The Effect of Positive Charge Distribution on the Cryoprotective Activity of Dehydrins
Abstract
1. Introduction
2. Materials and Methods
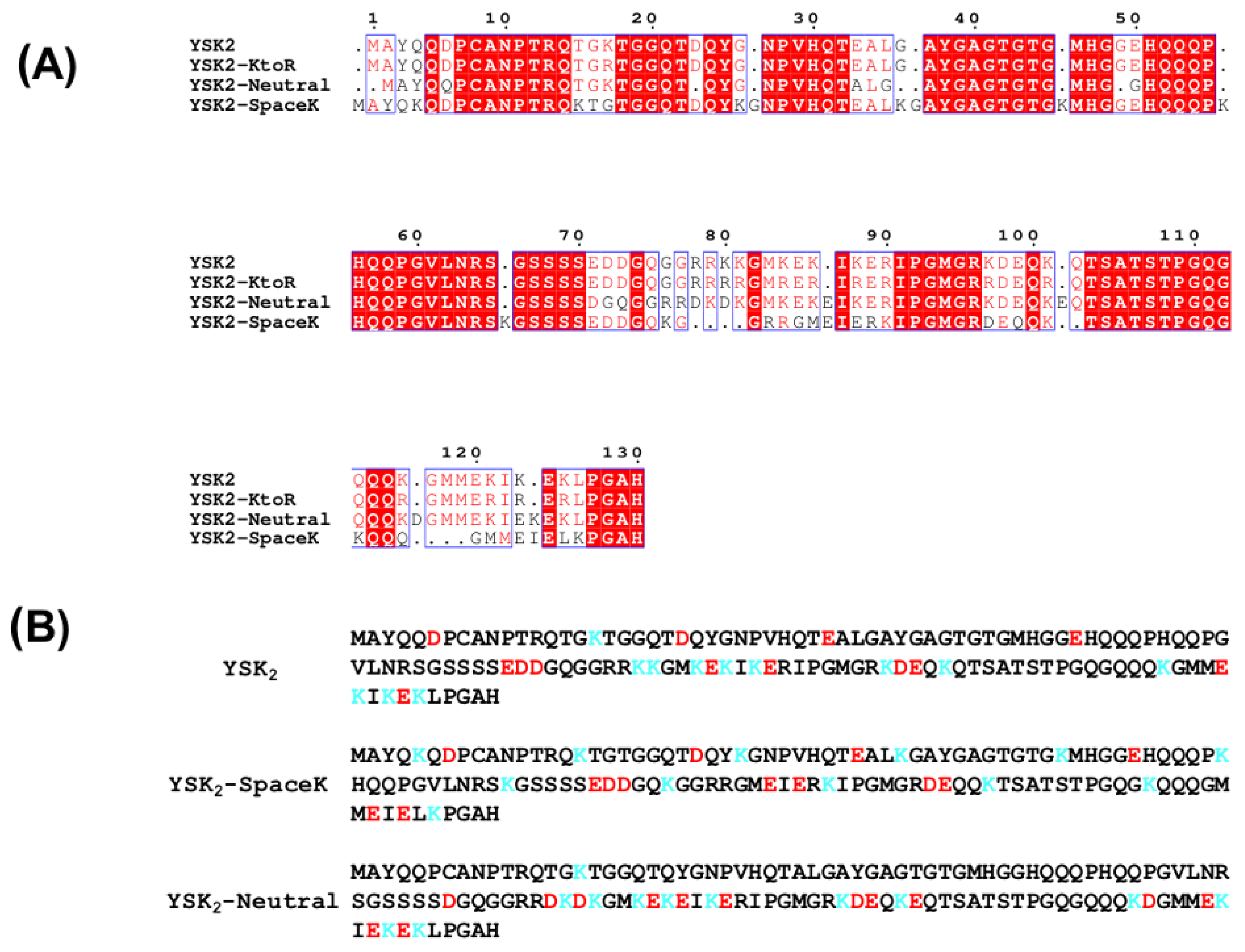
2.1. Dehydrin Construct Design and Cloning
2.2. Dehydrin Expression and Purification
2.3. Yfh1 Expression and Purification
2.4. LDH Cryoprotection
2.5. Circular Dichroism
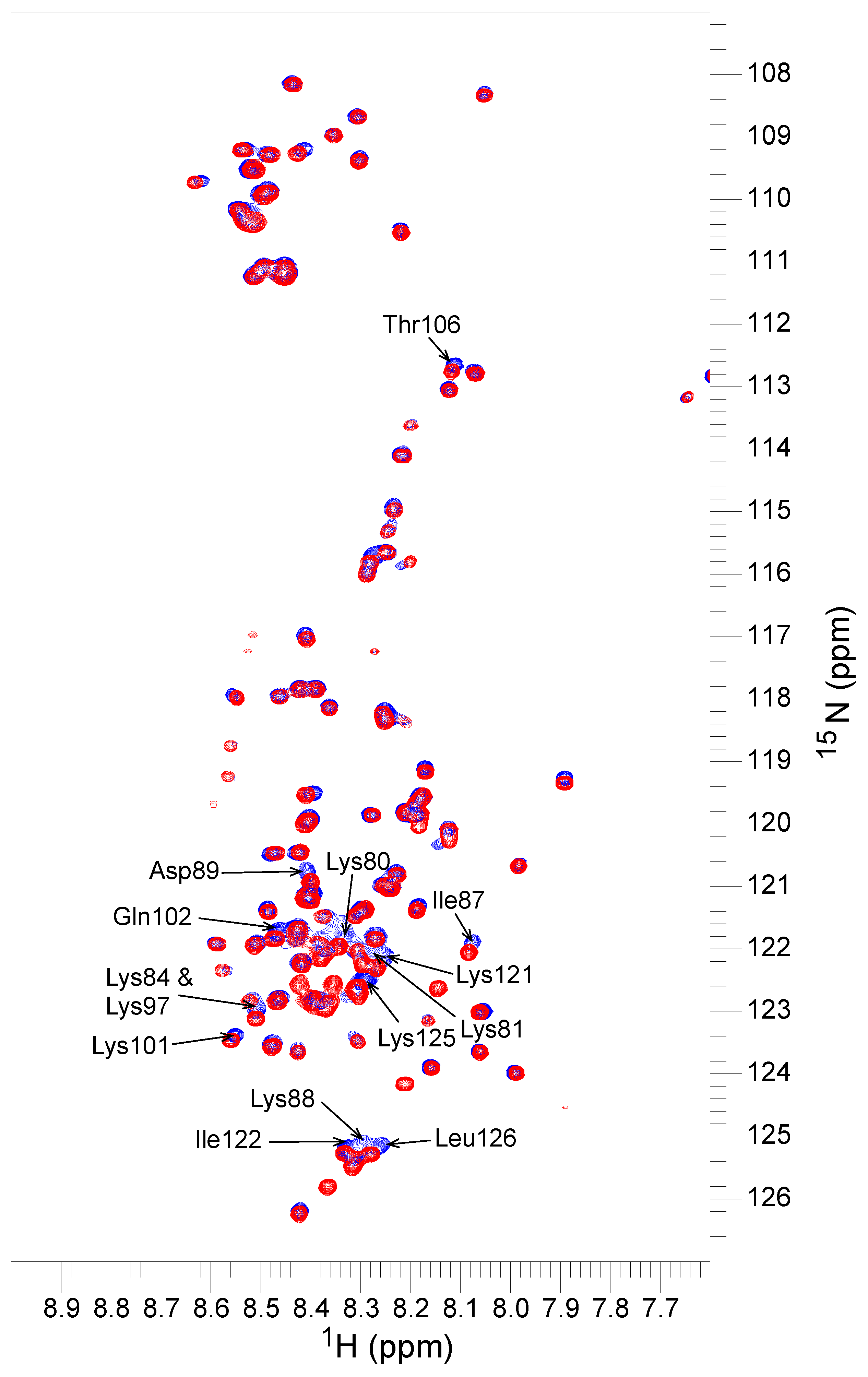
2.6. NMR Experiments
3. Results
3.1. Construct Design and Structure
3.2. LDH Cryoprotection
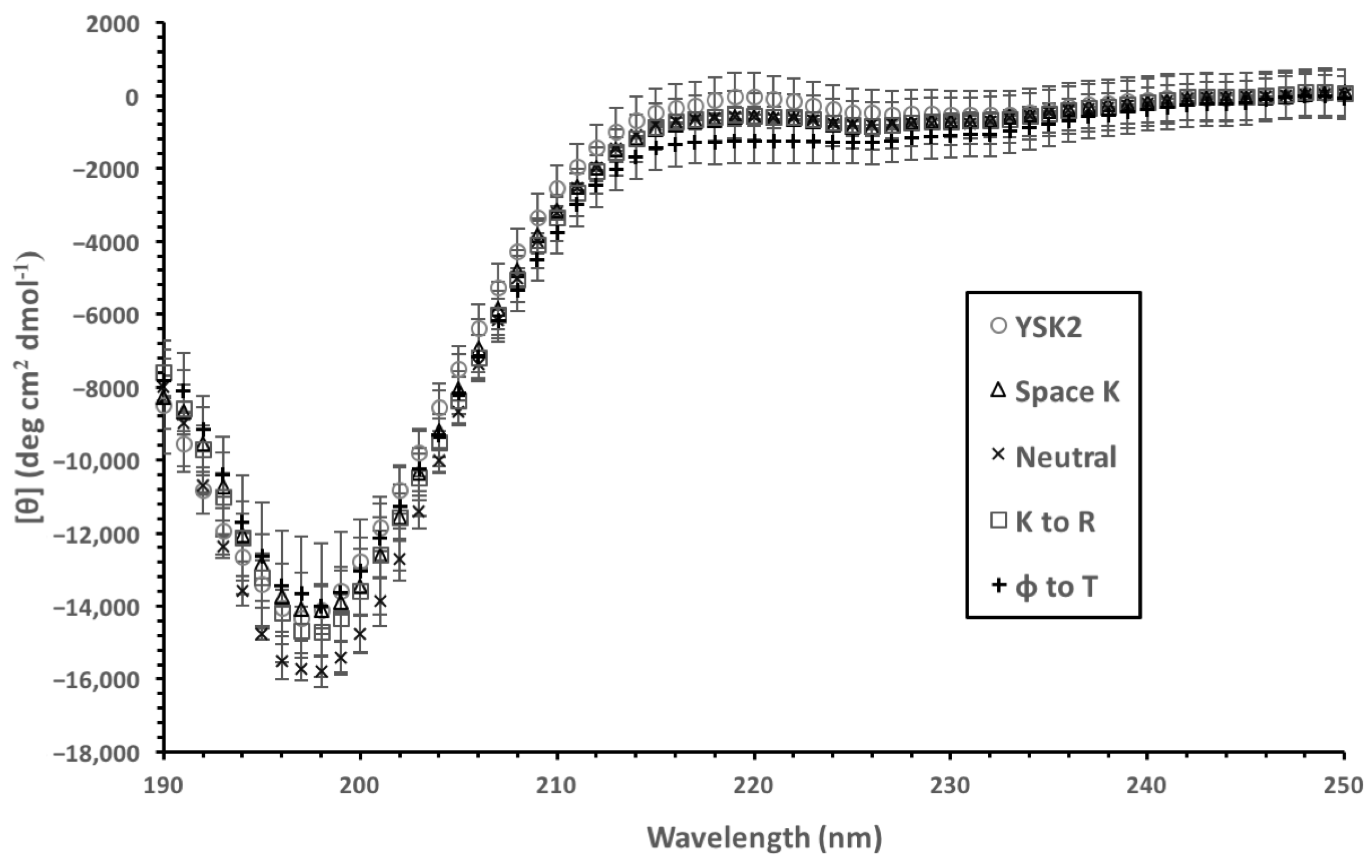
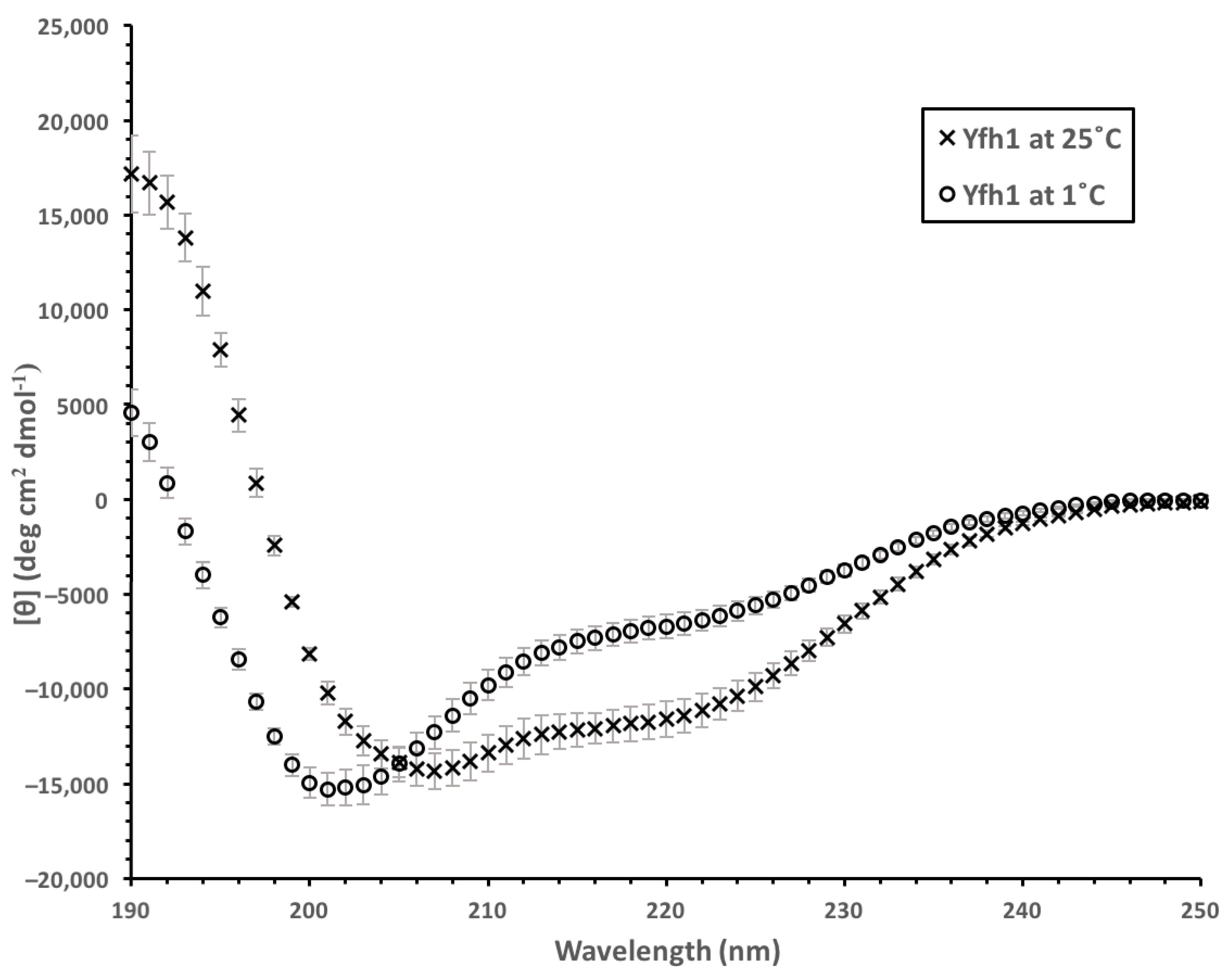
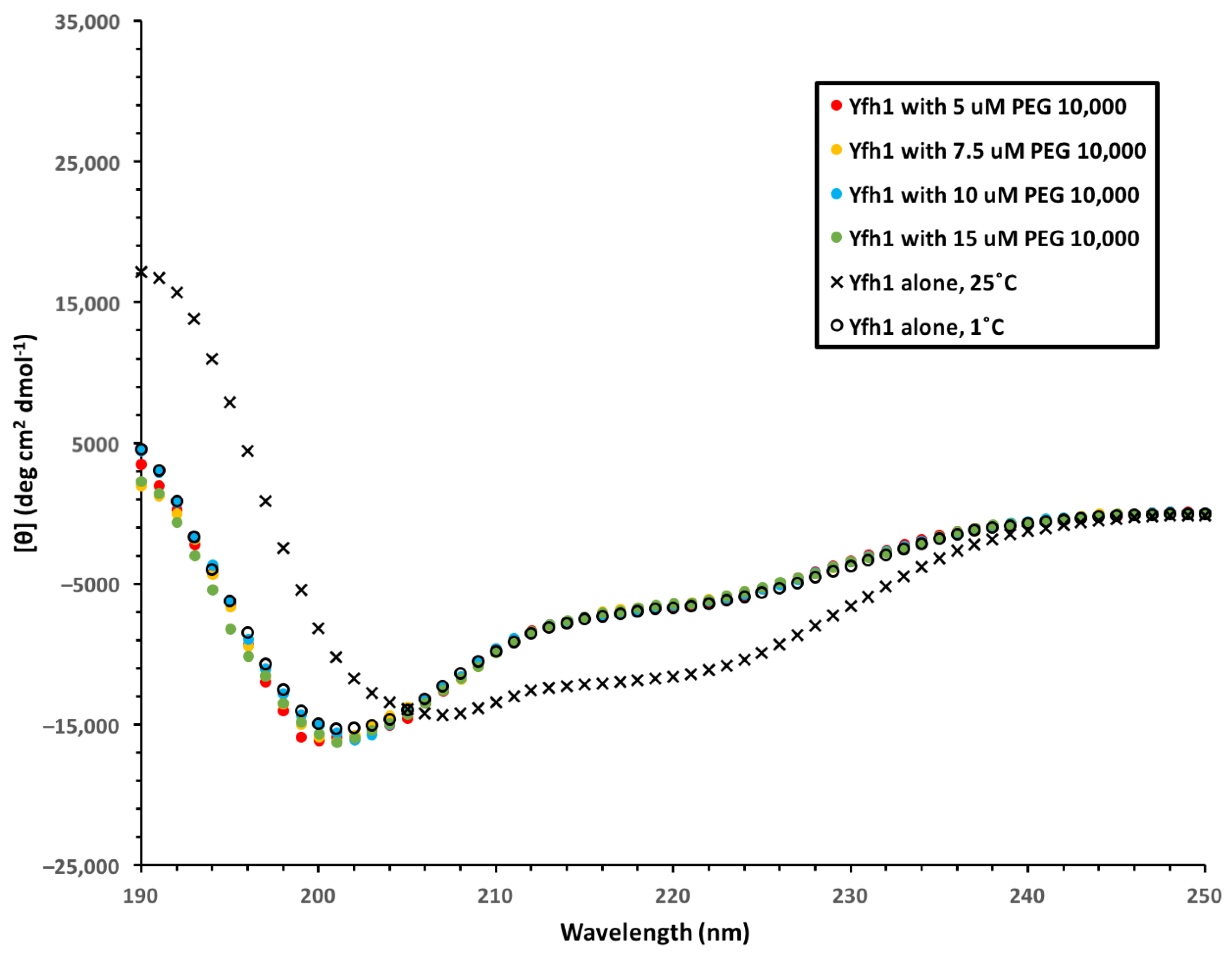
3.3. Yfh1 Secondary Structure
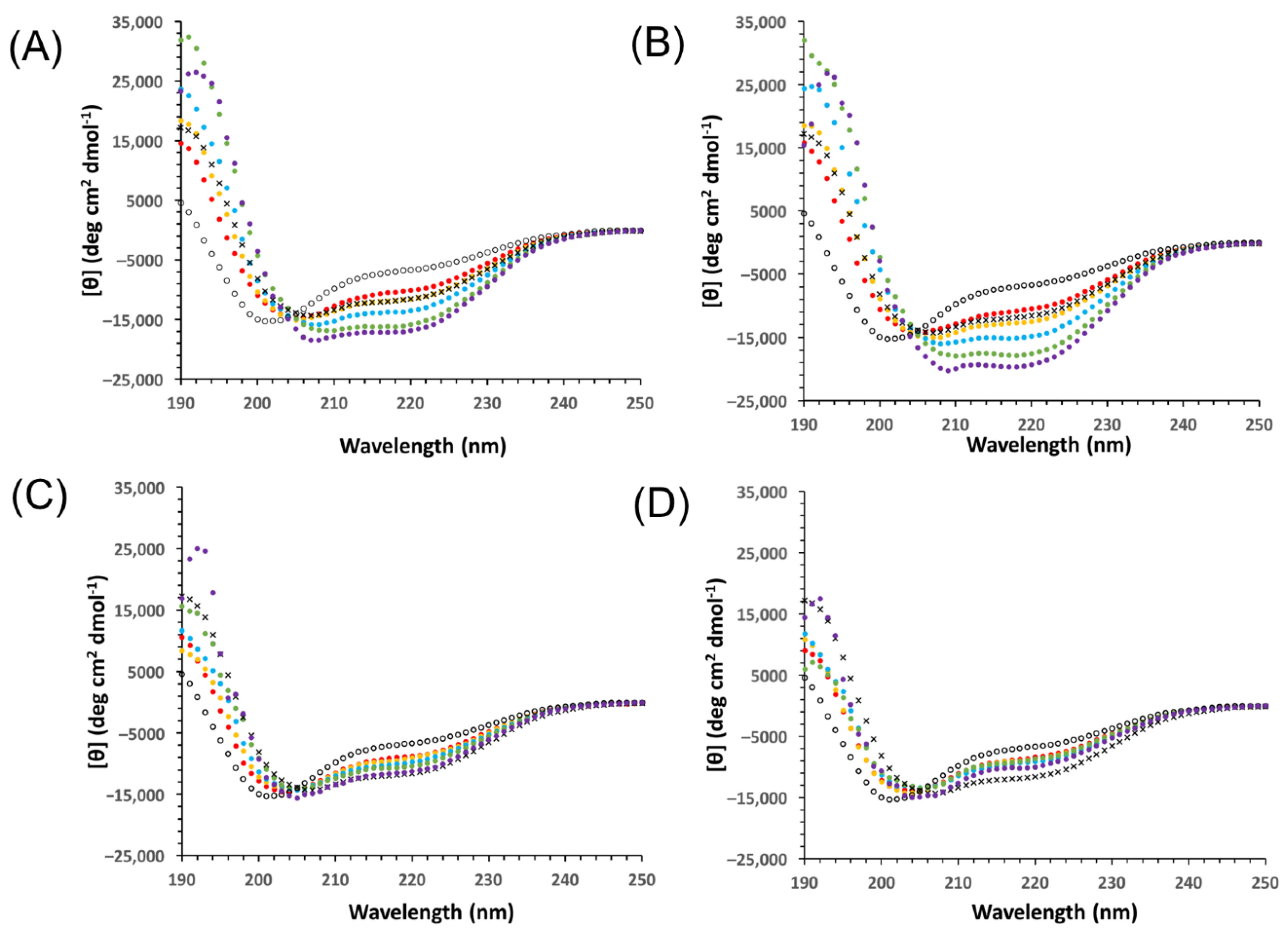
3.4. Yfh1 Secondary Structure in the Presence of Dehydrins
4. Discussion
4.1. Dehydrin Construct Design
4.2. Yfh1 Cryoprotection Is Affected by Charge Distribution
4.3. Yfh1 Is Not Protected by a Molecular Shield Effect
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steponkus, P.L.; Uemura, M.; Joseph, R.A.; Gilmour, S.J.; Thomashow, M.F. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 14570–14575. [Google Scholar] [CrossRef] [PubMed]
- Galau, G.A.; Hughes, D.W.; Dure, L., III. Abscisic acid induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol. 1986, 7, 155–170. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Die Nat. 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Targolli, J.; Huang, X.; Wu, R. Wheat LEA genes, PMA80 and PMA1959, enhance dehydration tolerance of transgenic rice (Oryza sativa L.). Mol. Breed. 