1. Introduction
Receptor transporter protein (RTP) is a family of receptor chaperone proteins that consists of four members (RTP1, RTP2, RTP3 and RTP4) that will transport G protein-coupled receptors to the cell surface membrane [
1]. RTP1 and 2 were first identified for their role in promoting cell-surface expression of odorant receptors (Saito et al., 2004), while RTP3 and 4 have been shown to increase surface expression of taste receptors in heterologous cells [
2]. In addition, we and others have reported that RTP4 has an important role as an opioid receptor chaperone. For instance, it mediates the maturation and trafficking of a heteromer consisting of a mu opioid receptor (MOPr) and delta opioid receptor (DOPr) (called an MOPr-DOPr heteromer) [
3]. However, the physiological and pathophysiological roles of the RTP4 chaperone remain to be assessed. Interestingly, the analysis of
Rtp expression in mouse brain [
4] reveals that RTP4 is the most abundant subtype of the RTP family in the brain. In addition, the mouse-brain database [
5] suggests that
Rtp4 is broadly expressed in neurons of various brain regions, including the hypothalamus, as well as in glia, including microglia, astrocyte, Schwann cells, satellite glia and enteric glia, creating an expectation of their role in brain function.
Prolonged or repeated MOPr activation is well known to lead to the development of antinociceptive tolerance to morphine [
6]. A previous study has shown that chronic morphine treatment upregulates MOPr-DOPr expression within various brain regions, including the hypothalamus [
7]. The disruption of the MOPr-DOPr heteromer by a TAT-peptide also suppresses morphine tolerance [
8], suggesting the possible involvement of the MOPr-DOPr heteromer. Recently, we have demonstrated that MOPr stimulation upregulates the gene expression of
Rtp4, a chaperone of the MOPr-DOPr heteromer, in both in vitro and in vivo systems [
9]. More importantly, chronic morphine treatment-induced RTP4 mRNA upregulation occurs in a hypothalamus-specific manner [
9]. Thus, in this study we directly tested the extent to which hypothalamic RTP4 plays in the development of morphine tolerance using conditional deletion of RTP4 using
Rtp4-floxed (
Rtp4flox) mice.
The hypothalamus consists of several nuclei, including the paraventricular nucleus (PVN), and all opioid receptor subtypes (MOPr, DOPr and KOPr) are expressed within these areas. The present study focused on the PVN region, since it plays an important role in the descending inhibitory system (i.e., PVN-induced descending antinociception) [
10]. Although the descending pathway has been considered to contribute to the antinociceptive tolerance to morphine [
11], the role of PVN has not been elucidated yet. The modulation of the neural circuit, including the PVN and ventral tegmental area, has been shown to inhibit the antinociceptive tolerance to morphine [
12], supporting the hypothesis of the possible involvement of PVN in the mechanism. In the present study, we targeted the PVN to locally decrease the level of RTP4 using adeno-associated viral (AAV)-mediated Cre-loxP recombination to achieve PVN-specific ablation of RTP4.
We find that chronic morphine treatment-induced morphine tolerance was diminished in PVN-specific RTP4 conditional knockout (cKO) mice. We also demonstrated that Gi activation and the MAPK pathway was involved in the mechanism of
Rtp4 gene induction after MOPr stimulation, by assessing the neuronal Neuro2A (N2A) cell line as our previous study [
9]. In addition, microglial cell line SIM-A9 cells also showed morphine-induced
Rtp4 upregulation, albeit requiring a higher concentration of morphine and Toll-like receptor 4 (TLR4) but not MOPr.
2. Materials and Methods
2.1. Chemicals
The following reagents were used: DAMGO ([D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt) (Sigma-Aldrich, Saint Louis, MO, USA); morphine hydrochloride (Takeda Pharmaceutical, Tokyo, Japan); pertussis toxin (PTX), an inhibitor of the G protein (Gi, Go and Gt) heterotrimer interaction with receptors (Tocris, Bio-Techne Japan, Tokyo, Japan); Actinomycin D (Act D), a transcriptional inhibitor (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan); U0126, a potent, selective inhibitor of MEK1 and 2 (Tocris, Bio-Techne Japan, Tokyo, Japan); and Pyridone 6, a potent pan-JAK inhibitor (Tocris, Bio-Techne Japan, Tokyo, Japan). Morphine was dissolved in saline for animal injection. For in vitro experiments, DAMGO, morphine and PTX were diluted with H2O, while the other reagents were diluted with dimethyl sulfoxide.
