NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy
Abstract
:1. Introduction
2. Materials and Methods
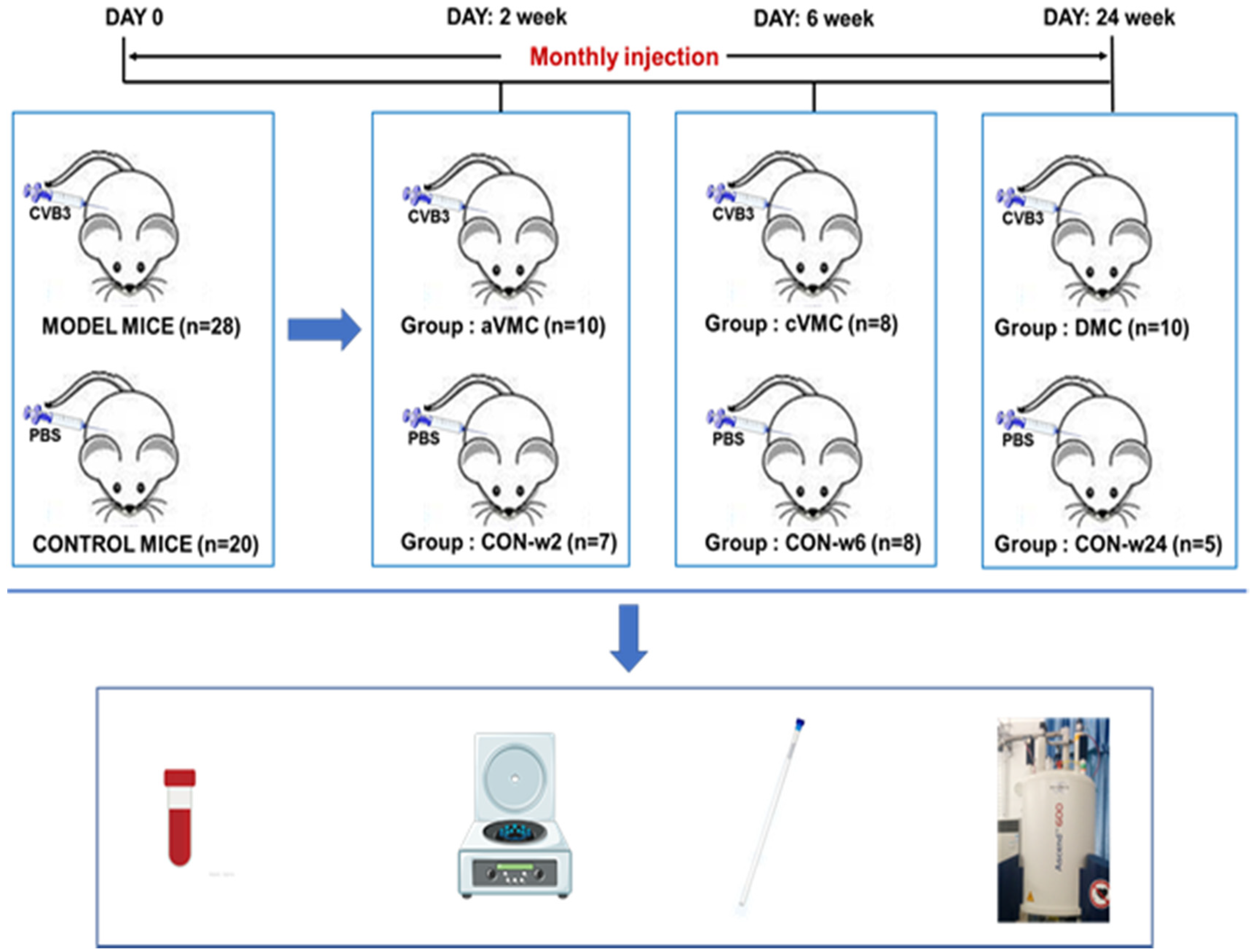
2.1. Mouse Models of Viral Myocarditis and Dilated Cardiomyopathy
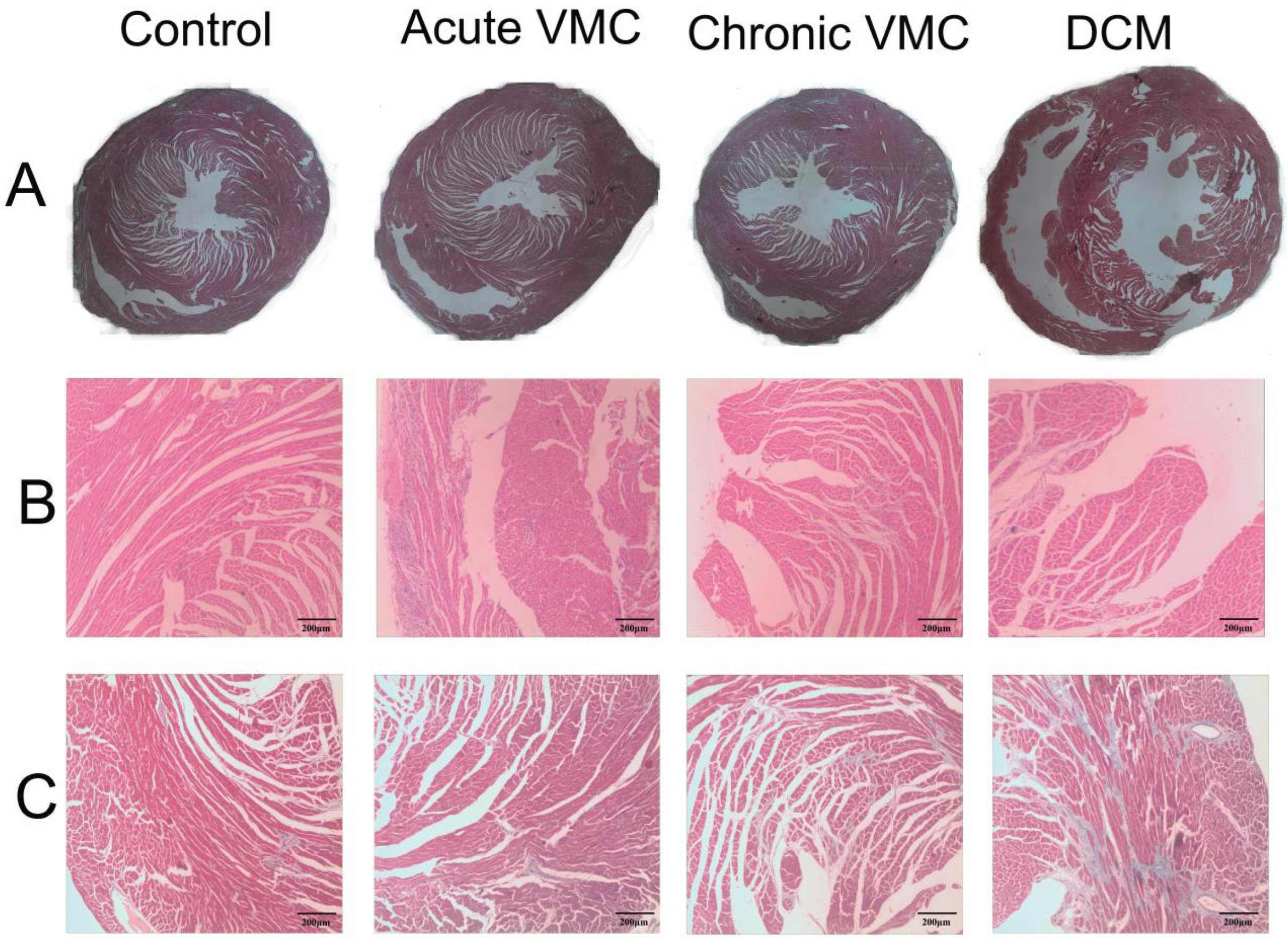
2.2. Histology
2.3. Plaque-Forming Assay
2.4. RNA Extraction and Real Time-PCR
2.5. Sample Preparation and 1H-NMR Spectroscopic Analysis
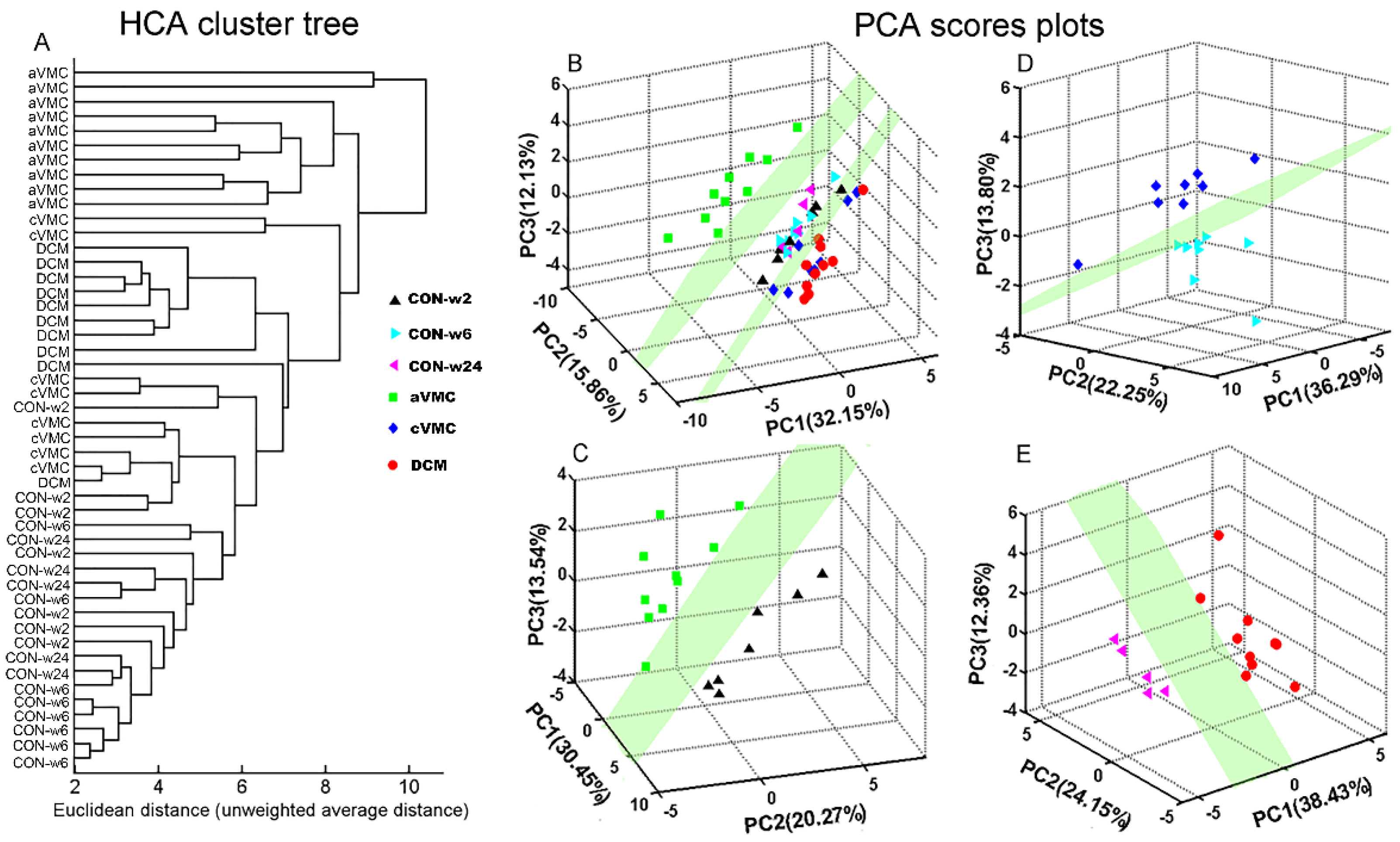
2.6. Multivariate Statistical Analysis
2.7. Identification of Significant Metabolites and Quantitative Comparison of Metabolite Levels
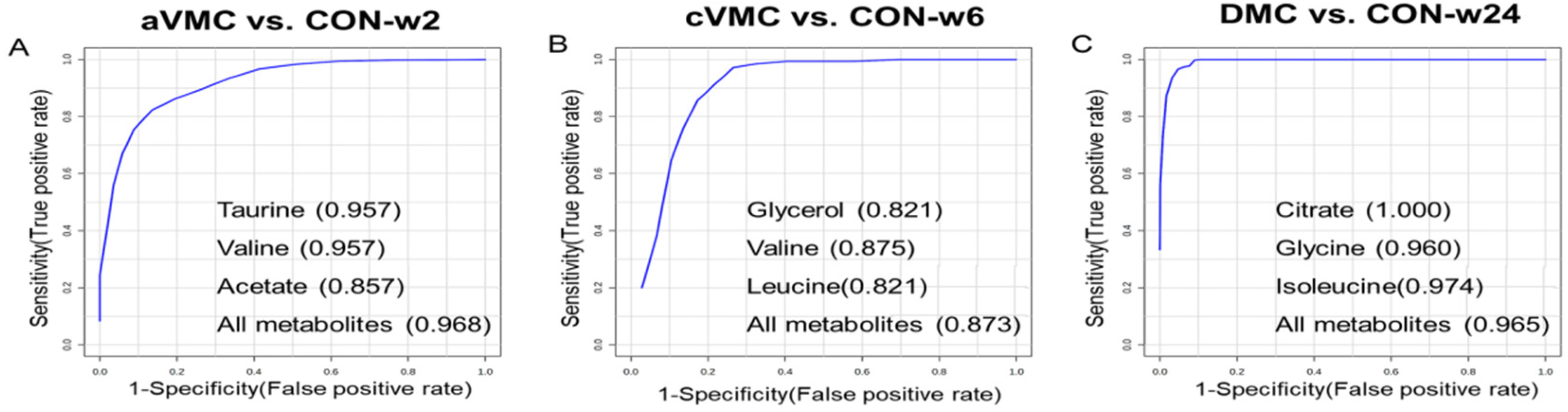
2.8. Multivariate Receiver Operating Characteristic (ROC) Curve Analysis
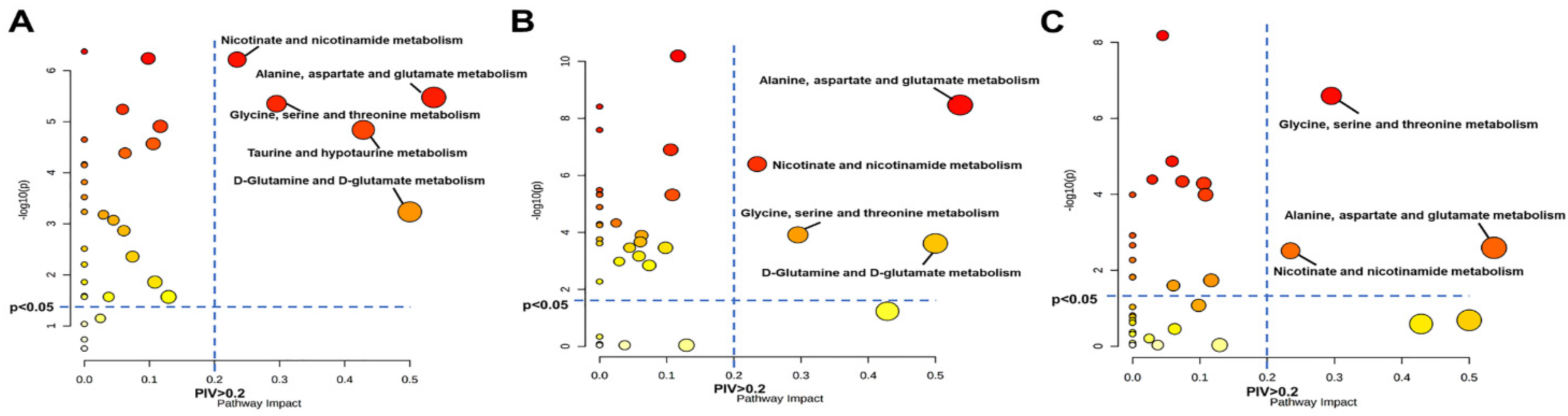
2.9. Metabolic Pathway Analysis
3. Results
3.1. Viral Myocarditis and Dilated Cardiomyopathy Induced by CVB3 in Mice
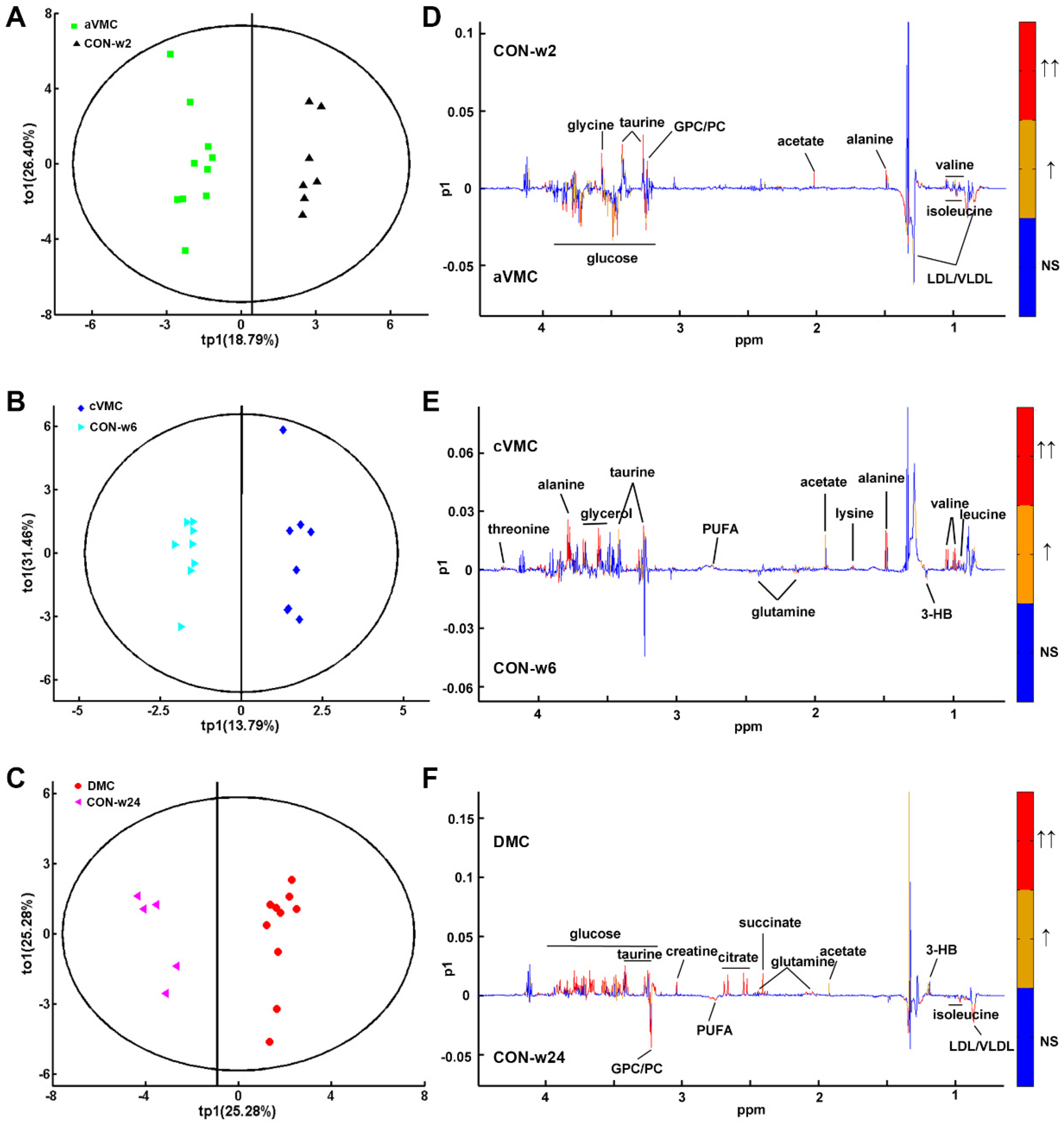
3.2. Metabolic Alterations in CVB3-Infected Groups Compared with Controls
3.3. Levels of the Metabolites Were Changed in the Three Pathological States
3.3.1. Amino Acid Metabolism
3.3.2. Carbohydrate Metabolism
3.3.3. Lipid Metabolism
3.3.4. Choline phosphorylation metabolism
3.4. Potential Biomarkers in the Progression from Acute VMC to DCM
3.5. Significantly Disturbed Metabolic Pathways in the Three Pathological Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| VMC | Viral myocarditis |
| DCM | Dilated cardiomyopathy |
| CVB3 | Coxsackievirus B3 |
| aVMC | Acute viral myocarditis |
| cVMC | Chronic viral myocarditis |
| CON | Normal controls |
| HF | Heart failure |
| PBS | Phosphate buffered saline |
| HCA | Hierarchical clustering analysis |
| PCA | Principal component analysis |
| RPTs | Response permutation tests |
| ROC | Receiver operating characteristic |
| PIV | Pathway impact values |
| LVEF | Left ventricle ejection fraction |
| BCAAs | Branch chain amino acids |