2002, 10, 71–82. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Puhakainen, T.; Hess, M.W.; Mäkelä, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of Multiple Dehydrin Genes Enhances Tolerance to Freezing Stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef]
- Ismail, A.M.; Hall, A.E.; Close, T.J. Chilling Tolerance during Emergence of Cowpea Associated with a Dehydrin and Slow Electrolyte Leakage. Crop Sci. 1997, 37, 1270–1277. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Guan, Z.; Wei, W.; Han, L.; Chai, T. Expression and function of two dehydrins under environmental stresses in Brassica juncea L. Mol. Breed. 2007, 21, 431–438. [Google Scholar] [CrossRef]
- Xing, X.; Liu, Y.; Kong, X.; Liu, Y.; Li, D. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 2011, 65, 109–118. [Google Scholar] [CrossRef]
- Bao, F.; Du, D.; An, Y.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Overexpression of Prunus mume Dehydrin Genes in Tobacco Enhances Tolerance to Cold and Drought. Front. Plant Sci. 2017, 8, 151. [Google Scholar] [CrossRef]
- Hincha, D.K.; Thalhammer, A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 2012, 40, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Graether, S.P. The Disordered Dehydrin and Its Role in Plant Protection: A Biochemical Perspective. Biomolecules 2022, 12, 294. [Google Scholar] [CrossRef]
