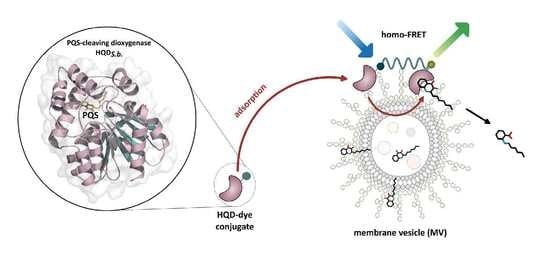
A PQS-Cleaving Quorum Quenching Enzyme Targets Extracellular Membrane Vesicles of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains
2.3. Enzyme Activity Assay and Protein Determination
2.4. Preparation of P. aeruginosa Culture Supernatants and Incubation with PQS Dioxygenases
2.5. Pull-Down Assay with PQS Dioxygenases as a Bait
2.6. Preparation of P. aeruginosa Extracellular Membrane Vesicles
2.7. Protein Labeling with Fluorescent Dyes
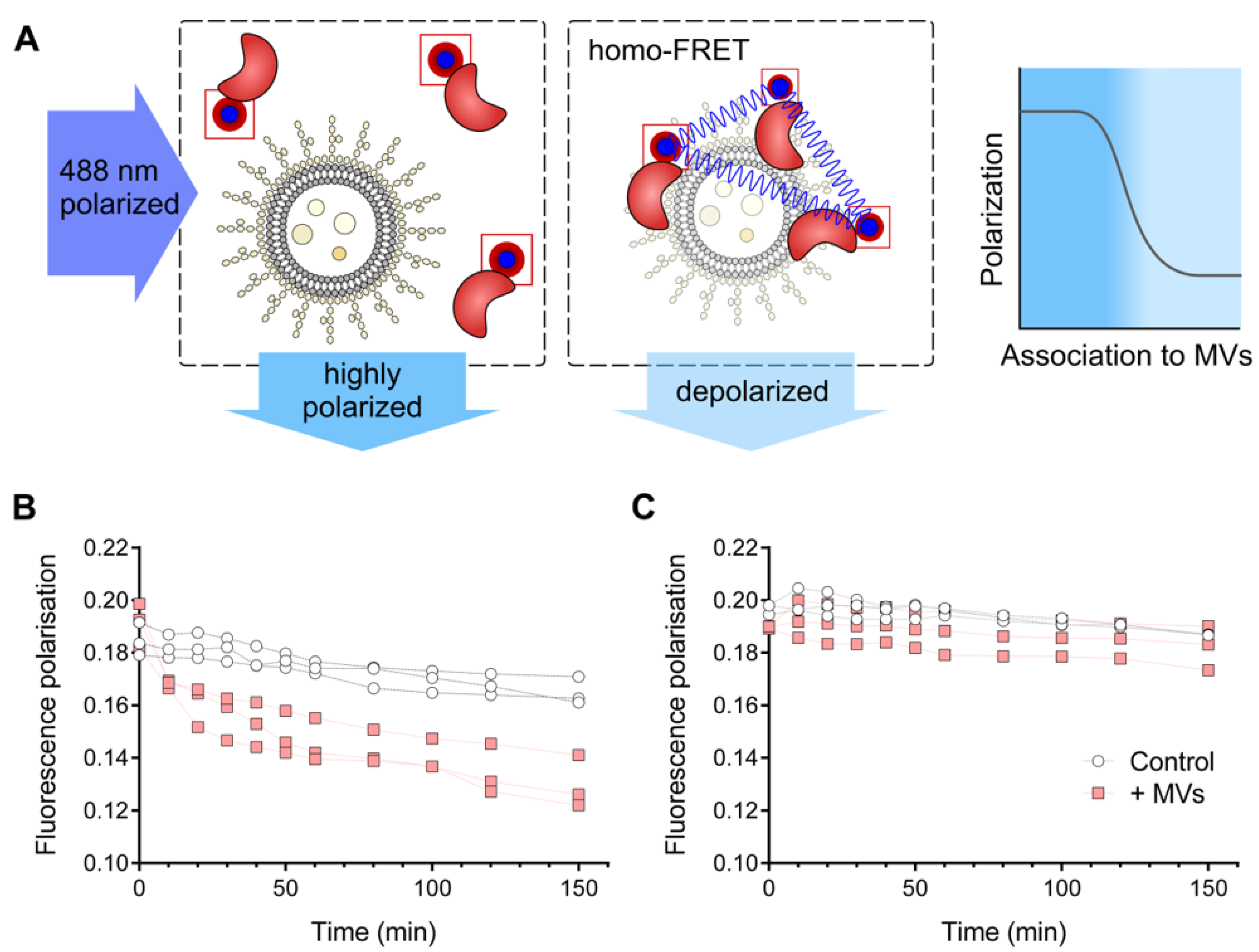
2.8. Homo-FRET-Based Interaction Assay
3. Results and Discussion
3.1. High-Molecular-Weight, Heat-Stable Factor(s) of P. aeruginosa Supernatant Stabilize(s) HQDS.b.
3.2. The HQDS.b. Protein Shows Apparent Interactions with P. aeruginosa Extracellular Proteins
3.3. HQDS.b. Is Stabilized by P. aeruginosa Extracellular Membrane Vesicles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Atkinson, S.; Williams, P. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef]
- Schuster, M.; Greenberg, E.P. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. [Google Scholar] [CrossRef]
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007, 14, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Mashburn, L.M.; Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 2005, 437, 422–425. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological Functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef]
- Kulkarni, H.M.; Jagannadham, M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 2014, 160, 2109–2121. [Google Scholar] [CrossRef]
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 1995, 177, 3998–4008. [Google Scholar] [CrossRef]
- Bomberger, J.M.; MacEachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Beveridge, T.J.; Kadurugamuwa, J.; Walther-Rasmussen, J.; Høiby, N. Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000, 45, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Martínez, O.F.; Cardoso, M.H.; Ribeiro, S.M.; Franco, O.L. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front. Cell. Infect. Microbiol. 2019, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2014, 201, 2–14. [Google Scholar] [CrossRef]
- Wullich, S.C.; Martín, A.A.S.; Fetzner, S. An α/β-hydrolase fold subfamily comprising Pseudomonas quinolone signal-cleaving dioxygenases. Appl. Environ. Microbiol. 2020, 86, e00279-20. [Google Scholar] [CrossRef]
- Pustelny, C.; Albers, A.; Büldt-Karentzopoulos, K.; Parschat, K.; Chhabra, S.R.; Cámara, M.; Williams, P.; Fetzner, S. Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 2009, 16, 1259–1267. [Google Scholar] [CrossRef]
- Birmes, F.S.; Wolf, T.; Kohl, T.A.; Rüger, K.; Bange, F.; Kalinowski, J.; Fetzner, S. Mycobacterium abscessus subsp. abscessus is capable of degrading Pseudomonas aeruginosa quinolone signals. Front. Microbiol. 2017, 8, 339. [Google Scholar] [CrossRef]
- Müller, C.; Birmes, F.S.; Rückert, C.; Kalinowski, J.; Fetzner, S. Rhodococcus erythropolis BG43 genes mediating Pseudomonas aeruginosa quinolone signal degradation and virulence factor attentuation. Appl. Environ. Microbiol. 2015, 81, 7720–7729. [Google Scholar] [CrossRef]