2.2. Animals
Male C57BL/6N mice (20–40 g, 12–24 weeks old) were purchased from CLEA Japan, Inc., and maintained on a 12-h light/dark cycle with rodent chow and water available ad libitum. Rtp4flox mice were generated, as described below, and male mutant mice (20–40 g, 12–24 weeks old) were used. All mice were maintained on a 12-h light/dark cycle with rodent chow and water available ad libitum, and they were housed in groups of four until testing. Animal studies were carried out according to protocols approved by the Nagasaki University Animal Care and Use Committee (approval number: 2002181596) and Institutional Review Board of Niigata University (approval number: SA00041).
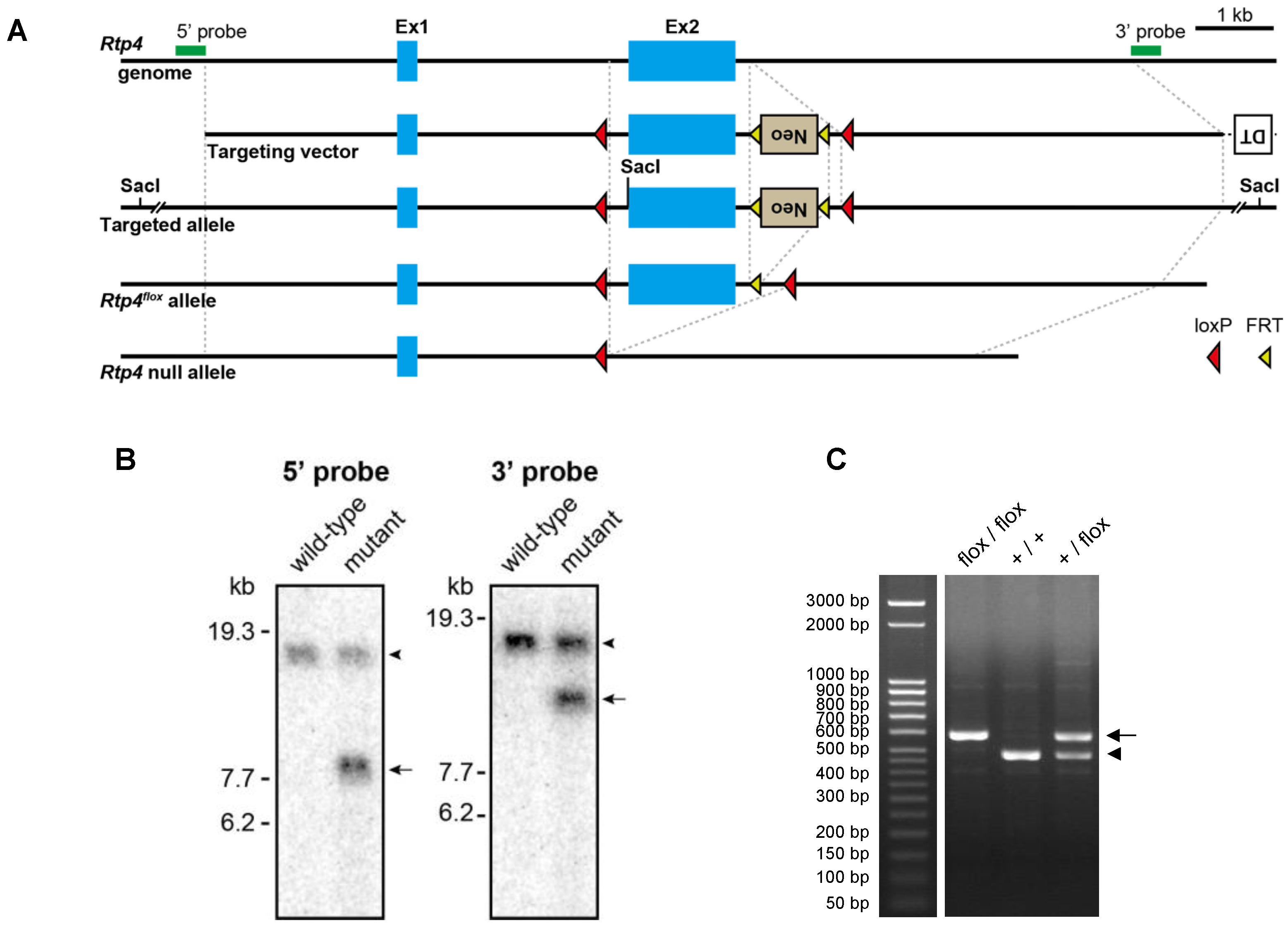
2.3. Generation of Rtp4flox Mice
To generate mice carrying floxed
Rtp4 alleles (
Rtp4flox), we constructed a gene-targeting vector containing loxP sites flanking exon 2 of the
Rtp4 gene and FRT-flanked neomycin (Neo) selection cassette. The plasmid vector was linearized and introduced into RENKA embryonic stem cells derived from the C57BL/6N strain and grown on feeder cells [
13]. Correctly targeted clones, validated by using Southern blot analysis, were injected into ICR blastocysts and transferred to pseudo-pregnant ICR females. The male chimeras were then crossed with female C57BL/6N mice. To excise the FRT-flanked Neo cassette, the F1 offspring with successful germline transmission of the targeted allele were crossed with Actb-FLP knock-in C57BL/6N mice [
14]. The resultant heterozygous
Rtp4flox/+ mice were intercrossed to obtain homozygous
Rtp4flox/flox C57BL/6N mice. Genotype was determined by using forward (CTAGCCTGGGAAGTTAAACCCTCG) and reverse (CCGGGTACATGTGGCACAAGATCA) PCR primers. To ablate the
Rtp4 gene in a PVN-specific manner, an AAV vector expressing CMV-driven Cre recombinase, fused to eGFP (Serotype: AAV9: Addgene.org/105545, Watertown, MA, USA) (AAV-Cre) or eGFP alone (Serotype: AAV9: Addgene.org/105530) (AAV-eGFP), as a control, was bilaterally injected into the PVN, as described below.