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome Analysis of Conserved Dehydrin Motifs in Vascular Plants. Front. Plant Sci. 2017, 8, 709. [Google Scholar] [CrossRef] [PubMed]
- Strimbeck, G.R. Hiding in plain sight: The F segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Riley, A.C.; Ashlock, D.A.; Graether, S.P. Evolution of the modular, disordered stress proteins known as dehydrins. PLoS ONE 2019, 14, e0211813. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 2001, 158, 1333–1339. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef]
- Tompa, P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005, 579, 3346–3354. [Google Scholar] [CrossRef]
- Graziano, G. On the molecular origin of cold denaturation of globular proteins. Phys. Chem. Chem. Phys. 2010, 12, 14245–14252. [Google Scholar] [CrossRef]
- Lin, C.; Thomashow, M.F. A cold-regulated Arabidopsis gene encodes A polypeptide having potent cryoprotective activity. Biochem. Biophys. Res. Commun. 1992, 183, 1103–1108. [Google Scholar] [CrossRef]
- Kazuoka, T.; Oeda, K. Purification and Characterization of COR85-Oligomeric Complex from Cold-Acclimated Spinach. Plant Cell Physiol. 1994, 35, 601–611. [Google Scholar] [CrossRef]
- Houde, M.; Daniel, C.; Lachapelle, M.; Allard, F.; Laliberte, S.; Sarhan, F. Immunolocalization of freezing-tolerance-associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J. 1995, 8, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Webb, R.; Balsamo, R.; Close, T.J.; Yu, X.M.; Griffith, M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant. 1999, 105, 600–608. [Google Scholar] [CrossRef]
