys. Acta 2015, 1852, 693–708. [Google Scholar] [CrossRef][Green Version]
- Berta, A.I.; Boesze-Battaglia, K.; Magyar, A.; Szel, A.; Kiss, A.L. Localization of caveolin-1 and c-src in mature and differentiating photoreceptors: Raft proteins co-distribute with rhodopsin during development. J. Mol. Histol. 2011, 42, 523–533. [Google Scholar] [CrossRef]
- Elliott, M.H.; Ghalayini, A.J. Phosphorylation of caveolin-1 in bovine rod outer segments in vitro by an endogenous tyrosine kinase. Adv. Exp. Med. Biol. 2008, 613, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Nazipova, A.A.; Denesyuk, A.I.; Bakunts, A.G.; Zinchenko, D.V.; Lipkin, V.M.; Uversky, V.N.; Permyakov, E.A. Recoverin as a redox-sensitive protein. J. Proteome Res. 2007, 6, 1855–1863. [Google Scholar] [CrossRef]
- Zernii, E.Y.; Nazipova, A.A.; Gancharova, O.S.; Kazakov, A.S.; Serebryakova, M.V.; Zinchenko, D.V.; Tikhomirova, N.K.; Senin, I.I.; Philippov, P.P.; Permyakov, E.A.; et al. Light-induced disulfide dimerization of recoverin under ex vivo and in vivo conditions. Free. Radic. Biol. Med. 2015, 83, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Zernii, E.Y.; Nazipova, A.A.; Nemashkalova, E.L.; Kazakov, A.S.; Gancharova, O.S.; Serebryakova, M.V.; Tikhomirova, N.K.; Baksheeva, V.E.; Vladimirov, V.I.; Zinchenko, D.V.; et al. Light-Induced Thiol Oxidation of Recoverin Affects Rhodopsin Desensitization. Front. Mol. Neurosci. 2018, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Ranaghan, M.J.; Kumar, R.P.; Chakrabarti, K.S.; Buosi, V.; Kern, D.; Oprian, D.D. A highly conserved cysteine of neuronal calcium-sensing proteins controls cooperative binding of Ca2+ to recoverin. J. Biol. Chem. 2013, 288, 36160–36167. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Zernii, E.Y.; Knyazeva, E.L.; Denesyuk, A.I.; Nazipova, A.A.; Kolpakova, T.V.; Zinchenko, D.V.; Philippov, P.P.; Permyakov, E.A.; Senin, I.I. Oxidation mimicking substitution of conservative cysteine in recoverin suppresses its membrane association. Amino Acids 2012, 42, 1435–1442. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Cherskaya, A.M.; Wasserman, L.A.; Khokhlova, T.I.; Senin, I.I.; Zargarov, A.A.; Zinchenko, D.V.; Zernii, E.Y.; Lipkin, V.M.; Philippov, P.P.; et al. Recoverin is a zinc-binding protein. J. Proteome Res. 2003, 2, 51–57. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Roman, A.Y.; Baksheeva, V.E.; Nazipova, A.A.; Shevelyova, M.P.; Vladimirov, V.I.; Buyanova, M.F.; Zinchenko, D.V.; Zamyatnin, A.A., Jr.; Devred, F.; et al. Functional Status of Neuronal Calcium Sensor-1 Is Modulated by Zinc Binding. Front. Mol. Neurosci. 2018, 11, 459. [Google Scholar] [CrossRef]
- Baksheeva, V.E.; Tsvetkov, P.O.; Zalevsky, A.O.; Vladimirov, V.I.; Gorokhovets, N.V.; Zinchenko, D.V.; Permyakov, S.E.; Devred, F.; Zernii, E.Y. Zinc Modulation of Neuronal Calcium Sensor Proteins: Three Modes of Interaction with Different Structural Outcomes. Biomolecules 2022, 12, 956. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Vologzhannikova, A.S.; Nemashkalova, E.L.; Kazakov, A.S.; Denesyuk, A.I.; Denessiouk, K.; Baksheeva, V.E.; Zamyatnin, A.A., Jr.; Zernii, E.Y.; Uversky, V.N.; et al. Experimental Insight into the Structural and Functional Roles of the ‘Black’ and ‘Gray’ Clusters in Recoverin, a Calcium Binding Protein with Four EF-Hand Motifs. Molecules 2019, 24, 2494. [Google Scholar] [CrossRef]
- Vladimirov, V.I.; Baksheeva, V.E.; Mikhailova, I.V.; Ismailov, R.G.; Litus, E.A.; Tikhomirova, N.K.; Nazipova, A.A.; Permyakov, S.E.; Zernii, E.Y.; Zinchenko, D.V. A Novel Approach to Bacterial Expression and Purification of Myristoylated Forms of Neuronal Calcium Sensor Proteins. Biomolecules 2020, 10, 1025. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000, 287, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Kavran, J.M.; Leahy, D.J. Coupling antibody to cyanogen bromide-activated sepharose. Methods Enzymol. 2014, 541, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Baksheeva, V.E.; Baldin, A.V.; Zalevsky, A.O.; Nazipova, A.A.; Kazakov, A.S.; Vladimirov, V.I.; Gorokhovets, N.V.; Devred, F.; Philippov, P.P.; Bazhin, A.V.; et al. Disulfide Dimerization of Neuronal Calcium Sensor-1: Implications for Zinc and Redox Signaling. Int. J. Mol. Sci. 2021, 22, 12602. [Google Scholar] [CrossRef]
- Senin, I.I.; Tikhomirova, N.K.; Churumova, V.A.; Grigoriev, I.I.; Kolpakova, T.A.; Zinchenko, D.V.; Philippov, P.P.; Zernii, E.Y. Amino acid sequences of two immune-dominant epitopes of recoverin are involved in Ca2+/recoverin-dependent inhibition of phosphorylation of rhodopsin. Biochemistry 2011, 76, 332–338. [Google Scholar] [CrossRef]