| PLS-DA | Partial least squares discriminant analysis |
| OPLS-DA | Orthogonal signal correction partial least-squares discriminant analysis |
References
- Gupta, S.; Markham, D.W.; Drazner, M.H.; Mammen, P.P. Fulminant myocarditis. Nat. Clin. Pract. Cardiovasc. Med. 2008, 5, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Blyszczuk, P.; Müller-Edenborn, B.; Valenta, T.; Osto, E.; Stellato, M.; Behnke, S.; Glatz, K.; Basler, K.; Lüscher, T.F.; Distler, O.; et al. Transforming growth factor-β-dependent Wnt secretion controls myofibroblast formation and myocardial fibrosis progression in experimental autoimmune myocarditis. Eur. Heart J. 2017, 38, 1413–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairweather, D.; Stafford, K.A.; Sung, Y.K. Update on coxsackievirus B3 myocarditis. Curr. Opin. Rheumatol. 2012, 24, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Kallwellis-Opara, A.; Dörner, A.; Poller, W.C.; Noutsias, M.; Kühl, U.; Schultheiss, H.P.; Pauschinger, M. Autoimmunological features in inflammatory cardiomyopathy. Clin. Res. Cardiol. 2007, 96, 469–480. [Google Scholar] [CrossRef]
- Gil, K.E.; Pawlak, A.; Gil, R.J.; Frontczak-Baniewicz, M.; Bil, J. The role of invasive diagnostics and its impact on the treatment of dilated cardiomyopathy: A systematic review. Adv. Med. Sci. 2016, 61, 331–343. [Google Scholar] [CrossRef]
- Stephenson, E.; Savvatis, K.; Mohiddin, S.A.; Marelli-Berg, F.M. T-cell immunity in myocardial inflammation: Pathogenic role and therapeutic manipulation. Br. J. Pharmacol. 2017, 174, 3914–3925. [Google Scholar] [CrossRef]
- Fairweather, D.; Rose, N.R. Coxsackievirus-induced myocarditis in mice: A model of autoimmune disease for studying immunotoxicity. Methods 2007, 41, 118–122. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Xie, Y.; Shen, E.; Li, G.G.; Yu, Y.; Zhang, C.B.; Yang, Y.; Zou, Y.; Ge, J.; Chen, R.; et al. Astragaloside IV attenuates myocardial fibrosis by inhibiting TGF-β1 signaling in coxsackievirus B3-induced cardiomyopathy. Eur. J. Pharmacol. 2011, 658, 168–174. [Google Scholar] [CrossRef]