- Bravo, L.A.; Gallardo, J.; Navarrete, A.; Olave, N.; Martínez, J.; Alberdi, M.; Close, T.J.; Corcuera, L.J. Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol. Plant. 2003, 118, 262–269. [Google Scholar] [CrossRef]
- Momma, M.; Kaneko, S.; Haraguchi, K.; Matsukura, U. Peptide mapping and assessment of cryoprotective activity of 26/27-kDa dehydrin from soybean seeds. Biosci. Biotech. Bioch. 2003, 67, 1832–1835. [Google Scholar] [CrossRef]
- Sanchez-Ballesta, M.T.; Rodrigo, M.J.; Lafuente, M.T.; Granell, A.A.; Zacarias, L. Dehydrin from Citrus, Which Confers in Vitro Dehydration and Freezing Protection Activity, Is Constitutive and Highly Expressed in the Flavedo of Fruit but Responsive to Cold and Water Stress in Leaves. J. Agric. Food Chem. 2004, 52, 1950–1957. [Google Scholar] [CrossRef]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 2005, 388, 151–157. [Google Scholar] [CrossRef]
- Kovacs, D.; Kalmár, É.; Torok, Z.; Tompa, P. Chaperone Activity of ERD10 and ERD14, Two Disordered Stress-Related Plant Proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef]
- Reyes, J.L.; Campos, F.; Wei, H.U.I.; Arora, R.; Yang, Y.; Karlson, D.T.; Covarrubias, A.A. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008, 31, 1781–1790. [Google Scholar] [CrossRef]
