From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy
Abstract
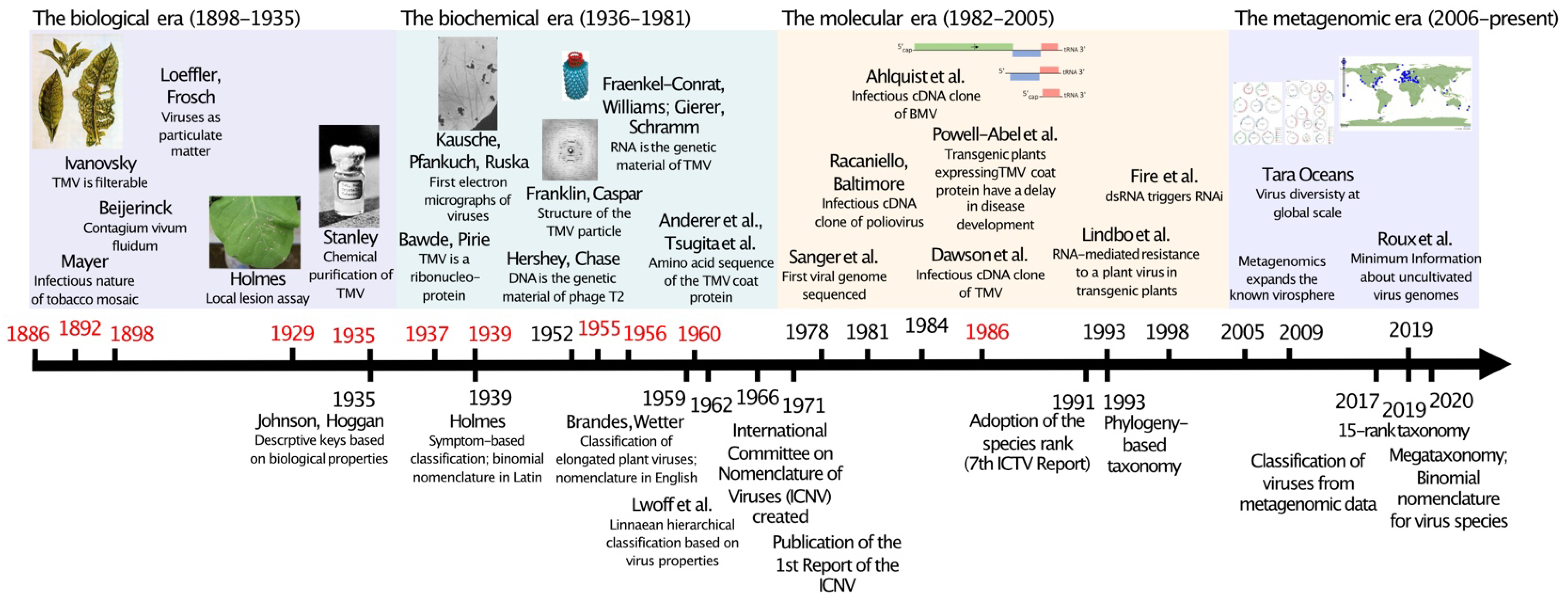
:1. Introduction
2. The Early Years of Virology and Initial Attempts at Classifying Viruses
3. The Biochemical Era and the Creation of the International Committee on Taxonomy of Viruses
4. The Molecular Era and the Concept of Virus Species
5. The Metagenomics Era—Unveiling the True Extent of the Virosphere
6. Tobacco Mosaic Virus: From Contagium vivum fluidum to Riboviria
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goodspeed, T.H. The Genus Nicotiana: Origins, Relationships and Evolution of Its Species in the Light of Their Distribution, Morphology and Cytogenetics; Chronica Botanica: Waltham, MA, USA, 1954; p. 536. [Google Scholar]
- Fischer, A.; Smith, E.F. The Fischer-Smith Controversy: Are There Bacterial Diseases of Plants? The American Phytopathological Society: Saint Paul, MN, USA, 1981. [Google Scholar]
- Mayer, A. Über die Mosaikkrankheit des Tabaks. Landwirtsch. Vers.-Stationen 1886, 32, 451–467. [Google Scholar]
- Ivanovsky, D. Über die Mosaikkrankheit der Tabakspflanze. Bull. L’Acad. Imp. Sci. St. Pétersbourg 1892, 35, 67–70. [Google Scholar]
- Beijerinck, M.W. Über ein Contagium vivum fluidum als Ursache der Fleckenkrankheit der Tabaksblätter. Verh. K. Akad. Weterschappen Amst. Afd. Natuurkunde 1898, 6, 3–21. [Google Scholar]
- Loeffler, F.; Frosch, P. Berichte der Kommission zur Erforschung der Maul- und Klauenseuche bei dem Institut für Infektionskrankheiten in Berlin. Zentrabl. Bacteriol. Parastenkunde Infektionkrankh. 1898, 23, 371–391. [Google Scholar]
- Stanley, W.M. Isolation of a crystalline protein possessing the properties of tobacco-mosaic virus. Science 1935, 81, 644–645. [Google Scholar] [CrossRef]
- Holmes, F.O. Local lesions in tobacco mosaic. Bot. Gaz. 1929, 87, 39–55. [Google Scholar] [CrossRef]
- Whitham, S.; Disnesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G. Making a virus visible: Francis O. Holmes and a biological assay for tobacco mosaic virus. J. Hist. Biol. 2014, 47, 107–145. [Google Scholar] [CrossRef]
- Johnson, J.; Hoggan, I.A. A descriptive key for plant viruses. Phytopathology 1935, 25, 328–343. [Google Scholar]
- Holmes, F.O. Proposal for extension of the binomial system of nomenclature to include viruses. Phytopathology 1939, 29, 431–436. [Google Scholar]
- Bawden, F.C.; Pirie, N.W.; Bernal, J.D.; Fankuchen, I. Liquid crystalline substances from virus-infected plants. Nature 1936, 138, 1051–1052. [Google Scholar] [CrossRef]
- Kausche, G.A.; Pfankuch, E.; Ruska, H. Die Sichtbarmachung von pflanzlichem Virus [TMV] im Übermikroskop. Naturwissenschaften 1939, 27, 292–299. [Google Scholar] [CrossRef]
- Von Borries, B.; Ruska, E.; Ruska, H. Bakterien und Virus in übermikroskopischer Aufnahme. Klin. Wochenschr. 1938, 17, 921–925. [Google Scholar] [CrossRef]
- Franklin, R.E. Structure of tobacco mosaic virus. Nature 1955, 175, 379–381. [Google Scholar] [CrossRef]
- Franklin, R.E. Structure of tobacco mosaic virus: Location of the ribonucleic acid in the tobacco mosaic virus particle. Nature 1956, 177, 928–930. [Google Scholar] [CrossRef]
- Caspar, D.L.D. Structure of tobacco mosaic virus: Radial density distribution in the tobacco mosaic virus particle. Nature 1956, 177, 928. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H. The role of nucleic acid in the reconstitution of active tobacco mosaic virus. J. Am. Chem. Soc. 1956, 78, 882–883. [Google Scholar] [CrossRef]
