LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Chemicals and Reagents
2.3. Depletion of High-Abundance Proteins in Serum Samples
2.4. N-Glycans Release, Purification, Reduction, and Permethylation
2.5. N-Glycan Profiling
2.6. Multiple Reaction Monitoring (MRM) Validation
2.7. Protein Analysis of the Depleted Serum Samples
3. Results
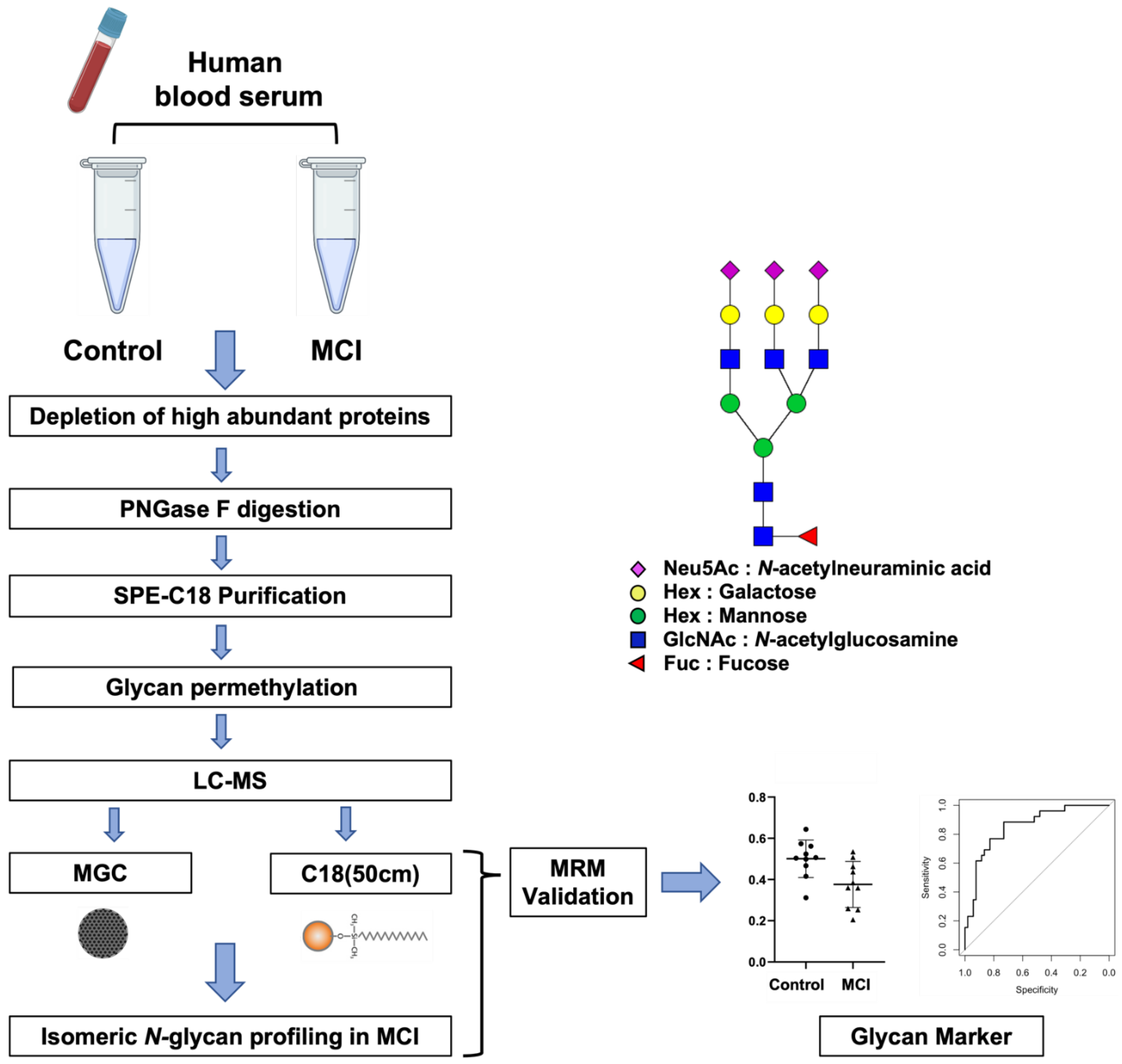
3.1. Analytical Workflow
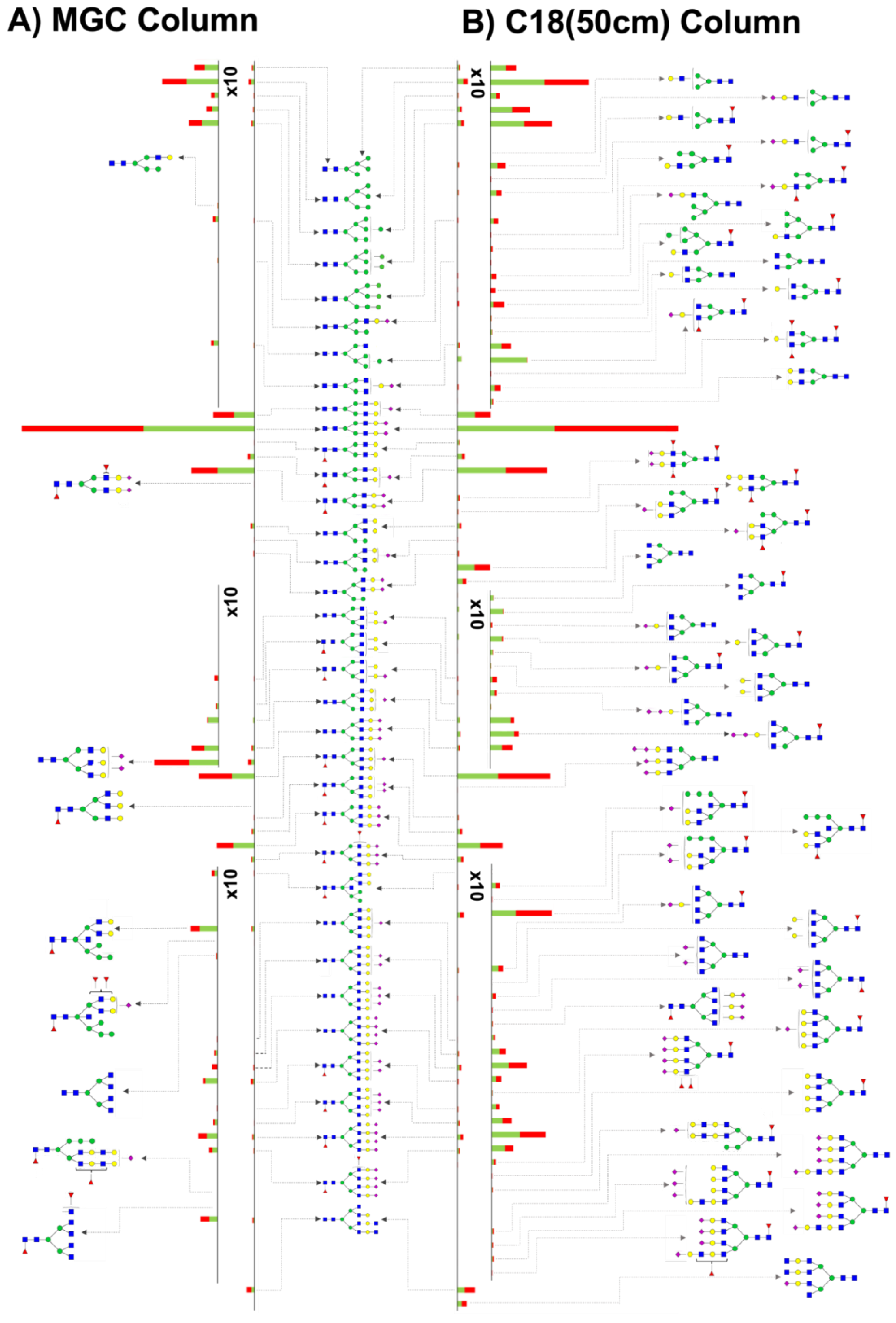
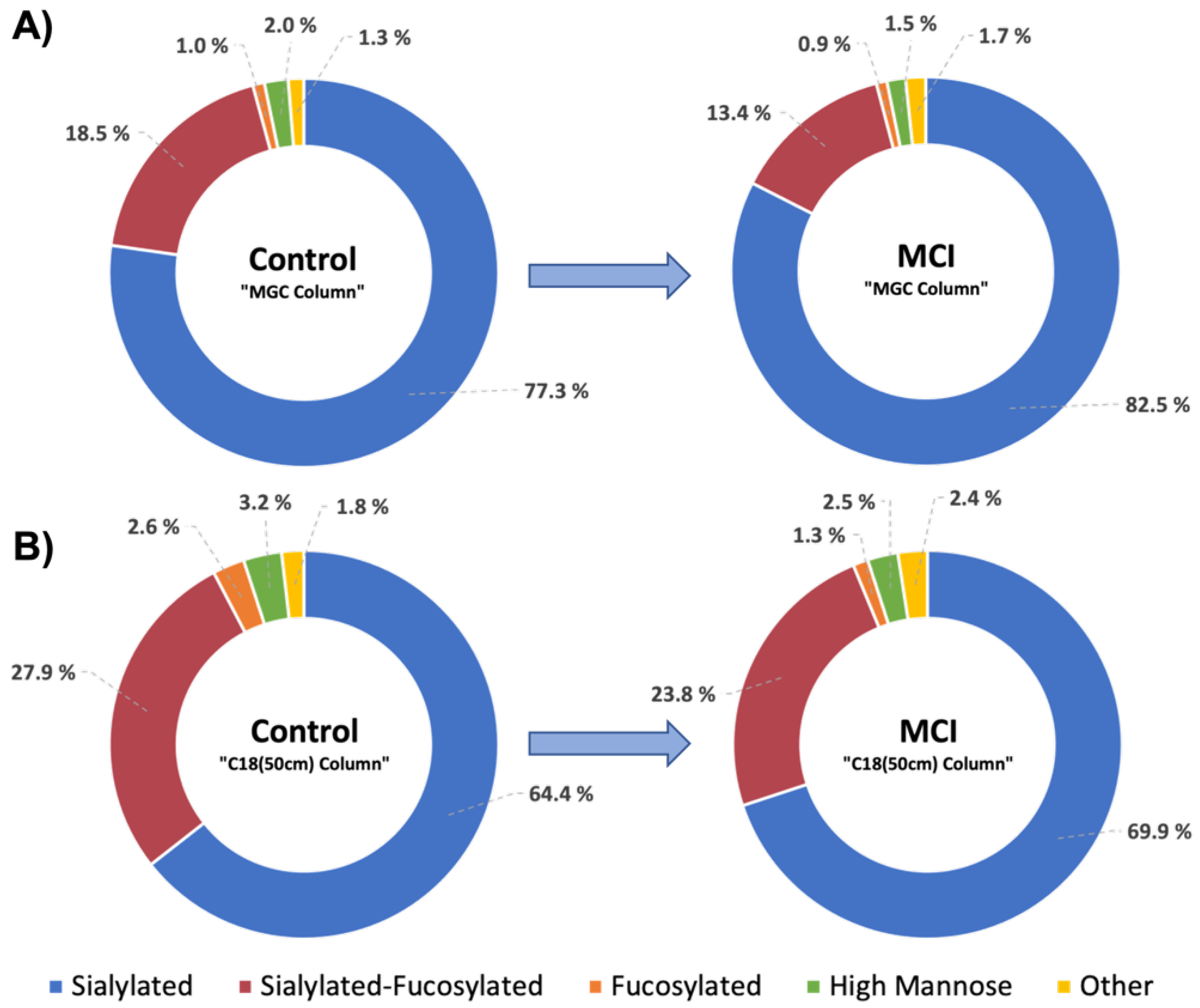
3.2. MGC-LC-MS and C18(50 cm)-LC-MS Comparison
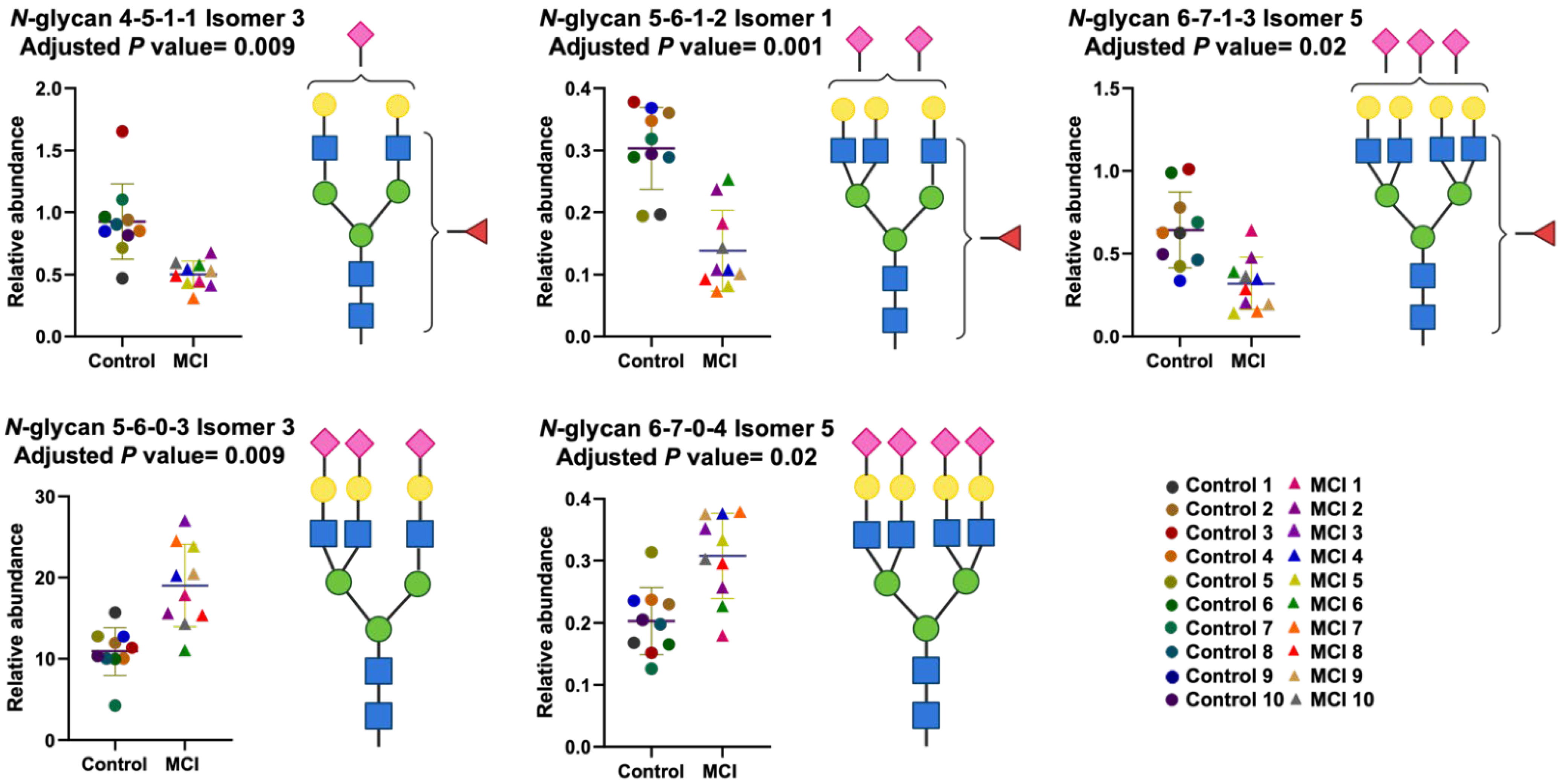
3.3. Differentially Expressed Relative Abundance of N-Glycans Derived from Low Abundant Serum Glycoproteins in Control and MCI Samples
3.4. Protein Analysis of the Depleted Serum Samples Extracted from Control and MCI Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2018, 4, 575–590. [Google Scholar] [CrossRef]
- Ganguli, M.; Snitz, B.E.; Saxton, J.A.; Chang, C.-C.H.; Lee, C.-W.; Vander Bilt, J.; Hughes, T.F.; Loewenstein, D.A.; Unverzagt, F.W.; Petersen, R.C. Outcomes of Mild Cognitive Impairment by Definition: A Population Study. Arch. Neurol. 2011, 68, 761–767. [Google Scholar] [CrossRef]
- Farias, S.T.; Mungas, D.; Reed, B.R.; Harvey, D.; Cahn-Weiner, D.; Decarli, C. MCI is associated with deficits in everyday functioning. Alzheimer Dis. Assoc. Disord. 2006, 20, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Albert, M.L. Amnestic MCI or prodromal Alzheimer’s disease? Lancet Neurol. 2004, 3, 246–248. [Google Scholar] [CrossRef]
- Solfrizzi, V.; Panza, F.; Colacicco, A.M.; D’Introno, A.; Capurso, C.; Torres, F.; Grigoletto, F.; Maggi, S.; Del Parigi, A.; Reiman, E.M.; et al. Vascular risk factors, incidence of MCI, and rates of progression to dementia. Neurology 2004, 63, 1882–1891. [Google Scholar] [CrossRef] [PubMed]