- Hughes, S.; Graether, S.P. Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 2010, 20, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-Segments of the Wheat Dehydrin DHN-5 are Essential for the Protection of Lactate Dehydrogenase and β-Glucosidase Activities In Vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz DMTralman-Baker, E.; Patel, S.N.; Graether, S.P. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liu, M.; Chen, Y.; Wang, J.; Lu, C. Heterologous expression of the dehydrin-like protein gene AmCIP from Ammopiptanthus mongolicus enhances viability of Escherichia coli and tobacco under cold stress. Plant Growth Regul. 2015, 79, 71–80. [Google Scholar] [CrossRef]
- Hara, M.; Endo, T.; Kamiya, K.; Kameyama, A. The role of hydrophobic amino acids of K-segments in the cryoprotection of lactate dehydrogenase by dehydrins. J. Plant Physiol. 2017, 210, 18–23. [Google Scholar] [CrossRef]
- Palmer, S.R.; De Villa, R.; Graether, S.P. Sequence composition versus sequence order in the cryoprotective function of an intrinsically disordered stress-response protein. Protein Sci. 2019, 28, 1448–1459. [Google Scholar] [CrossRef]
- Ohkubo, T.; Kameyama, A.; Kamiya, K.; Kondo, M.; Hara, M. F-segments of Arabidopsis dehydrins show cryoprotective activities for lactate dehydrogenase depending on the hydrophobic residues. Phytochemistry 2020, 173, 112300. [Google Scholar] [CrossRef]
- Pastore, A.; Martin, S.R.; Politou, A.; Kondapalli, K.C.; Stemmler, T.; Temussi, P.A. Unbiased cold denaturation: Low-and high-temperature unfolding of yeast frataxin under physiological conditions. J. Am. Chem. Soc. 2007, 129, 5374–5375. [Google Scholar] [CrossRef]