- Gierer, A.; Schramm, G. Infectivity of ribonucleic acid from tobacco mosaic virus. Nature 1956, 177, 702–703. [Google Scholar] [CrossRef]
- Anderer, F.A.; Uhlig, H.; Weber, E.; Schramm, G. Primary structure of the protein of tobacco mosaic virus. Nature 1960, 186, 922–925. [Google Scholar] [CrossRef]
- Tsugita, A.; Gish, D.T.; Young, J.; Fraenkel-Conrat, H.; Knight, C.A.; Stanley, W.M. The complete amino acid sequence of the protein of tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 1960, 46, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Brandes, J.; Wetter, C. Classification of elongated plant viruses on the basis of particle morphology. Virology 1959, 8, 99–115. [Google Scholar] [CrossRef]
- Lwoff, A.; Horne, R.; Tournier, P. A system of viruses. Cold Spring Harb. Sym. 1962, 27, 51–55. [Google Scholar] [CrossRef]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. 50 years of the International Committee on Taxonomy of Viruses: Progress and prospects. Arch. Virol. 2017, 162, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Wildy, P. Classification and Nomenclature of Viruses: First Report of the International Committee on Nomenclature of Viruses; S. Karger: Basel, Switzerland, 1971. [Google Scholar]
- Sanger, F.; Coulson, A.R.; Friedmann, T.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Fiddes, J.C.; Hutchison, C.A., 3rd; Slocombe, P.M.; Smith, M. The nucleotide sequence of bacteriophage phiX174. J. Mol. Biol. 1978, 125, 225–246. [Google Scholar] [CrossRef]
- Racaniello, V.R.; Baltimore, D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 1981, 214, 916–919. [Google Scholar] [CrossRef]
- Ahlquist, P.; French, R.; Janda, M.; Loesch-Fries, L.S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA 1984, 81, 7066–7070. [Google Scholar] [CrossRef]
- Dawson, W.O.; Beck, D.L.; Knorr, D.A.; Grantham, G.L. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc. Natl. Acad. Sci. USA 1986, 83, 1832–1836. [Google Scholar] [CrossRef]
- Meshi, T.; Ishikawa, M.; Motoyoshi, F.; Semba, K.; Okada, Y. In vitro transcription of infectious RNAs from full-length cDNAs of tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 1986, 83, 5043–5047. [Google Scholar] [CrossRef]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef]
- Wolf, S.; Deom, C.M.; Beachy, R.N.; Lucas, W.J. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 1989, 246, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Powell-Abel, P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef]
- Goldbach, R. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 1987, 4, 197–202. [Google Scholar] [PubMed]
- Franssen, H.; Leunissen, J.; Goldbach, R.; Lomonossoff, G.; Zimmern, D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984, 3, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Haseloff, J.; Goelet, P.; Zimmern, D.; Ahlquist, P.; Dasgupta, R.; Kaesberg, P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. USA 1984, 81, 4358–4362. [Google Scholar] [CrossRef]
- Padidam, M.; Beachy, R.N.; Fauquet, C.M. Classification and identification of geminiviruses using sequence comparisons. J. Gen. Virol. 1995, 76, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Antoniw, J.F.; Fauquet, C.M. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 2005, 150, 459–479. [Google Scholar] [CrossRef] [PubMed]