2.4. Stereotactic Surgery
For intracranial injections, mice (12–24 weeks old) were deeply anesthetized with a combination anesthetic (0.75 mg/kg medetomidine, 4.0 mg/kg midazolam and 5.0 mg/kg butorphanol) before surgery. The scalp was shaved, Povidone-Iodine (Isodine
®, Mundipharma K.K., Tokyo, Japan) was applied, and an incision was made along the midline of the scalp. The skull was scraped clean of periosteum. A micro drill (Tamiya, Inc., Shizuoka, Japan) was used to create a hole approximately 0.4 mm bilateral and 0.7 mm caudal to the bregma (bilateral PVN). A 300-nL aliquot of virus particle (2.85 × 10
9 genome copies/side) was given per hemisphere at 400-nL/min using a syringe pump (KDS-310PLUS) (kdScientific, Holliston, MA, USA). The needle was left to stand for 10 min and then removed within 2 min. The incision was closed with a soft silk suture and antibacterial ointment was applied to the wound. The animals were allowed to recover on a disposable heating pad until they recovered by using 0.75 mg/kg of atipamezole and were returned to their home cages in the animal facility for 4 weeks until the measurement. Cannula placement into the bilateral PVN was verified with trypan blue (4%). According to the previous research related to the in vivo virus genome introduction, the viral genome has been reported to remain episomal in the nucleus but maintains sustained expression in terminally differentiated cells for several weeks to months [
15,
16]. To obtain stable expression of Cre-recombinase by Cre-AAV injection, we decided to perform the evaluations in this study 4 weeks after injection of the AAV-Cre-eGFP or AAV-eGFP into the PVN.
2.5. Drug Administration
Morphine hydrochloride was dissolved in saline. Mice were treated with morphine hydrochloride (10 mg/kg) subcutaneously once a day for 11 days.