- Livernois, A.M.; Hnatchuk, D.J.; Findlater, E.E.; Graether, S.P. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Anal. Biochem. 2009, 392, 70–76. [Google Scholar] [CrossRef]
- Morrow, J.A.; Segall, M.L.; Lund-Katz, S.; Phillips, M.C.; Knapp, M.; Rupp, B.; Weisgraber, K.H. Differences in Stability among the Human Apolipoprotein E Isoforms Determined by the Amino-Terminal Domain. Biochemistry 2000, 39, 11657–11666. [Google Scholar] [CrossRef]
- Findlater, E.E.; Graether, S.P. NMR assignments of the intrinsically disordered K2 and YSK2 dehydrins. Biomol. NMR Assign. 2009, 3, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Bioinform. 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Venyaminov, S.; Baikalov, I.; Shen, Z.; Wu, C.; Yang, J. Circular Dichroic Analysis of Denatured Proteins: Inclusion of Denatured Proteins in the Reference Set. Anal. Biochem. 1993, 214, 17–24. [Google Scholar] [CrossRef]
- Tamiya, T.; Okahashi, N.; Sakuma, R.; Aoyama, T.; Akahane, T.; Matsumoto, J.J. Freeze denaturation of enzymes and its prevention with additives. Cryobiology 1985, 22, 446–456. [Google Scholar] [CrossRef]
- Holzwarth, G.; Doty, P. The Ultraviolet Circular Dichroism of Polypeptides1. J. Am. Chem. Soc. 1965, 87, 218–228. [Google Scholar] [CrossRef]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.K.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol. BioSyst. 2011, 8, 210–219. [Google Scholar] [CrossRef]