- Mallo, S.C.; Pereiro, A.X.; Campos-Magdaleno, M.; Nieto-Vieites, A.; Lojo-Seoane, C.; Facal, D.; Ismail, Z.; Juncos-Rabadán, O. Neuropsychiatric symptoms in subjective cognitive complaints (SCC) and mild cognitive impairment (MCI): Detecting changes over time with the Mild Behavioral Impairment Checklist (MBI-C). Alzheimer’s Dement. 2020, 16, e041963. [Google Scholar] [CrossRef]
- Zhang, D.; Shen, D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS ONE 2012, 7, e33182. [Google Scholar] [CrossRef]
- Platero, C.; Tobar, M.C. Predicting Alzheimer’s conversion in mild cognitive impairment patients using longitudinal neuroimaging and clinical markers. Brain Imaging Behav. 2021, 15, 1728–1738. [Google Scholar] [CrossRef]
- Davatzikos, C.; Bhatt, P.; Shaw, L.M.; Batmanghelich, K.N.; Trojanowski, J.Q. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol. Aging 2011, 32, 2322-e19. [Google Scholar] [CrossRef]
- Dennis, J.W.; Granovsky, M.; Warren, C.E. Protein glycosylation in development and disease. Bioessays 1999, 21, 412–421. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, S.; Zhu, J.; Lubman, D.M.; Mechref, Y. LC-MS/MS isomeric profiling of permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma (HCC) and cirrhotic patients. Electrophoresis 2017, 38, 2160–2167. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.G.; Veillon, L.; Mechref, Y. N-Glycan Profile of Cerebrospinal Fluids from Alzheimer’s Disease Patients Using Liquid Chromatography with Mass Spectrometry. J. Proteome Res. 2019, 18, 3770–3779. [Google Scholar] [CrossRef]
- Gutierrez Reyes, C.D.; Jiang, P.; Donohoo, K.; Atashi, M.; Mechref, Y.S. Glycomics and glycoproteomics: Approaches to address isomeric separation of glycans and glycopeptides. J. Sep. Sci. 2021, 44, 403–425. [Google Scholar] [CrossRef]
- Cho, B.G.; Gutierrez Reyes, C.D.; Mechref, Y. N-Glycomics of Cerebrospinal Fluid: Method Comparison. Molecules 2021, 26, 1712. [Google Scholar] [CrossRef]
- Palmigiano, A.; Barone, R.; Sturiale, L.; Sanfilippo, C.; Bua, R.O.; Romeo, D.A.; Messina, A.; Capuana, M.L.; Maci, T.; Le Pira, F.; et al. CSF N-glycoproteomics for early diagnosis in Alzheimer’s disease. J. Proteom. 2016, 131, 29–37. [Google Scholar] [CrossRef]