- Guang-Yi, C.; Li-Sha, G.; Yue-Chun, L. Role of Heart Rate Reduction in the Management of Myocarditis. Curr. Pharm. Des. 2018, 24, 365–378. [Google Scholar] [CrossRef]
- Ong, S.; Rose, N.R.; Čiháková, D. Natural killer cells in inflammatory heart disease. Clin. Immunol. 2017, 175, 26–33. [Google Scholar] [CrossRef] [Green Version]
- McGarrah, R.W.; Crown, S.B.; Zhang, G.F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122, 1238–1258. [Google Scholar] [CrossRef]
- Ussher, J.R.; Elmariah, S.; Gerszten, R.E.; Dyck, J.R. The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2850–2870. [Google Scholar] [CrossRef] [PubMed]
- Halade, G.V.; Kain, V.; Tourki, B.; Jadapalli, J.K. Lipoxygenase drives lipidomic and metabolic reprogramming in ischemic heart failure. Metab. Clin. Exp. 2019, 96, 22–32. [Google Scholar] [CrossRef]
- Diguet, N.; Trammell, S.A.J.; Tannous, C.; Deloux, R.; Piquereau, J.; Mougenot, N.; Gouge, A.; Gressette, M.; Manoury, B.; Blanc, J.; et al. Nicotinamide Riboside Preserves Cardiac Function in a Mouse Model of Dilated Cardiomyopathy. Circulation 2018, 137, 2256–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, O.J.; Heckmann, M.B.; Ding, L.; Rapti, K.; Rangrez, A.Y.; Gerken, T.; Christiansen, N.; Rennefahrt, U.E.E.; Witt, H.; González Maldonado, S.; et al. Comprehensive plasma and tissue profiling reveals systemic metabolic alterations in cardiac hypertrophy and failure. Cardiovasc. Res. 2019, 115, 1296–1305. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, H.; Gao, J.; Zhu, M.; Wang, Y.; Jiang, C.; Xu, M. Metabolic disorder in the progression of heart failure. Sci. China. Life Sci. 2019, 62, 1153–1167. [Google Scholar] [CrossRef]