- Sanfelice, D.; Morandi, E.; Pastore, A.; Niccolai, N.; Temussi, P.A. Cold Denaturation Unveiled: Molecular Mechanism of the Asymmetric Unfolding of Yeast Frataxin. ChemPhysChem 2015, 16, 3599–3602. [Google Scholar] [CrossRef]
- Kiraga, J.; Mackiewicz, P.; Mackiewicz, D.; Kowalczuk, M.; Biecek, P.; Polak, N.; Smolarczyk, K.; Dudek, M.R.; Cebrat, S. The relationships between the isoelectric point and: Length of proteins, taxonomy and ecology of organisms. BMC Genom. 2007, 8, 163. [Google Scholar] [CrossRef]
- White, S.H. Amino acid preferences of small proteins: Implications for protein stability and evolution. J. Mol. Biol. 1992, 227, 991–995. [Google Scholar] [CrossRef]
- Tesei, G.; Vazdar, M.; Jensen, M.R.; Cragnell, C.; Mason, P.E.; Heyda, J.; Skepö, M.; Jungwirth, P.; Lund, M. Self-association of a highly charged arginine-rich cell-penetrating peptide. Proc. Natl. Acad. Sci. USA 2017, 114, 11428–11433. [Google Scholar] [CrossRef] [PubMed]
- Vazdar, M.; Uhlig, F.; Jungwirth, P. Like-Charge Ion Pairing in Water: An Ab Initio Molecular Dynamics Study of Aqueous Guanidinium Cations. J. Phys. Chem. Lett. 2012, 3, 2021–2024. [Google Scholar] [CrossRef]
- Ferrari, L.; Stucchi, R.; Konstantoulea, K.; van de Kamp, G.; Kos, R.; Geerts, W.J.; van Bezouwen, L.S.; Förster, F.G.; Altelaar, M.; Hoogenraad, C.C.; et al. Arginine π-stacking drives binding to fibrils of the Alzheimer protein Tau. Nat. Commun. 