- Lundström, S.L.; Yang, H.; Lyutvinskiy, Y.; Rutishauser, D.; Herukka, S.K.; Soininen, H.; Zubarev, R.A. Blood plasma IgG Fc glycans are significantly altered in Alzheimer’s disease and progressive mild cognitive impairment. J. Alzheimers Dis. 2014, 38, 567–579. [Google Scholar] [CrossRef]
- Giau, V.V.; Bagyinszky, E.; An, S.S.A. Potential Fluid Biomarkers for the Diagnosis of Mild Cognitive Impairment. Int. J. Mol. Sci. 2019, 20, 4149. [Google Scholar] [CrossRef]
- Schedin-Weiss, S.; Gaunitz, S.; Sui, P.; Chen, Q.; Haslam, S.M.; Blennow, K.; Winblad, B.; Dell, A.; Tjernberg, L.O. Glycan biomarkers for Alzheimer disease correlate with T-tau and P-tau in cerebrospinal fluid in subjective cognitive impairment. FEBS J. 2020, 287, 3221–3234. [Google Scholar] [CrossRef] [PubMed]
- Haukedal, H.; Freude, K.K. Implications of Glycosylation in Alzheimer’s Disease. Front. Neurosci. 2021, 14, 625348. [Google Scholar] [CrossRef] [PubMed]
- Llop, E.; Ardá, A.; Zacco, E.; O’Flaherty, R.; García-Ayllón, M.S.; Aureli, M.; Frenkel-Pinter, M.; Reis, C.A.; Greiner-Tollersrud, O.K.; Cuchillo-Ibáñez, I. Proceedings of workshop: “Neuroglycoproteins in health and disease”, INNOGLY cost action. Glycoconj. J. 2022, 39, 579–586. [Google Scholar] [CrossRef]
- Peng, W.; Gutierrez Reyes, C.D.; Gautam, S.; Yu, A.; Cho, B.G.; Goli, M.; Donohoo, K.; Mondello, S.; Kobeissy, F.; Mechref, Y. MS-based glycomics and glycoproteomics methods enabling isomeric characterization. Mass Spectrom. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Akasaka-Manya, K.; Manya, H.; Sakurai, Y.; Wojczyk, B.S.; Kozutsumi, Y.; Saito, Y.; Taniguchi, N.; Murayama, S.; Spitalnik, S.L.; Endo, T. Protective effect of N -glycan bisecting GlcNAc residues on β-amyloid production in Alzheimer’s disease. Glycobiology 2009, 20, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Kitazume, S.; Taniguchi, N. N-glycan and Alzheimer’s disease. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, M.L.; Colombo, M.; McGurnaghan, S.J.; Blackbourn, L.A.K.; Vučković, F.; Pučić Baković, M.; Trbojević-Akmačić, I.; Lauc, G.; Agakov, F.; Agakova, A.S.; et al. N-Glycan Profile and Kidney Disease in Type 1 Diabetes. Diabetes Care 2018, 41, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, M. Advanced protein glycosylation in diabetes and aging. Annu. Rev. Med. 1995, 46, 223–234. [Google Scholar] [CrossRef]
- Peng, W.; Goli, M.; Mirzaei, P.; Mechref, Y. Revealing the Biological Attributes of N-Glycan Isomers in Breast Cancer Brain Metastasis Using Porous Graphitic Carbon (PGC) Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS). J. Proteome Res. 2019, 18, 3731–3740. [Google Scholar] [CrossRef]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol 2015, 10, 473–510. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Albert, M.S.; DeKosky, S.T.; Dickson, D.; Dubois, B.; Feldman, H.H.; Fox, N.C.; Gamst, A.; Holtzman, D.M.; Jagust, W.J.; Petersen, R.C.; et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 270–279. [Google Scholar] [CrossRef]
- Gautam, S.; Banazadeh, A.; Cho, B.G.; Goli, M.; Zhong, J.; Mechref, Y. Mesoporous Graphitized Carbon Column for Efficient Isomeric Separation of Permethylated Glycans. Anal. Chem. 2021, 93, 5061–5070. [Google Scholar] [CrossRef]
- Wang, J.; Dong, X.; Yu, A.; Huang, Y.; Peng, W.; Mechref, Y. Isomeric separation of permethylated glycans by extra-long reversed-phase liquid chromatography (RPLC)-MS/MS. Analyst 2022, 147, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- Donohoo, K.B.; Wang, J.; Goli, M.; Yu, A.; Peng, W.; Hakim, M.A.; Mechref, Y. Advances in mass spectrometry-based glycomics-An update covering the period 2017–2021. Electrophoresis 2021, 43, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Huang, Y.; Cho, B.G.; Zhong, J.; Gautam, S.; Peng, W.; Williamson, S.D.; Banazadeh, A.; Torres-Ulloa, K.Y.; Mechref, Y. Advances in mass spectrometry-based glycomics. Electrophoresis 2018, 39, 3063–3081. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.H.; Wang, M.; Di Poto, C.; Hu, Y.; Zhou, S.; Zhao, Y.; Varghese, R.S.; Luo, Y.; Tadesse, M.G.; Ziada, D.H.; et al. LC-MS profiling of N-Glycans derived from human serum samples for biomarker discovery in hepatocellular carcinoma. J. Proteome Res. 2014, 13, 4859–4868. [Google Scholar] [CrossRef]
- Meng, Z.; Veenstra, T.D. Chapter 26—Mass Spectrometry–Based Approach for Protein Biomarker Verification. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Issaq, H.J., Veenstra, T.D., Eds.; Academic Press: Boston, MA, USA, 2013; pp. 407–424. [Google Scholar] [CrossRef]
- Parker, C.E.; Borchers, C.H. Mass spectrometry based biomarker discovery, verification, and validation--quality assurance and control of protein biomarker assays. Mol. Oncol. 2014, 8, 840–858. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, Y.; DeSantos-Garcia, J.L.; Mechref, Y. Quantitation of permethylated N-glycans through multiple-reaction monitoring (MRM) LC-MS/MS. J. Am. Soc. Mass Spectrom. 2015, 26, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Method of the Year 2012. Nat. Methods 2013, 10, 1. [CrossRef]
- Tu, C.; Rudnick, P.A.; Martinez, M.Y.; Cheek, K.L.; Stein, S.E.; Slebos, R.J.C.; Liebler, D.C. Depletion of abundant plasma proteins and limitations of plasma proteomics. J. Proteome Res. 2010, 9, 4982–4991. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, Y.; Mechref, Y. High-temperature LC-MS/MS of permethylated glycans derived from glycoproteins. Electrophoresis 2016, 37, 1506–1513. [Google Scholar] [CrossRef]
- Kurz, S.; Sheikh, M.O.; Lu, S.; Wells, L.; Tiemeyer, M. Separation and Identification of Permethylated Glycan Isomers by Reversed Phase NanoLC-NSI-MSn. Mol. Cell. Proteom. 2021, 20, 100045. [Google Scholar] [CrossRef]
- Shajahan, A.; Supekar, N.; Heiss, C.; Azadi, P. High-Throughput Automated Micro-permethylation for Glycan Structure Analysis. Anal. Chem. 2019, 91, 1237–1240. [Google Scholar] [CrossRef] [PubMed]