2.6. Tail-Flick Test
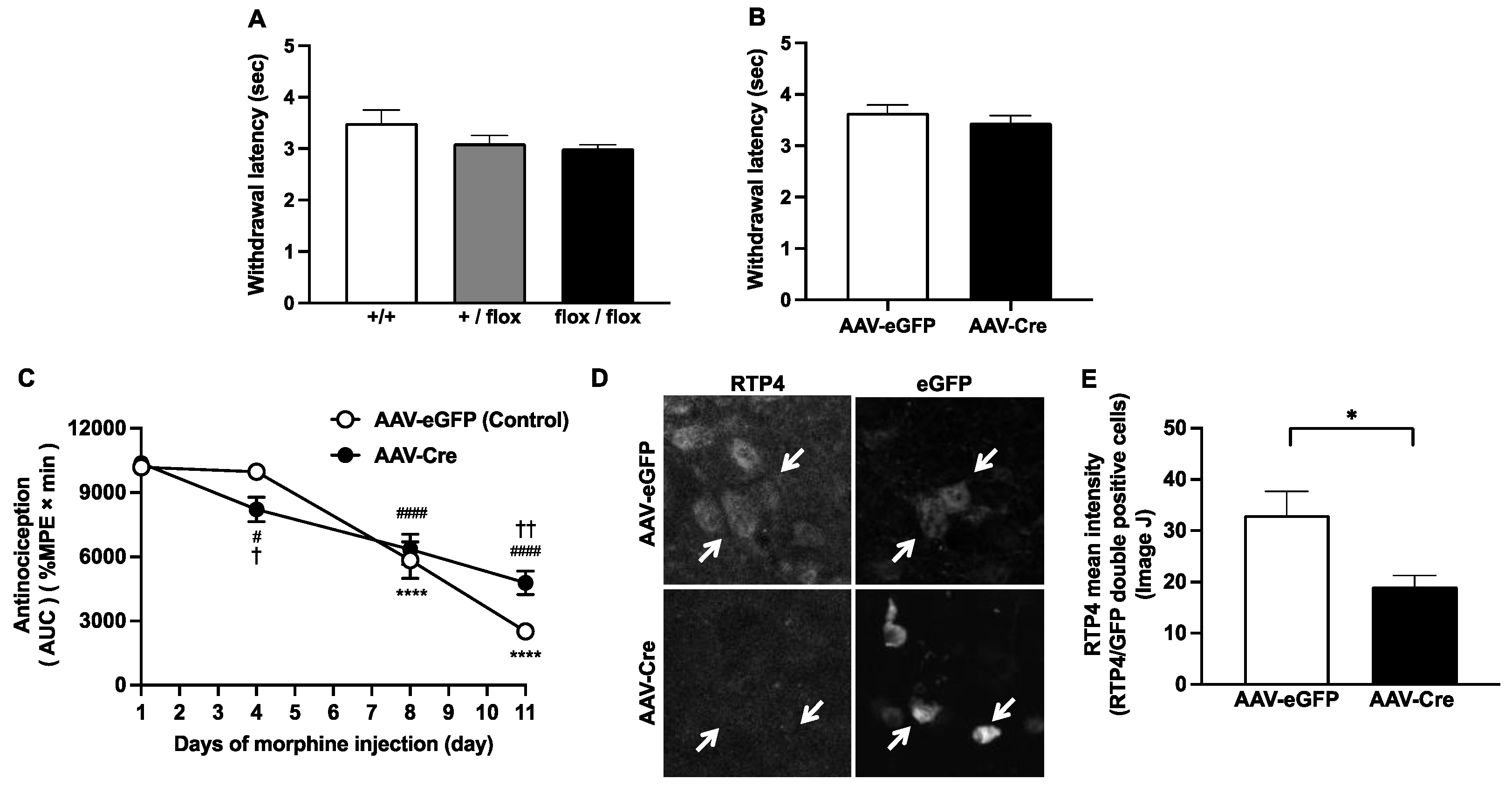
Four weeks after the AAV injection, morphine-induced antinociception was evaluated by using the tail-flick test [
6]. Using a tail-flick apparatus (IITC Life Science, Woodland Hills, CA, USA), the intensity of the heat source was set at 16–18, which resulted in the basal tail-flick latency occurring between 2 and 3 s for most of the animals. The tail-flick latency was recorded before (0 min, baseline latency) and at every 30 min after morphine injection (30, 60, 90, and 120 min); %MPE was calculated for each mouse at each time point on the indicated days (Days 4, 8 and 11) of morphine treatment according to the following formula: %MPE = ((latency after drug − baseline latency)/(10 − baseline latency)) × 100. Cutoff latency was selected at 10 s to minimize tissue damage. The area under the %MPE vs. time curves (area under the curve (AUC)) for each treatment condition is shown in Figure 2C.