- Ortiz, R.; Diaz, J.; Diaz, N.; Lobos-Gonzalez, L.; Cardenas, A.; Contreras, P.; Diaz, M.I.; Otte, E.; Cooper-White, J.; Torres, V.; et al. Extracellular matrix-specific Caveolin-1 phosphorylation on tyrosine 14 is linked to augmented melanoma metastasis but not tumorigenesis. Oncotarget 2016, 7, 40571–40593. [Google Scholar] [CrossRef]
- Fernandez, I.; Ying, Y.; Albanesi, J.; Anderson, R.G. Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. USA 2002, 99, 11193–11198. [Google Scholar] [CrossRef]
- Zimnicka, A.M.; Husain, Y.S.; Shajahan, A.N.; Sverdlov, M.; Chaga, O.; Chen, Z.; Toth, P.T.; Klomp, J.; Karginov, A.V.; Tiruppathi, C.; et al. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol. Biol. Cell 2016, 27, 2090–2106. [Google Scholar] [CrossRef]
- Manzerra, P.; Behrens, M.M.; Canzoniero, L.M.; Wang, X.Q.; Heidinger, V.; Ichinose, T.; Yu, S.P.; Choi, D.W. Zinc induces a Src family kinase-mediated up-regulation of NMDA receptor activity and excitotoxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 11055–11061. [Google Scholar] [CrossRef]
- Vogel, U.; Sandvig, K.; van Deurs, B. Expression of caveolin-1 and polarized formation of invaginated caveolae in Caco-2 and MDCK II cells. J. Cell Sci. 1998, 111 Pt 6, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Vepa, S.; Scribner, W.M.; Natarajan, V. Activation of protein phosphorylation by oxidants in vascular endothelial cells: Identification of tyrosine phosphorylation of caveolin. Free Radic. Biol. Med. 1997, 22, 25–35. [Google Scholar] [CrossRef]
- Castro, J.; Bittner, C.X.; Humeres, A.; Montecinos, V.P.; Vera, J.C.; Barros, L.F. A cytosolic source of calcium unveiled by hydrogen peroxide with relevance for epithelial cell death. Cell Death Differ 2004, 11, 468–478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Donovan, M.; Carmody, R.J.; Cotter, T.G. Light-induced photoreceptor apoptosis in vivo requires neuronal nitric-oxide synthase and guanylate cyclase activity and is caspase-3-independent. J. Biol. Chem. 2001, 276, 23000–23008. [Google Scholar] [CrossRef]
- Maret, W.; Vallee, B.L. Thiolate ligands in metallothionein confer redox activity on zinc clusters. Proc. Natl. Acad. Sci. USA 1998, 95, 3478–3482. [Google Scholar] [CrossRef]