- Ubina, T.; Magallanes, M.; Srivastava, S.; Warden, C.; Yee, J.-K.; Salvaterra, P.M. A human embryonic stem cell model of Aβ-dependent chronic progressive neurodegeneration. bioRxiv 2018, arXiv:10.1101/419978. [Google Scholar]
- Klein, A.; Carre, Y.; Louvet, A.; Michalski, J.-C.; Morelle, W. Immunoglobulins are the major glycoproteins involved in the modifications of total serum N-glycome in cirrhotic patients. PROTEOMICS–Clin. Appl. 2010, 4, 379–393. [Google Scholar] [CrossRef]
- Barone, R.; Sturiale, L.; Palmigiano, A.; Zappia, M.; Garozzo, D. Glycomics of pediatric and adulthood diseases of the central nervous system. J. Proteom. 2012, 75, 5123–5139. [Google Scholar] [CrossRef]
- Chen, C.C.; Engelborghs, S.; Dewaele, S.; Le Bastard, N.; Martin, J.J.; Vanhooren, V.; Libert, C.; De Deyn, P.P. Altered serum glycomics in Alzheimer disease: A potential blood biomarker? Rejuvenation Res. 2010, 13, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Parekh, R.; Roitt, I.; Isenberg, D.; Dwek, R.; Rademacher, T. Age-related galactosylation of the N-linked oligosaccharides of human serum IgG. J. Exp. Med. 1988, 167, 1731–1736. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. MVAPACK: A complete data handling package for NMR metabolomics. ACS Chem. Biol. 2014, 9, 1138–1144. [Google Scholar] [CrossRef]
- Mondello, S.; Sandner, V.; Goli, M.; Czeiter, E.; Amrein, K.; Kochanek, P.M.; Gautam, S.; Cho, B.G.; Morgan, R.; Nehme, A.; et al. Exploring serum glycome patterns after moderate to severe traumatic brain injury: A prospective pilot study. EClinicalMedicine 2022, 50, 101494. [Google Scholar] [CrossRef]
- van Kooyk, Y.; Rabinovich, G.A. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 2008, 9, 593–601. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Blixt, O.; Collins, B.E.; van den Nieuwenhof, I.M.; Crocker, P.R.; Paulson, J.C. Sialoside Specificity of the Siglec Family Assessed Using Novel Multivalent Probes: Identification of potent inhibitors of myelin-associated glycoprotein *. J. Biol. Chem. 2003, 278, 31007–31019. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.; Panin, V.M. The role of protein N-glycosylation in neural transmission. Glycobiology 2014, 24, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Nimtz, M.; Wurster, U.; Conradt, H.S. Carbohydrate Structures of β-Trace Protein from Human Cerebrospinal Fluid: Evidence for “Brain-Type”N-Glycosylation. J. Neurochem. 1994, 63, 2185–2196. [Google Scholar] [CrossRef]
- Lee, S.U.; Grigorian, A.; Pawling, J.; Chen, I.J.; Gao, G.; Mozaffar, T.; McKerlie, C.; Demetriou, M. N-glycan processing deficiency promotes spontaneous inflammatory demyelination and neurodegeneration. J. Biol. Chem. 2007, 282, 33725–33734. [Google Scholar] [CrossRef] [PubMed]
- Mkhikian, H.; Grigorian, A.; Li, C.F.; Chen, H.L.; Newton, B.; Zhou, R.W.; Beeton, C.; Torossian, S.; Tatarian, G.G.; Lee, S.U.; et al. Genetics and the environment converge to dysregulate N-glycosylation in multiple sclerosis. Nat. Commun. 2011, 2, 334. [Google Scholar] [CrossRef] [PubMed]
- Kleene, R.; Schachner, M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004, 5, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Helm, J.; Hirtler, L.; Altmann, F. Towards Mapping of the Human Brain N-Glycome with Standardized Graphitic Carbon Chromatography. Biomolecules 2022, 12, 85. [Google Scholar] [CrossRef]
- Pollio, G.; Hoozemans, J.J.M.; Andersen, C.A.; Roncarati, R.; Rosi, M.C.; van Haastert, E.S.; Seredenina, T.; Diamanti, D.; Gotta, S.; Fiorentini, A.; et al. Increased expression of the oligopeptidase THOP1 is a neuroprotective response to Aβ toxicity. Neurobiol. Dis. 2008, 31, 145–158. [Google Scholar] [CrossRef]
- Sebastiani, P.; Monti, S.; Morris, M.; Gurinovich, A.; Toshiko, T.; Andersen, S.L.; Sweigart, B.; Ferrucci, L.; Jennings, L.L.; Glass, D.J.; et al. A serum protein signature of APOE genotypes in centenarians. Aging Cell 2019, 18, e13023. [Google Scholar] [CrossRef]