2.7. Immunohistochemistry
On the day following the last morphine injection (i.e., 12th day), as described above, mice were deeply anesthetized with a combination anesthetic (0.75 mg/kg medetomidine, 4.0 mg/kg midazolam, and 5.0 mg/kg butorphanol) before surgery and transcardially perfused 2 min with ice-cold PBS and then for 8 min with ice-cold 4% paraformaldehyde (Wako, Osaka, Japan) for 8 min at a rate of 5–6 mL/min. Brains were extracted, post-fixed in 4% PFA at 4 °C overnight, and transferred to 10% (2 h), 20% (24 h) and then 30% sucrose in PBS for 24 h at 4 °C. Brains were frozen in O.C.T. compound (SAKURA, Tokyo, Japan) and stored at −80 °C. For virus expression and RTP4 expression verification, 20–25 µm coronal cryo-sections were cut using a cryostat (Leica CM3050) (Leica biosystems, Tokyo, Japan), dried and stored at −80 °C until use. For immunostaining, brain sections were gently washed 3 × 5 min in PBS and blocked with blocking solution (PBS containing 3% bovine serum albumin and 0.3% Triton X-100) for 1 h at room temperature (RT). Sections were then incubated in rabbit anti-RTP4 antibody (1:200) (MyBioSource, San Diego, CA, USA) in blocking solution at 4 °C overnight. Sections were rinsed 3 × 5 min with PBS and incubated secondary antibody (1:500) (Alexa-594-conjugated Goat anti-rabbit IgG) (Life Technologies Corp, Carlsbad, CA, USA) in blocking solution for 2 h at RT. Sections were washed 3 × 5 min with PBS, mounted on slides, and coverslipped with ProLongTM Diamond Antifade Mountant with DAPI (Thermo Fischer Scientific, Waltham, MA, USA). They were stored at −30 °C until analysis.
2.8. Imaging and Quantification by ImageJ Software
The histochemical analysis of the stained brain sections (from 2.7.) were performed on images acquired using a confocal laser scanning microscope (LSM-800) (Carl Zeiss Meditec Co., Ltd., Tokyo, Japan) (40 × objective). The signal intensity of RTP4 in the eGFP-co-localized area (cells) was calculated using macros in Image J software.
2.9. N2AMOPr Cell Culture and Transfection
In vitro experiments were performed as described previously [
9]. Briefly, Neuro 2A (N2A) cells were grown in complete growth medium (E-MEM with 10% FBS and 1% P/S). Cells were transfected with Flag-MOPr (N2A
MOPr cells) using Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Scientific Inc., Waltham, MA, USA). Twenty-four hours after transfection, cells were seeded into 24-well plates for further experiments.
2.10. Treatment of N2AMOPr Cells
The experiments were performed as described previously [
9]. Briefly, 24 h after plasmid transfection, N2A
MOPr cells were seeded into a 24-well plate (2 × 10
5 cells/ well) in complete growth medium and incubated overnight at 37 °C with 5% CO
2. Next day, media were replaced with 500 µL of the medium containing DAMGO or vehicle at the final concentration of 10 µM and incubated for 24 h. In one set of experiments, the cells were pretreated with inhibitors or vehicle at the final concentration of 100 ng/mL (PTX), 300 pM (ActD), 10 µM (U0126), 300 nM (Cal C) or 10 pM (Pyridone 6) for 24 h (PTX) or 30 min (others) followed by co-treatment with DAMGO and inhibitors for 24 h, as indicated in the figure legends. After ligand treatment, N2A
MOPr cells were washed once with cold phosphate-buffered saline (PBS) and then collected with 300 µL of cold RLT buffer (QIAGEN, Hilden, Germany). The cell lysates were transferred into an RNase-free centrifuge tube and stored at −80 °C until they were further processed for RT-qPCR.
2.11. SIM-A9 Cell Culture and Treatment
SIM-A9 cells (ATCC, Manassas, VA, USA) were grown in complete growth medium (D-MEM with 10% FBS, 5% HS, and 1% P/S) at 37 °C with a 5% CO2 condition. SIM-A9 cells (8 × 104) in culture medium were seeded into 24-well plates one day before the experiment. The next day, media were replaced with 500 µL of the complete growth medium containing morphine or vehicle at a final concentration of 10 µM to 1 mM and incubated for 3 to 48 h at 37 °C with 5% CO2. In one set of experiments, cells were pretreated with inhibitors or vehicle at a final concentration of 1 to 100 µM (naltrexone; NTX) or 1 to 10 µg/mL of TLR4/MD-2 monoclonal antibody (Thermo Fischer Scientific, Waltham, MA, USA) as indicated in the figures for 30 min followed by co-treatment with morphine and reagents for 24 h. After ligand treatment, SIM-A9 cells were collected the same as N2A cells.