- Maret, W.; Krezel, A. Cellular zinc and redox buffering capacity of metallothionein/thionein in health and disease. Mol. Med. 2007, 13, 371–375. [Google Scholar] [CrossRef]
- Simon, L.; Campos, A.; Leyton, L.; Quest, A.F.G. Caveolin-1 function at the plasma membrane and in intracellular compartments in cancer. Cancer Metastasis Rev. 2020, 39, 435–453. [Google Scholar] [CrossRef]
- Li, W.P.; Liu, P.; Pilcher, B.K.; Anderson, R.G. Cell-specific targeting of caveolin-1 to caveolae, secretory vesicles, cytoplasm or mitochondria. J. Cell Sci. 2001, 114, 1397–1408. [Google Scholar] [CrossRef]
- Couet, J.; Sargiacomo, M.; Lisanti, M.P. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J. Biol. Chem. 1997, 272, 30429–30438. [Google Scholar] [CrossRef]
- Yamamoto, M.; Toya, Y.; Schwencke, C.; Lisanti, M.P.; Myers, M.G., Jr.; Ishikawa, Y. Caveolin is an activator of insulin receptor signaling. J. Biol. Chem. 1998, 273, 26962–26968. [Google Scholar] [CrossRef]
- Rathor, N.; Zhuang, R.; Wang, J.Y.; Donahue, J.M.; Turner, D.J.; Rao, J.N. Src-mediated caveolin-1 phosphorylation regulates intestinal epithelial restitution by altering Ca(2+) influx after wounding. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G650–G658. [Google Scholar] [CrossRef]
- Ugarte, M.; Osborne, N.N. The localization of free zinc varies in rat photoreceptors during light and dark adaptation. Exp. Eye Res. 1999, 69, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Oh, P.; Schnitzer, J.E. Segregation of heterotrimeric G proteins in cell surface microdomains. G(q) binds caveolin to concentrate in caveolae, whereas G(i) and G(s) target lipid rafts by default. Mol. Biol. Cell 2001, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Hayer, A.; Stoeber, M.; Ritz, D.; Engel, S.; Meyer, H.H.; Helenius, A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell Biol. 2010, 191, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Beatty, S.; Koh, H.; Phil, M.; Henson, D.; Boulton, M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv. Ophthalmol. 2000, 45, 115–134. [Google Scholar] [CrossRef]
- Blasiak, J.; Pawlowska, E.; Chojnacki, J.; Szczepanska, J.; Chojnacki, C.; Kaarniranta, K. Zinc and Autophagy in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2020, 21, 4994. [Google Scholar] [CrossRef]