- Eriksson, U.; Ricci, R.; Hunziker, L.; Kurrer, M.O.; Oudit, G.Y.; Watts, T.H.; Sonderegger, I.; Bachmaier, K.; Kopf, M.; Penninger, J.M. Dendritic cell-induced autoimmune heart failure requires cooperation between adaptive and innate immunity. Nat. Med. 2003, 9, 1484–1490. [Google Scholar] [CrossRef]
- Querejeta, R.; Varo, N.; López, B.; Larman, M.; Artiñano, E.; Etayo, J.C.; Martínez Ubago, J.L.; Gutierrez-Stampa, M.; Emparanza, J.I.; Gil, M.J.; et al. Serum carboxy-terminal propeptide of procollagen type I is a marker of myocardial fibrosis in hypertensive heart disease. Circulation 2000, 101, 1729–1735. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; An, Y.; Li, N.; Liu, B.; Wang, Y.; Tang, H. Multiple univariate data analysis reveals the inulin effects on the high-fat-diet induced metabolic alterations in rat myocardium and testicles in the preobesity state. J. Proteome Res. 2013, 12, 3480–3495. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Rosen Vollmar, A.K.; Rattray, N.J.W. Normalizing Untargeted Periconceptional Urinary Metabolomics Data: A Comparison of Approaches. Metabolites 2019, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Tso, V.K.; Slupsky, C.M.; Fedorak, R.N. Metabolomics and detection of colorectal cancer in humans: A systematic review. Future Oncol. 2010, 6, 1395–1406. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Box, G.E. A general distribution theory for a class of likelihood criteria. Biometrika 1949, 36, 317–346. [Google Scholar] [CrossRef] [PubMed]
- Cloarec, O.; Dumas, M.E.; Trygg, J.; Craig, A.; Barton, R.H.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 2005, 77, 517–526. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Huang, C. Nuclear magnetic resonance-based tissue metabolomic analysis clarifies molecular mechanisms of gastric carcinogenesis. Cancer Sci. 2020, 111, 3195–3209. [Google Scholar] [CrossRef]