2.12. RT-qPCR
RT-qPCR was performed as described previously [
9]. We extracted RNA-containing aqueous solution using the TRIzol reagent according to the manufacturer’s protocol (Thermo Fisher Scientific Inc.) before the total RNA purification. Total RNA was purified using the RNeasy Mini kit (QIAGEN Inc., Germantown, MD, USA) according to the manufacturer’s protocol. cDNA was synthesized using the PrimeScript
TM RT Master Mix (Takara, Shiga, Japan) according to the manufacturer’s protocol. Real-time PCR was performed using the Power SYBR Green qPCR Master Mix (Applied Biosystems, Foster City, CA, USA). The PCR template source was 4 µL of 10-times-diluted first-strand cDNA. Amplification was performed with an ABI PRISM 7900HT sequence-detection system (Applied Biosystems) or StepOne Real Time PCR system (Applied Biosystems). After an initial denaturation step at 95 °C for 10 min, amplification was performed using 45 cycles of denaturation (95 °C for 15 s), annealing (55 °C for 30 s) and extension (72 °C for 30 s). We amplified GAPDH, a housekeeping gene, as a control. The data were analyzed using the sequence-detection system software (version 2.2.1, for ABI PRISM 7900HT Software Applied Biosystems or version 2.3, for StepOne Software, Applied Biosystems) as described in Data Analysis. The software generates the baseline subtracted amplification plot of the normalized reporter values (ΔRn) versus cycle number. The amplification threshold was set at 6–7 of the ΔRn linear dynamic range (50–60% of maximum ΔRn). The fractional cycle at which the intersection of amplification threshold and the plot occurs is defined as the threshold cycle (Ct-value) for the plot. Samples that gave a Ct-value within 45 cycles were considered to be positive for mRNA expression. Then, quantitative analysis was performed using the ΔΔCT method, as described previously [
9,
17].
The forward (F) and reverse (R) primers were as follows:
GAPDH-F; TGAAGGTCGGTGTGAACG
GAPDH-R; CAATCTCCACTTTGCCACTG
RTP4-F; GGAGCCTGCATTTGGATAAG
RTP4-R; GCAGCATCTGGAACACTGG
TLR4-F; GCCTTTCAGGGAATTAAGCTCC
TLR4-R; AGATCAACCGATGGACGTGTAA
OPRM1-F; TCGCCTCCAACATCAGTTAG
OPRM1-R; TTTAGGGGATTGCCTTGATC
OPRD1-F; TCCCCATAACACAAATGCTG
OPRD1-R; CCGCCTTGAGATAACATCG
2.13. Data and Statistical Analysis
The data are expressed as the mean ± SEM. Student’s t-test or one-way ANOVA with a multiple-comparison test (Dunnett’s test or Tukey’s test) were used to analyze the data. A difference was considered to be significant at p < 0.05. All behavioral experiments were randomized, performed by a blinded researcher and then unblinded before statistical analysis. The results for each experiment were expressed as the mean ± SEM. The AUC was calculated for each animal with the baseline defined as zero. A normal distribution of the data was verified before performing parametric statistical analysis. Wherever appropriate, data were analyzed using two-way ANOVA, followed by Tukey’s post-hoc tests or multiple unpaired t-tests with significance set at p < 0.05. All calculations were performed using GraphPad Prism 9 software (GraphPad Software, Inc., San Diego, CA, USA) and EZR (version 1.40, Saitama Medical Center, Jichi Medical University, Saitama, Japan).
4. Discussion
In this study, we show that a moderate decrease in RTP4 expression in the PVN has a significant inhibitory effect on morphine tolerance. In our previous report,
Rtp4 was shown to be upregulated in the hypothalamus after repeated administration of morphine [
9]. Here, we determined the antinociceptive effect of morphine in locally RTP4-reduced mice in the hypothalamus that were generated by using a combination of novel
Rtp4flox/flox mice and local injection of Cre-expressing AAV into the PVN region of the brain.
Interestingly, AAV-Cre injection into the PVN of
Rtp4flox/flox mice partially but significantly delayed the development of antinociceptive tolerance to morphine after 11 days of morphine administration, as shown in
Figure 2C. These results suggest that RTP4 in PVN cells partly contribute to the mechanism of development of antinociceptive tolerance to morphine. Since there were no differences in pain sensitivity per se against heat stimulation in the tail-flick test with or without AAV-Cre injection, the downregulation of the
Rtp4 gene via the Cre-Lox system seems to simply affect the mechanism of development of antinociceptive tolerance to morphine. On the other hand, on Day 4 of repeated administration of morphine, rather, the AAV-Cre group exerted significantly lower levels of morphine antinociception than the control group. These results suggest that downregulation of RTP4 per se may induce some “compensatory effects” that “facilitate the morphine tolerance” if it is the case that RTP4 positively contribute to facilitating the development of antinociceptive tolerance to morphine.