- Wood, J.P.; Osborne, N.N. Zinc and energy requirements in induction of oxidative stress to retinal pigmented epithelial cells. Neurochem. Res. 2003, 28, 1525–1533. [Google Scholar] [CrossRef]
- Julien, S.; Biesemeier, A.; Kokkinou, D.; Eibl, O.; Schraermeyer, U. Zinc deficiency leads to lipofuscin accumulation in the retinal pigment epithelium of pigmented rats. PLoS ONE 2011, 6, e29245. [Google Scholar] [CrossRef]
- Sethna, S.; Chamakkala, T.; Gu, X.; Thompson, T.C.; Cao, G.; Elliott, M.H.; Finnemann, S.C. Regulation of Phagolysosomal Digestion by Caveolin-1 of the Retinal Pigment Epithelium Is Essential for Vision. J. Biol. Chem. 2016, 291, 6494–6506. [Google Scholar] [CrossRef]
- Jiang, Y.; Lin, X.; Tang, Z.; Lee, C.; Tian, G.; Du, Y.; Yin, X.; Ren, X.; Huang, L.; Ye, Z.; et al. Critical role of caveolin-1 in ocular neovascularization and multitargeted antiangiogenic effects of cavtratin via JNK. Proc. Natl. Acad. Sci. USA 2017, 114, 10737–10742. [Google Scholar] [CrossRef]
- Yuan, X.; Gu, X.; Crabb, J.S.; Yue, X.; Shadrach, K.; Hollyfield, J.G.; Crabb, J.W. Quantitative proteomics: Comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol. Cell. Proteom. MCP 2010, 9, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Senabouth, A.; Daniszewski, M.; Lidgerwood, G.E.; Liang, H.H.; Hernandez, D.; Mirzaei, M.; Keenan, S.N.; Zhang, R.; Han, X.; Neavin, D.; et al. Transcriptomic and proteomic retinal pigment epithelium signatures of age-related macular degeneration. Nat. Commun. 2022, 13, 4233. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Yamada, K.; Suzumura, A.; Kataoka, K.; Takayama, K.; Sugimoto, M.; Terasaki, H.; Kaneko, H. Caveolin-1 Promotes Cellular Senescence in Exchange for Blocking Subretinal Fibrosis in Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2020, 61, 21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shin, J.; Park, H. Structural characterization for N-terminal domain of caveolin-1. Korean J. Biol. Sci. 2003, 7, 207–211. [Google Scholar] [CrossRef]
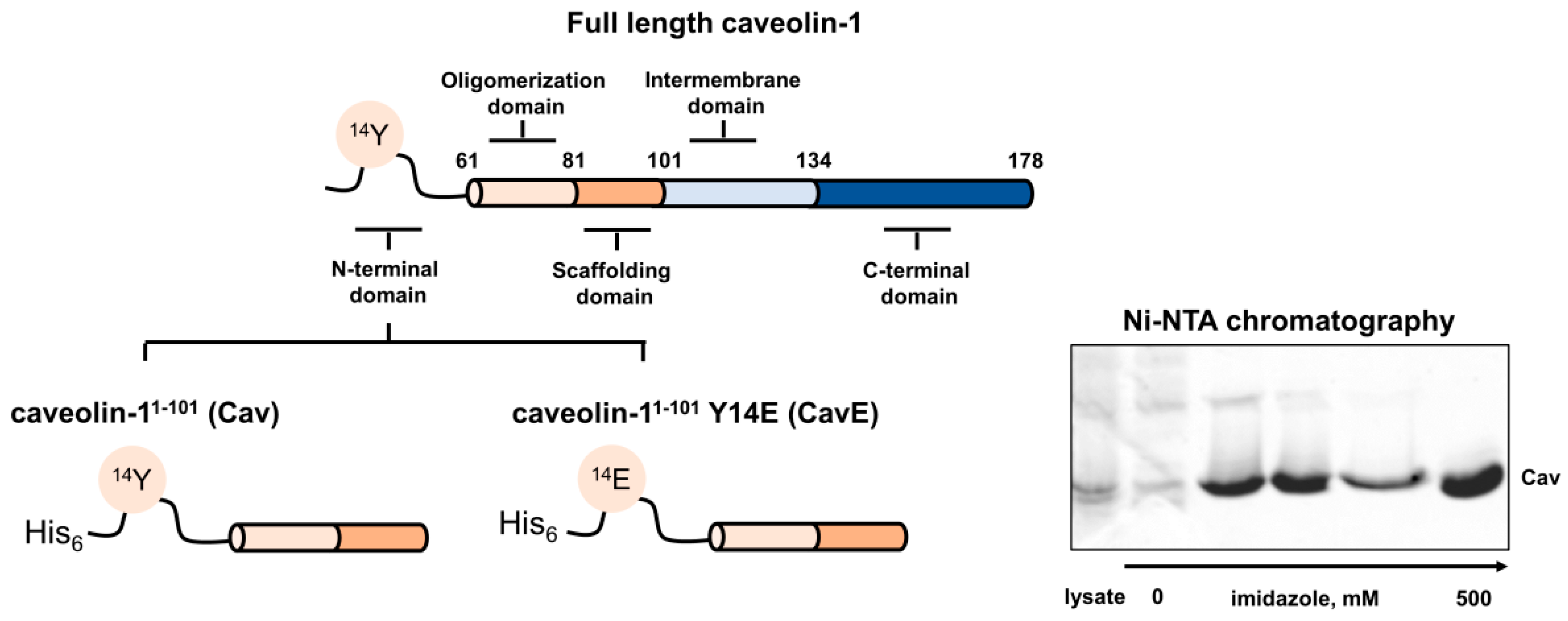
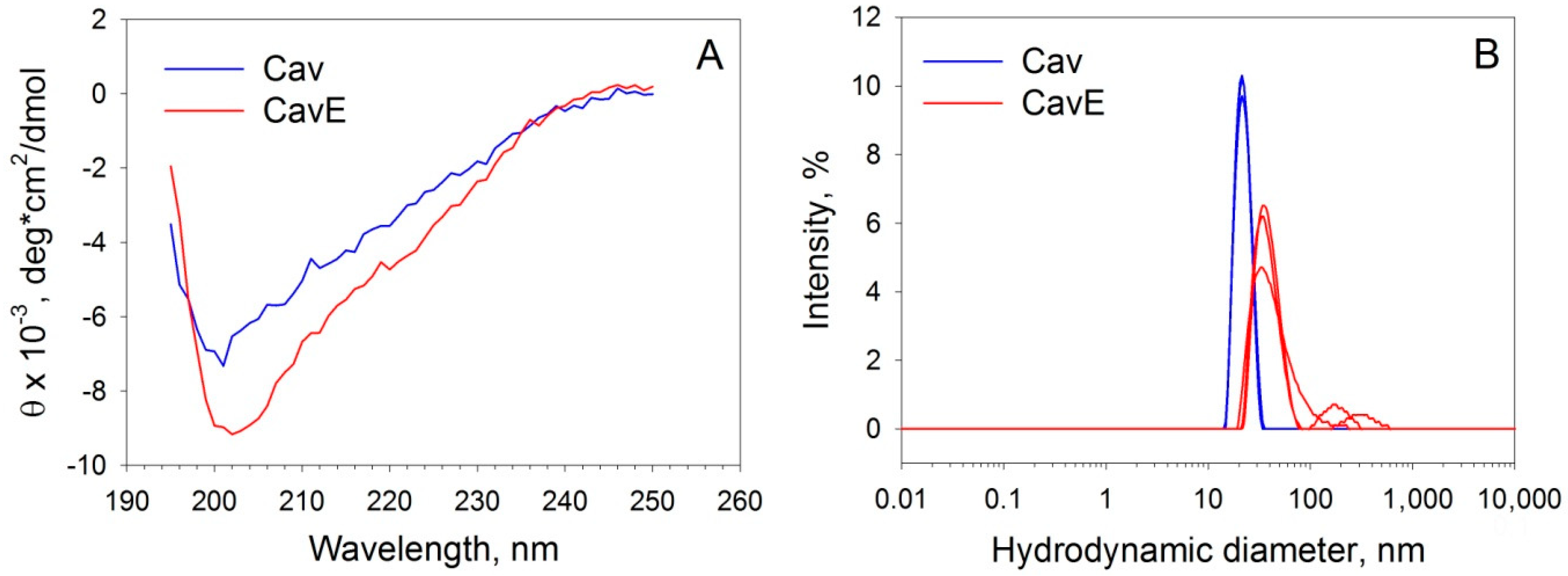


| Caveolin-1 Form | CD | DLS | ||||
|---|---|---|---|---|---|---|
| α-Helix,% | β-Sheet, % | Turn, % | Random Coil, % | dh, nm | Mr, kDa | |