- Goeman, J.J.; Bühlmann, P. Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics 2007, 23, 980–987. [Google Scholar] [CrossRef] [Green Version]
- Lopaschuk, G.D.; Folmes, C.D.; Stanley, W.C. Cardiac energy metabolism in obesity. Circ. Res. 2007, 101, 335–347. [Google Scholar] [CrossRef] [Green Version]
- Maneikyte, J.; Bausys, A.; Leber, B. Dietary Glycine Prevents FOLFOX Chemotherapy-Induced Heart Injury: A Colorectal Cancer Liver Metastasis Treatment Model in Rats. Nutrients 2020, 12, 2634. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Karwi, Q.G.; Ho, K.L.; Pherwani, S.; Ketema, E.B. Ketone metabolism in the failing heart. Biochim. Et Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158813. [Google Scholar] [CrossRef] [PubMed]
- Greenwell, A.A.; Gopal, K.; Ussher, J.R. Myocardial Energy Metabolism in Non-ischemic Cardiomyopathy. Front. Physiol. 2020, 11, 570421. [Google Scholar] [CrossRef]
- Metzsch, C.; Liao, Q.; Steen, S.; Algotsson, L. Myocardial glycerol release, arrhythmias and hemodynamic instability during regional ischemia-reperfusion in an open chest pig model. Acta Anaesthesiol. Scand. 2006, 50, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, P.; Nordström, J.; Harms, H.J.; Lubberink, M.; Gadler, F.; Sörensen, J.; Mörner, S. Positron emission tomography ((15)O-water, (11)C-acetate, (11)C-HED) risk markers and nonsustained ventricular tachycardia in hypertrophic cardiomyopathy. IJC Heart Vasc. 2020, 26, 100452. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiong, Z.; Yan, W.; Gao, E.; Cheng, H.; Wu, G.; Liu, Y.; Zhang, L.; Li, C.; Wang, S.; et al. Branched chain amino acids exacerbate myocardial ischemia/reperfusion vulnerability via enhancing GCN2/ATF6/PPAR-α pathway-dependent fatty acid oxidation. Theranostics 2020, 10, 5623–5640. [Google Scholar] [CrossRef]