- Arranz San Martín, A.; Vogel, J.; Wullich, S.C.; Quax, W.J.; Fetzner, S. Enzyme-mediated quenching of the Pseudomonas quinolone signal (PQS): A comparison between naturally occurring and engineered PQS-cleaving dioxygenases. Biomolecules 2022, 12, 170. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]
- Klebensberger, J.; Lautenschlager, K.; Bressler, D.; Wingender, J.; Philipp, B. Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ. Microbiol. 2007, 9, 2247–2259. [Google Scholar] [CrossRef] [PubMed]
- Tettmann, B.; Niewerth, C.; Kirschhöfer, F.; Neidig, A.; Dötsch, A.; Brenner-Weiss, G.; Fetzner, S.; Overhage, J. Enzyme-mediated quenching of the Pseudomonas quinolone signal (PQS) promotes biofilm formation of Pseudomonas aeruginosa by increasing iron availability. Front. Microbiol. 2016, 7, 1978. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Zor, T.; Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Elbing, K.; Brent, R. Media preparation and bacteriological tools. Curr. Protoc. Mol. Biol. 2002, 59, 1.1.1–1.1.7. [Google Scholar] [CrossRef] [PubMed]
- Bauman, S.J.; Kuehn, M.J. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006, 8, 2400–2408. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, I.A.; Kuehn, M.J. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2971–2981. [Google Scholar] [CrossRef]
- Bader, A.N.; Hofman, E.G.; Voortman, J.; Van Bergen En Henegouwen, P.M.P.; Gerritsen, H.C. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 2009, 97, 2613–2622. [Google Scholar] [CrossRef]
- Moerke, N.J. Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr. Protoc. Chem. Biol. 2009, 1, 1–15. [Google Scholar] [CrossRef]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638. [Google Scholar] [CrossRef]
- Shuai, Y.; Xinyi, C.; Zhenyu, J.; Aiguo, X.; Lei, N.; Rongrong, Z.; Fan, J. Differential production of Psl in planktonic cells leads to two distinctive attachment phenotypes in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2018, 84, e00700-18. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Choi, S.J.; Lee, J.; Choi, J.P.; Rho, S.; Park, S.H.; Kim, Y.K.; Hwang, D.; Gho, Y.S. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 2011, 11, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Schooling, S.R.; Dutcher, J.R.; Barber, J. Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J. Proteome Res. 2015, 14, 4207–4222. [Google Scholar] [CrossRef] [PubMed]
- Koeppen, K.; Barnaby, R.; Jackson, A.A.; Gerber, S.A.; Hogan, D.A.; Stanton, B.A. Tobramycin reduces key virulence determinants in the proteome of Pseudomonas aeruginosa outer membrane vesicles. PLoS ONE 2019, 14, e0211290. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef]
- Lewenza, S.; Gardy, J.L.; Brinkman, F.S.L.; Hancock, R.E.W. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 2005, 15, 321–329. [Google Scholar] [CrossRef]
- Schaar, V.; de Vries, S.P.W.; Perez Vidakovics, M.L.A.; Bootsma, H.J.; Larsson, L.; Hermans, P.W.M.; Bjartell, A.; Mörgelin, M.; Riesbeck, K. Multicomponent Moraxella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell. Microbiol. 2011, 13, 432–449. [Google Scholar] [CrossRef]
- Schulz, E.; Goes, A.; Garcia, R.; Panter, F.; Koch, M.; Müller, R.; Fuhrmann, K.; Fuhrmann, G. Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J. Control. Release 2018, 290, 46–55. [Google Scholar] [CrossRef]