2020, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.W.; Boddington, K.F.; Warnica, J.M.; Atkinson, J.; McKenna, S.; Madge, J.; Barker, C.H.; Graether, S.P. Structural and Functional Insights into the Cryoprotection of Membranes by the Intrinsically Disordered Dehydrins. J. Biol. Chem. 2015, 290, 26900–26913. [Google Scholar] [CrossRef] [PubMed]
- Hasek, J. Poly(ethylene glycol) interactions with proteins. Z. Kristallogr. Suppl. 2006, 23, 613–618. [Google Scholar] [CrossRef]
- Rawat, S.; Suri, C.R.; Sahoo, D.K. Molecular mechanism of polyethylene glycol mediated stabilization of protein. Biochem. Biophys. Res. Commun. 2010, 392, 561–566. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, M.A.; Graether, S.P. The Effect of Positive Charge Distribution on the Cryoprotective Activity of Dehydrins. Biomolecules 2022, 12, 1510. https://doi.org/10.3390/biom12101510
Smith MA, Graether SP. The Effect of Positive Charge Distribution on the Cryoprotective Activity of Dehydrins. Biomolecules. 2022; 12(10):1510. https://doi.org/10.3390/biom12101510
Chicago/Turabian StyleSmith, Margaret A., and Steffen P. Graether. 2022. "The Effect of Positive Charge Distribution on the Cryoprotective Activity of Dehydrins" Biomolecules 12, no. 10: 1510. https://doi.org/10.3390/biom12101510
APA StyleSmith, M. A., & Graether, S. P. (2022). The Effect of Positive Charge Distribution on the Cryoprotective Activity of Dehydrins. Biomolecules, 12(10), 1510. https://doi.org/10.3390/biom12101510