- Kimura, Y.; Okumura, T.; Kazama, S.; Shibata, N.; Oishi, H.; Arao, Y.; Kuwayama, T.; Kato, H.; Yamaguchi, S.; Hiraiwa, H.; et al. Usefulness of Plasma Branched-Chain Amino Acid Analysis in Predicting Outcomes of Patients with Nonischemic Dilated Cardiomyopathy. Int. Heart J. 2020, 61, 739–747. [Google Scholar] [CrossRef]
- Li, W.; Yang, J.; Lyu, Q.; Wu, G.; Lin, S.; Yang, Q. Taurine attenuates isoproterenol-induced H9c2 cardiomyocytes hypertrophy by improving antioxidative ability and inhibiting calpain-1-mediated apoptosis. Mol. Cell. Biochem. 2020, 469, 119–132. [Google Scholar] [CrossRef]
- Bkaily, G.; Jazzar, A.; Normand, A.; Simon, Y.; Al-Khoury, J.; Jacques, D. Taurine and cardiac disease: State of the art and perspectives. Can. J. Physiol. Pharmacol. 2020, 98, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Shao, H.; Dong, L.; Feng, Y.; Wang, C.; Tong, H. The protective effect of L-glutamine against acute Cantharidin-induced Cardiotoxicity in the mice. BMC Pharmacol. Toxicol. 2020, 21, 71. [Google Scholar] [CrossRef]
- Strifler, G.; Tuboly, E.; Görbe, A.; Boros, M.; Pécz, D.; Hartmann, P. Targeting Mitochondrial Dysfunction with L-Alpha Glycerylphosphorylcholine. PLoS ONE 2016, 11, e0166682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, P.; Fet, N.; Garab, D.; Szabó, A.; Kaszaki, J.; Srinivasan, P.K.; Tolba, R.H.; Boros, M. L-α-glycerylphosphorylcholine reduces the microcirculatory dysfunction and nicotinamide adenine dinucleotide phosphate-oxidase type 4 induction after partial hepatic ischemia in rats. J. Surg. Res. 2014, 189, 32–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tőkés, T.; Tuboly, E.; Varga, G.; Major, L.; Ghyczy, M.; Kaszaki, J.; Boros, M. Protective effects of L-alpha-glycerylphosphorylcholine on ischaemia-reperfusion-induced inflammatory reactions. Eur. J. Nutr. 2015, 54, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuboly, E.; Gáspár, R.; Ibor, M.O.; Gömöri, K.; Kiss, B.; Strifler, G.; Hartmann, P.; Ferdinandy, P.; Bartekova, M.; Boros, M.; et al. L-Alpha-glycerylphosphorylcholine can be cytoprotective or cytotoxic in neonatal rat cardiac myocytes: A double-edged sword phenomenon. Mol. Cell. Biochem. 2019, 460, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| CON-w2 | CON-w6 | CON-w24 | aVMC | cVMC | DCM | |
|---|---|---|---|---|---|---|
| Amino acid metabolism | ||||||
| leucine | 0.814 ± 0.114 | 0.834 ± 0.060 | 0.866 ± 0.102 | 0.866 ± 0.123 | 0.917 ± 0.063↑ | 0.677 ± 0.090↓↓ |
| isoleucine | 0.148 ± 0.024 | 0.144 ± 0.015 | 0.153 ± 0.022 | 0.125 ± 0.050 | 0.173 ± 0.028↑ | 0.124 ± 0.031 |
| valine | 0.397 ± 0.035 | 0.396 ± 0.034 | 0.420 ± 0.083 | 0.267 ± 0.078↓↓↓ | 0.514 ± 0.085↑↑ | 0.363 ± 0.088 |
| threonine | 0.256 ± 0.077 | 0.227 ± 0.061 | 0.213 ± 0.056 | 0.261 ± 0.094 | 0.248 ± 0.066 | 0.220 ± 0.052 |
| glycine | 0.333 ± 0.020 | 0.339 ± 0.067 | 0.327 ± 0.013 | 0.284 ± 0.037↓↓ | 0.346 ± 0.041 | 0.448 ± 0.087↑↑ |
| lysine | 0.508 ± 0.071 | 0.499 ± 0.057 | 0.505 ± 0.051 | 0.465 ± 0.138 | 0.589 ± 0.070↑ | 0.520 ± 0.083 |
| alanine | 0.314 ± 0.020 | 0.351 ± 0.044 | 0.348 ± 0.034 | 0.285 ± 0.055 | 0.372 ± 0.046 | 0.401 ± 0.061 |
| taurine | 1.622 ± 0.192 | 1.511 ± 0.103 | 1.537 ± 0.104 | 1.246 ± 0.148↓↓↓ | 1.641 ± 0.096↑ | 1.926 ± 0.143↑↑↑ |
| glutamine | 0.767 ± 0.142 | 0.723 ± 0.055 | 0.761 ± 0.091 | 0.906 ± 0.245 | 0.647 ± 0.072↓ | 1.007 ± 0.168↑↑ |
| Carbohydrate metabolism | ||||||
| creatine | 0.384 ± 0.070 | 0.364 ± 0.039 | 0.382 ± 0.074 | 0.320 ± 0.074 | 0.326 ± 0.048 | 0.498 ± 0.092↑ |
| acetate | 0.231 ± 0.057 | 0.223 ± 0.072 | 0.189 ± 0.037 | 0.154 ± 0.025↓↓ | 0.311 ± 0.084↑ | 0.282 ± 0.073↑ |
| glucose | 3.421 ± 0.425 | 3.066 ± 0.353 | 3.342 ± 0.113 | 2.971 ± 0.399↓ | 2.596 ± 0.285↓ | 3.855 ± 0.324↑↑↑ |