| Cav | 7.2 ± 3.7 | 30.1 ± 5.6 | 19.8 ± 3.1 | 40.3 ± 8.3 | 21.4 ± 4.6 | 866 ± 206 |
| CavE | 7.4 ± 2.9 | 26.2 ± 5.2 | 21.1 ± 3.8 | 44.7 ± 9.1 | 35.1 ± 13.6 | 2700 ± 1400 |
| Recoverin Form | Cav KD, nM (koff, s−1) | CavE KD, nM (koff, s−1) | ||
|---|---|---|---|---|
| 2 mM Ca2+ | 2 mM EGTA | 2 mM Ca2+ | 2 mM EGTA | |
| Rec | n/d 1 | 490 ± 160 ((1.05 ± 0.18) × 10−4) | n/d | 1110 ± 66 ((8.81 ± 2.59) × 10−5) |
| dRec | n/d | 48.6 ± 1.1 ((2.44 ± 0.59) × 10−5) | n/d | 218 ± 87 ((8.08 ± 0.27) × 10−5) |
| Rec-C39D | 376 ± 173 ((1.86 ± 0.13) × 10−4) | 140 ± 44 ((1.17 ± 0.76) × 10−4) | 629 ± 257 ((2.02 ± 0.21) × 10−4) | 329 ± 145 ((1.72 ± 0.15) × 10−4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vladimirov, V.I.; Shchannikova, M.P.; Baldin, A.V.; Kazakov, A.S.; Shevelyova, M.P.; Nazipova, A.A.; Baksheeva, V.E.; Nemashkalova, E.L.; Frolova, A.S.; Tikhomirova, N.K.; et al. Redox Regulation of Signaling Complex between Caveolin-1 and Neuronal Calcium Sensor Recoverin. Biomolecules 2022, 12, 1698. https://doi.org/10.3390/biom12111698
Vladimirov VI, Shchannikova MP, Baldin AV, Kazakov AS, Shevelyova MP, Nazipova AA, Baksheeva VE, Nemashkalova EL, Frolova AS, Tikhomirova NK, et al. Redox Regulation of Signaling Complex between Caveolin-1 and Neuronal Calcium Sensor Recoverin. Biomolecules. 2022; 12(11):1698. https://doi.org/10.3390/biom12111698
Chicago/Turabian StyleVladimirov, Vasiliy I., Margarita P. Shchannikova, Alexey V. Baldin, Alexey S. Kazakov, Marina P. Shevelyova, Aliya A. Nazipova, Viktoriia E. Baksheeva, Ekaterina L. Nemashkalova, Anastasia S. Frolova, Natalia K. Tikhomirova, and et al. 2022. "Redox Regulation of Signaling Complex between Caveolin-1 and Neuronal Calcium Sensor Recoverin" Biomolecules 12, no. 11: 1698. https://doi.org/10.3390/biom12111698
APA StyleVladimirov, V. I., Shchannikova, M. P., Baldin, A. V., Kazakov, A. S., Shevelyova, M. P., Nazipova, A. A., Baksheeva, V. E., Nemashkalova, E. L., Frolova, A. S., Tikhomirova, N. K., Philippov, P. P., Zamyatnin, A. A., Jr., Permyakov, S. E., Zinchenko, D. V., & Zernii, E. Y. (2022). Redox Regulation of Signaling Complex between Caveolin-1 and Neuronal Calcium Sensor Recoverin. Biomolecules, 12(11), 1698. https://doi.org/10.3390/biom12111698