Although the present study did not address whether the downregulation of RTP4 as a receptor chaperone protein could affect the expression of the MOPr-DOPr heteromer in vivo, this point will be shown in the future. Interestingly, a partial (~50%) but not complete RTP4 protein knockdown in the PVN was found to give a moderate phenotype. Improvement (i.e., the complete knockout of RTP4 in local brain region) will be expected by using a higher titter of AAV-Cre or neuron-selective pAAV in the future. In this study, we measured the expression levels of RTP4 in the eGFP-positive cells by immunohistochemistry, but a more definitive and quantitative measure of protein (Western blot) or mRNA (PCR) levels would also be valuable by extracting the eGFP-positive cells, while this remains the limitation in this study. Since RTP4 was shown to be broadly expressed in whole brain [
4], RTP4 in other brain regions and/or in other nuclei of the hypothalamus may also contribute to the mechanism, and thus it needs further determination.
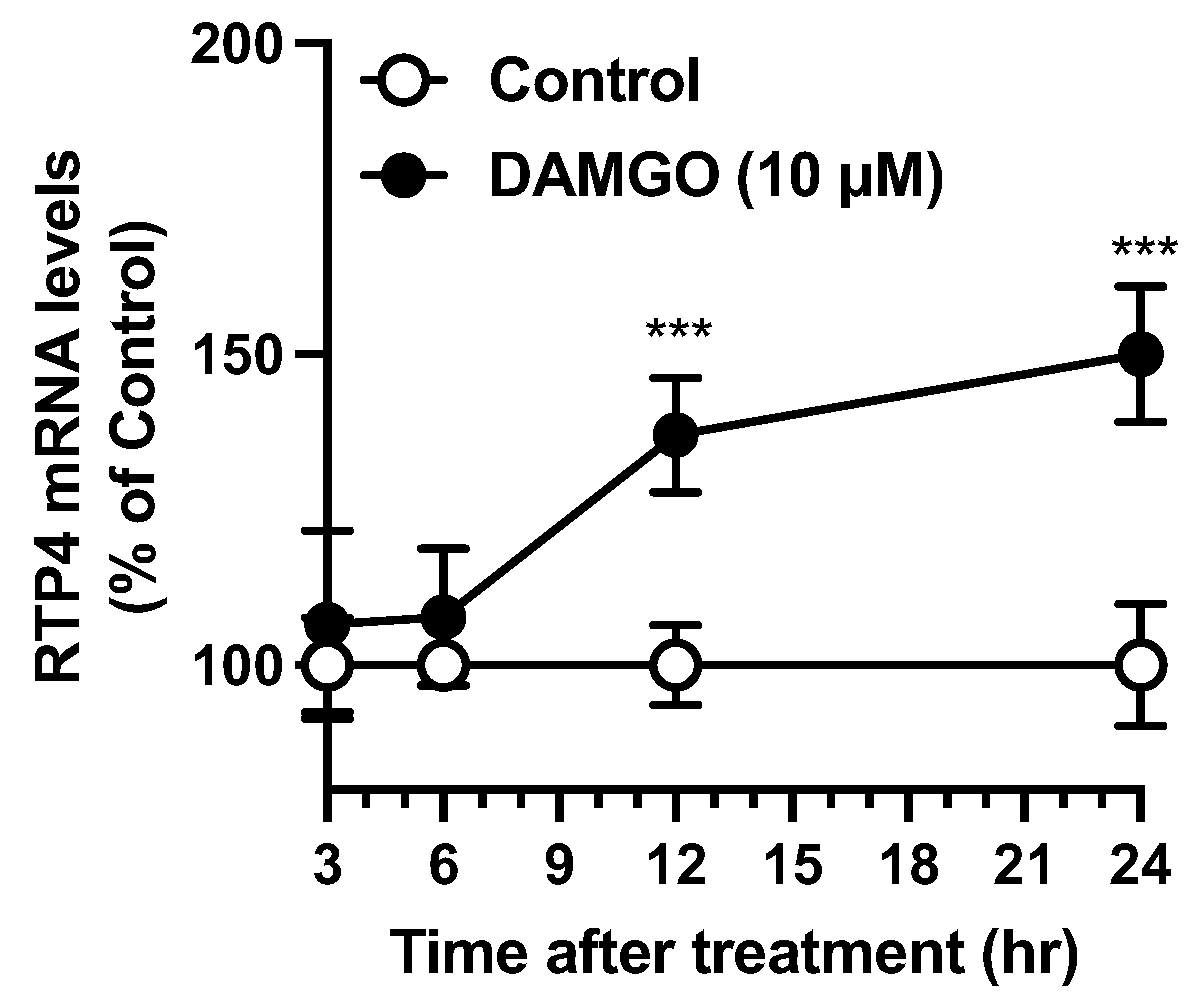
The in vitro results in the present study suggest that at least the neuronal cells could respond to MOPr stimulation and thus upregulate
Rtp4 gene via the Gi and MAPK pathway. Note that we have previously confirmed that the DAMGO-induced upregulation of the RTP4 mRNA levels was completely suppressed by MOPr antagonist NTX [
9], suggesting the involvement of MOPr in the mechanism. Regarding the transcription of the
Rtp4 gene, as RTP4 is known as one of the interferon (IFN)-stimulated genes (ISGs), whose expression is induced by the activation of the inflammatory or immune system [
18], it may be mediated by the signaling molecules underlying IFN receptors, such as the JAK-STAT pathway, which leads to rapid transcriptional activation of ISGs [
19,
20]. However, pyridone 6, a pan-type JAK inhibitor, did not affect the DAMGO-induced RTP4 gene induction in N2A
MOPr cells (
Figure 4E), suggesting that some distinct cellular responses were exerted under MOPr activation from those under IFNr activation.