| lactate | 4.266 ± 0.772 | 4.529 ± 0.389 | 4.632 ± 0.767 | 4.356 ± 0.475 | 4.215 ± 0.511 | 5.468 ± 0.513↑ |
| succinate | 0.195 ± 0.108 | 0.196 ± 0.075 | 0.160 ± 0.062 | 0.221 ± 0.076 | 0.166 ± 0.025 | 0.328 ± 0.128↑ |
| citrate | 0.224 ± 0.060 | 0.253 ± 0.042 | 0.243 ± 0.033 | 0.242 ± 0.078 | 0.241 ± 0.029 | 0.586 ± 0.158↑↑↑ |
| Lipid metabolism | ||||||
| LDL/VLDL | 7.579 ± 0.462 | 7.941 ± 0.724 | 7.240 ± 0.601 | 8.955 ± 0.801↑↑↑ | 8.696 ± 1.911 | 5.607 ± 0.546↓↓↓ |
| PUFA | 2.254 ± 0.382 | 2.109 ± 0.321 | 2.086 ± 0.204 | 2.216 ± 0.471 | 2.495 ± 0.355↑ | 1.526 ± 0.284↓↓ |
| 3-HB | 0.440 ± 0.109 | 0.460 ± 0.057 | 0.482 ± 0.161 | 0.462 ± 0.100 | 0.405 ± 0.161 | 0.760 ± 0.247↑ |
| glycerol | 1.430 ± 0.133 | 1.478 ± 0.239 | 1.449 ± 0.050 | 1.399 ± 0.378 | 1.700 ± 0.113↑ | 2.098 ± 0.320↑↑↑ |
| Choline phosphorylation metabolism | ||||||
| GPC | 4.104 ± 0.377 | 4.459 ± 0.700 | 4.269 ± 0.351 | 3.470 ± 0.538↓ | 4.544 ± 0.440 | 3.712 ± 0.363↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Q.; Gu, J.; Lu, R.; Huang, C.; Hu, X.; Wu, W.; Lin, D. NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Biomolecules 2022, 12, 112. https://doi.org/10.3390/biom12010112
Kong Q, Gu J, Lu R, Huang C, Hu X, Wu W, Lin D. NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Biomolecules. 2022; 12(1):112. https://doi.org/10.3390/biom12010112
Chicago/Turabian StyleKong, Qing, Jinping Gu, Ruohan Lu, Caihua Huang, Xiaomin Hu, Weifeng Wu, and Donghai Lin. 2022. "NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy" Biomolecules 12, no. 1: 112. https://doi.org/10.3390/biom12010112
APA StyleKong, Q., Gu, J., Lu, R., Huang, C., Hu, X., Wu, W., & Lin, D. (2022). NMR-Based Metabolomic Analysis of Sera in Mouse Models of CVB3-Induced Viral Myocarditis and Dilated Cardiomyopathy. Biomolecules, 12(1), 112. https://doi.org/10.3390/biom12010112