- Schulz, E.; Karagianni, A.; Koch, M.; Fuhrmann, G. Hot EVs—How temperature affects extracellular vesicles. Eur. J. Pharm. Biopharm. 2020, 146, 55–63. [Google Scholar] [CrossRef]
- Palmieri, E.; Arato, V.; Oldrini, D.; Ricchetti, B.; Aruta, M.G.; Pansegrau, W.; Marchi, S.; Giusti, F.; Ferlenghi, I.; Rossi, O.; et al. Stability of outer membrane vesicles-based vaccines, identifying the most appropriate methods to detect changes in vaccine potency. Vaccines 2021, 9, 229. [Google Scholar] [CrossRef]
- Wullich, S.C.; Kobus, S.; Wienhold, M.; Hennecke, U.; Smits, S.H.J.; Fetzner, S. Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the α/β-hydrolase fold family. J. Struct. Biol. 2019, 207, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef]
- Chen, H.; Puhl, H.L.; Koushik, S.V.; Vogel, S.S.; Ikeda, S.R. Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys. J. 2006, 91, L39–L41. [Google Scholar] [CrossRef]
- Florez, C.; Raab, J.E.; Cooke, A.C.; Schertzer, J.W. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 2017, 8, 1034–1051. [Google Scholar] [CrossRef] [PubMed]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef] [PubMed]
- Sueki, A.; Stein, F.; Savitski, M.M.; Selkrig, J.; Typas, A. Systematic localization of Escherichia coli membrane proteins. mSystems 2020, 5, e00808-19. [Google Scholar] [CrossRef]
- Papanastasiou, M.; Orfanoudaki, G.; Koukaki, M.; Kountourakis, N.; Sardis, M.F.; Aivaliotis, M.; Karamanou, S.; Economou, A. The Escherichia coli peripheral inner membrane proteome. Mol. Cell. Proteomics 2013, 12, 599–610. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Avila-Calderón, E.D.; Ruiz-Palma, M.d.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer membrane vesicles of Gram-negative bacteria: An outlook on biogenesis. Front. Microbiol. 2021, 12, 557902. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.; Howe, J.; Garidel, P.; Richter, W.; Steiniger, F.; Roessle, M.; Brandenburg, K.; Whiteley, M. Interaction of quorum signals with outer membrane lipids: Insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 2008, 69, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Faddetta, T.; Renzone, G.; Vassallo, A.; Rimini, E.; Nasillo, G.; Buscarino, G.; Agnello, S.; Licciardi, M.; Botta, L.; Scaloni, A.; et al. Streptomyces coelicolor vesicles: Many molecules to be delivered. Appl. Environ. Microbiol. 2022, 88, e01881-21. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arranz San Martín, A.; Drees, S.L.; Fetzner, S. A PQS-Cleaving Quorum Quenching Enzyme Targets Extracellular Membrane Vesicles of Pseudomonas aeruginosa. Biomolecules 2022, 12, 1656. https://doi.org/10.3390/biom12111656
Arranz San Martín A, Drees SL, Fetzner S. A PQS-Cleaving Quorum Quenching Enzyme Targets Extracellular Membrane Vesicles of Pseudomonas aeruginosa. Biomolecules. 2022; 12(11):1656. https://doi.org/10.3390/biom12111656
Chicago/Turabian StyleArranz San Martín, Alba, Steffen Lorenz Drees, and Susanne Fetzner. 2022. "A PQS-Cleaving Quorum Quenching Enzyme Targets Extracellular Membrane Vesicles of Pseudomonas aeruginosa" Biomolecules 12, no. 11: 1656. https://doi.org/10.3390/biom12111656
APA StyleArranz San Martín, A., Drees, S. L., & Fetzner, S. (2022). A PQS-Cleaving Quorum Quenching Enzyme Targets Extracellular Membrane Vesicles of Pseudomonas aeruginosa. Biomolecules, 12(11), 1656. https://doi.org/10.3390/biom12111656