| Disease Diagnosis | Control 1 | MCI 2 |
|---|---|---|
| Number | 10 | 10 |
| Gender (M/F) | 8/2 | 7/3 |
| Age (mean ± SD) | 73 ± 10 | 74 ± 8 |
| Smoker (Yes/No) | 3/7 | 4/6 |
| Diabetes (Yes/No) | 0/10 | 3/7 |
| Glycan Composition | p Value 1 | Adjusted p Value 2 |
|---|---|---|
| 5-6-1-2 Isomer 1 | 0.00002 | 0.001 |
| 5-6-0-3 Isomer 3 | 0.0004 | 0.009 |
| 4-5-1-1 Isomer 3 | 0.0006 | 0.009 |
| 6-7-0-4 Isomer 5 | 0.001 | 0.02 |
| 6-7-1-3 Isomer 5 | 0.002 | 0.02 |
| 4-5-1-2 Isomer 5 | 0.008 | 0.06 |
| 4-5-1-1 Isomer 1 | 0.008 | 0.06 |
| 6-7-1-2 | 0.01 | 0.08 |
| 4-5-1-2 Isomer 4 | 0.01 | 0.07 |
| 4-5-2-2 | 0.02 | 0.08 |
| 6-7-1-3 Isomer 6 | 0.02 | 0.09 |
| 5-6-1-0 | 0.02 | 0.09 |
| 6-7-1-3 Isomer 4 | 0.03 | 0.09 |
| 5-6-1-2 Isomer 3 | 0.03 | 0.1 |
| 5-6-0-3 Isomer 4 | 0.03 | 0.09 |
| 5-6-1-2 Isomer 2 | 0.04 | 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes, C.D.G.; Hakim, M.A.; Atashi, M.; Goli, M.; Gautam, S.; Wang, J.; Bennett, A.I.; Zhu, J.; Lubman, D.M.; Mechref, Y. LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients. Biomolecules 2022, 12, 1657. https://doi.org/10.3390/biom12111657
Reyes CDG, Hakim MA, Atashi M, Goli M, Gautam S, Wang J, Bennett AI, Zhu J, Lubman DM, Mechref Y. LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients. Biomolecules. 2022; 12(11):1657. https://doi.org/10.3390/biom12111657
Chicago/Turabian StyleReyes, Cristian D. Gutierrez, Md. Abdul Hakim, Mojgan Atashi, Mona Goli, Sakshi Gautam, Junyao Wang, Andrew I. Bennett, Jianhui Zhu, David M. Lubman, and Yehia Mechref. 2022. "LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients" Biomolecules 12, no. 11: 1657. https://doi.org/10.3390/biom12111657
APA StyleReyes, C. D. G., Hakim, M. A., Atashi, M., Goli, M., Gautam, S., Wang, J., Bennett, A. I., Zhu, J., Lubman, D. M., & Mechref, Y. (2022). LC-MS/MS Isomeric Profiling of N-Glycans Derived from Low-Abundant Serum Glycoproteins in Mild Cognitive Impairment Patients. Biomolecules, 12(11), 1657. https://doi.org/10.3390/biom12111657