- Ávila, A.C.; De Haan, P.; Kormelink, R.; Resende, R.O.; Goldbach, R.W.; Peters, D. Classification of tospoviruses based on the phylogeny of nucleocapsid sequences. J. Gen. Virol. 1993, 74, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.A.; Lohuis, D.; Goldbach, R.; Dijkstra, J. Sequence data to settle the taxonomic position of bean common mosaic virus and blackeye cowpea mosaic virus isolates. J. Gen. Virol. 1993, 74, 2243–2249. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V.; Fauquet, C.M.; Bishop, D.H.L.; Carstens, E.; Estes, M.K.; Lemon, S.M.; Maniloff, J.; Mayo, M.A.; McGeoch, D.J.; Pringle, C.R.; et al. Virus Taxonomy: Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2000; p. 1162. [Google Scholar]
- Ansorge, W.J. Next-generation DNA sequencing techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Roossinck, M.J. Plant virus metagenomics: Biodiversity and ecology. Annu. Rev. Genet. 2012, 46, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant virus metagenomics: Advances in virus discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Krupovic, M.; Mushegian, A.; Kropinski, A.M.; Siddell, S.G.; Varsani, A.; Adams, M.J.; Davison, A.J.; Dutilh, B.E.; Harrach, B.; et al. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2020, 165, 2737–2748. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. Metaviromics: A tectonic shift in understanding virus evolution. Virus Res. 2018, 246, A1–A3. [Google Scholar] [CrossRef]
- Dolja, V.V.; Koonin, E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018, 244, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Metagenomics of plant and fungal viruses reveals an abundance of persistent lifestyles. Front. Microbiol. 2014, 5, 767. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Silas, S.; Wang, Y.; Wu, S.; Bocek, M.; Kazlauskas, D.; Krupovic, M.; Fire, A.; Dolja, V.V.; Koonin, E.V. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 2020, 5, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Zayed, A.A.; Wainaina, J.M.; Dominguez-Huerta, G.; Pelletier, E.; Guo, J.; Mohssen, M.; Tian, F.; Pratama, A.A.; Bolduc, B.; Zablocki, O.; et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 2022, 376, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Shi, M.; Holmes, E.C. Using Metagenomics to Characterize an Expanding Virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, Y.Z.; Holmes, E.C. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018, 243, 83–90. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zerbini, F.M.; Kitajima, E.W. From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy. Biomolecules 2022, 12, 1363. https://doi.org/10.3390/biom12101363
Zerbini FM, Kitajima EW. From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy. Biomolecules. 2022; 12(10):1363. https://doi.org/10.3390/biom12101363
Chicago/Turabian StyleZerbini, F. Murilo, and Elliot W. Kitajima. 2022. "From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy" Biomolecules 12, no. 10: 1363. https://doi.org/10.3390/biom12101363
APA StyleZerbini, F. M., & Kitajima, E. W. (2022). From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy. Biomolecules, 12(10), 1363. https://doi.org/10.3390/biom12101363