The MAPK pathway includes a range of proteins, such as p38, ERK and JNK, involved in many faces of cellular regulation, from gene expression to cell death [
21,
22]. Our results showed that ERK-MAPK is involved in the mechanism of
Rtp4 induction, since U0126, which inhibits MEK1 and MEK2, and thus ERK activation, suppressed the DAMGO-induced upregulation of RTP4 mRNA levels (
Figure 4C). The opioid receptor is primarily controlled by interactions with G proteins and/or beta-arrestin [
23,
24,
25] and both pathways can activate MAPK. This study revealed Gi inhibition by PTX completely blocked the DAMGO-induced
Rtp4 induction, suggesting the importance of the Gi protein-mediated pathway for
Rtp4 gene induction, while it needs further determination on the contribution of the beta-arrestin-mediated pathway. In addition, the present results indicate the insignificant effect of inhibitors such as Cal C or Pyridone 6 on the RTP4 mRNA-inducing effect of DAMGO. However, as shown in
Figure 4D,E, that the significant RTP4 mRNA-inducing effect of DAMGO also disappeared in the presence of inhibitors may suggest the weak but not statistically significant involvement of PKC or JAK in the mechanism of the RTP4 mRNA-inducing effect of DAMGO, although it needs further determination.
Not only the neuronal cell, but also the involvement of glial cell, an essential contributor to the brain function, should be addressed, and our results revealed that microglial cells may respond to morphine only at a higher dose via TLR4 but not to MOPr stimulation (
Figure 5). Importantly, we confirmed that SIM-A9 cells surely possesses MOPr (OPRM1) mRNA while their expression levels are 200-times lower than that of TLR4 (
Table S1). As previously reported, microglial cells can be activated by morphine administration, although it is still controversial whether they can be stimulated via MOPr per se or not [
26,
27,
28]. Many studies have reported that morphine will be able to bind to TLR4 and the involvement of TLR4 in response to morphine in glial cells [
22], consistent with the results in the present study. Importantly, not only the neuronal cells, but also RTP4 has been shown to be highly expressed in macrophages, immunoreactive cells, according to a database [
4], suggesting the role of RTP4 in “microglia”, an immune cell equivalent to macrophage in the brain. Since chronic opioids can cause microglia and astrocyte activation, and interfering with glial function has been shown to reduce tolerance [
26,
29,
30], the involvement of crosstalk between the opioid pathway and glial or immune system may contribute to the mechanism of antinociceptive tolerance to morphine [
31]. Although the database suggests that RTP4 is expressed in most of the cell types, including neurons, astrocytes, microglia and Schwann cells, as described above, it would be important to reveal the cell-type specificity for the endogenous expression of RTP4 in PVN. Future studies addressing this using primary culture or immunohistochemical analysis of the brain sections from naive and morphine-tolerant mice are needed.
An increasing number of studies indicate that RTP4 can serve as one of the important markers of diagnosis or prognosis of various diseases, such as cutaneous melanoma, breast cancer, type 2 diabetes and viral infection [
18,
32,
33,
34]. Especially since ISGs including RTP4 possesses immuno-reactive properties, such as anti-virus activity [
18], it may be involved in the changes in the immune response under chronic morphine treatment [
31], although it needs further determination in the future. In addition, further investigation to distinguish the cell type (i.e., neurons or glia) where
Rtp4 will be upregulated after chronic morphine treatment in vivo (i.e., in the hypothalamus) should be addressed in the future.