An Arsenite Relay between PSMD14 and AIRAP Enables Revival of Proteasomal DUB Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Transfection, Lysis and Protein Purification
2.2. Proteasomal Psmd14 DUB Assay
2.3. Sic1 Ubiquitination
2.4. Antibodies
2.5. Recombinant AIRAP Purification and PAO Binding Assay
2.6. ICP-OES and MST Measurements
3. Results
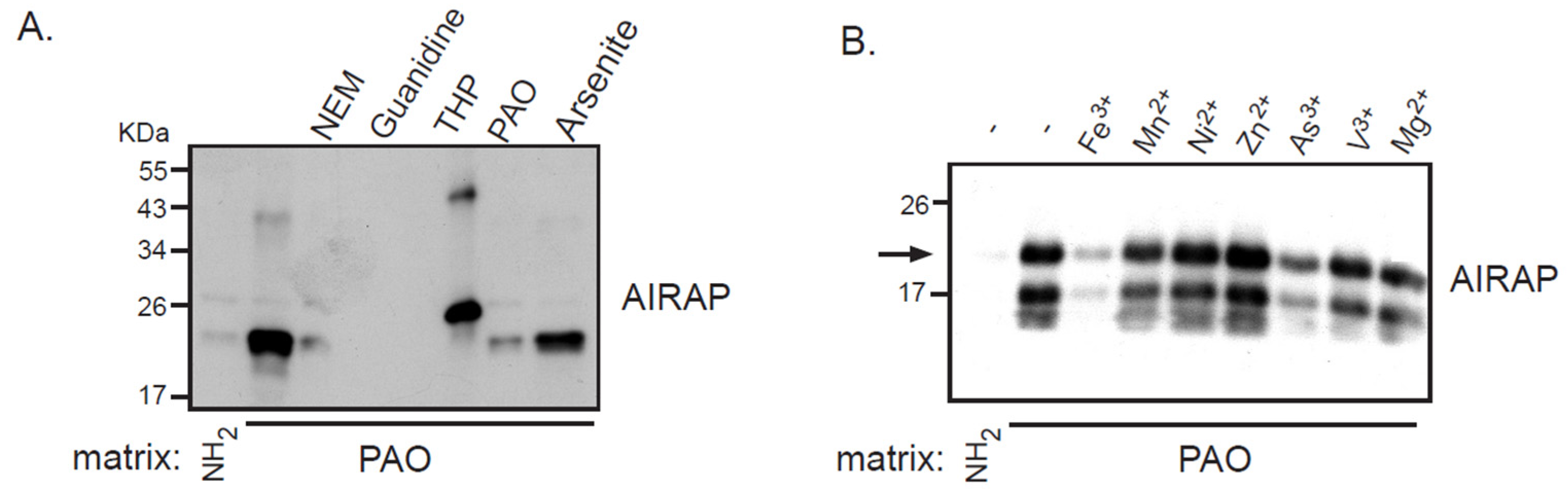
3.1. Recombinant AIRAP Binding to Arsenite
3.2. AIRAP Binding to Arsenite In Vivo
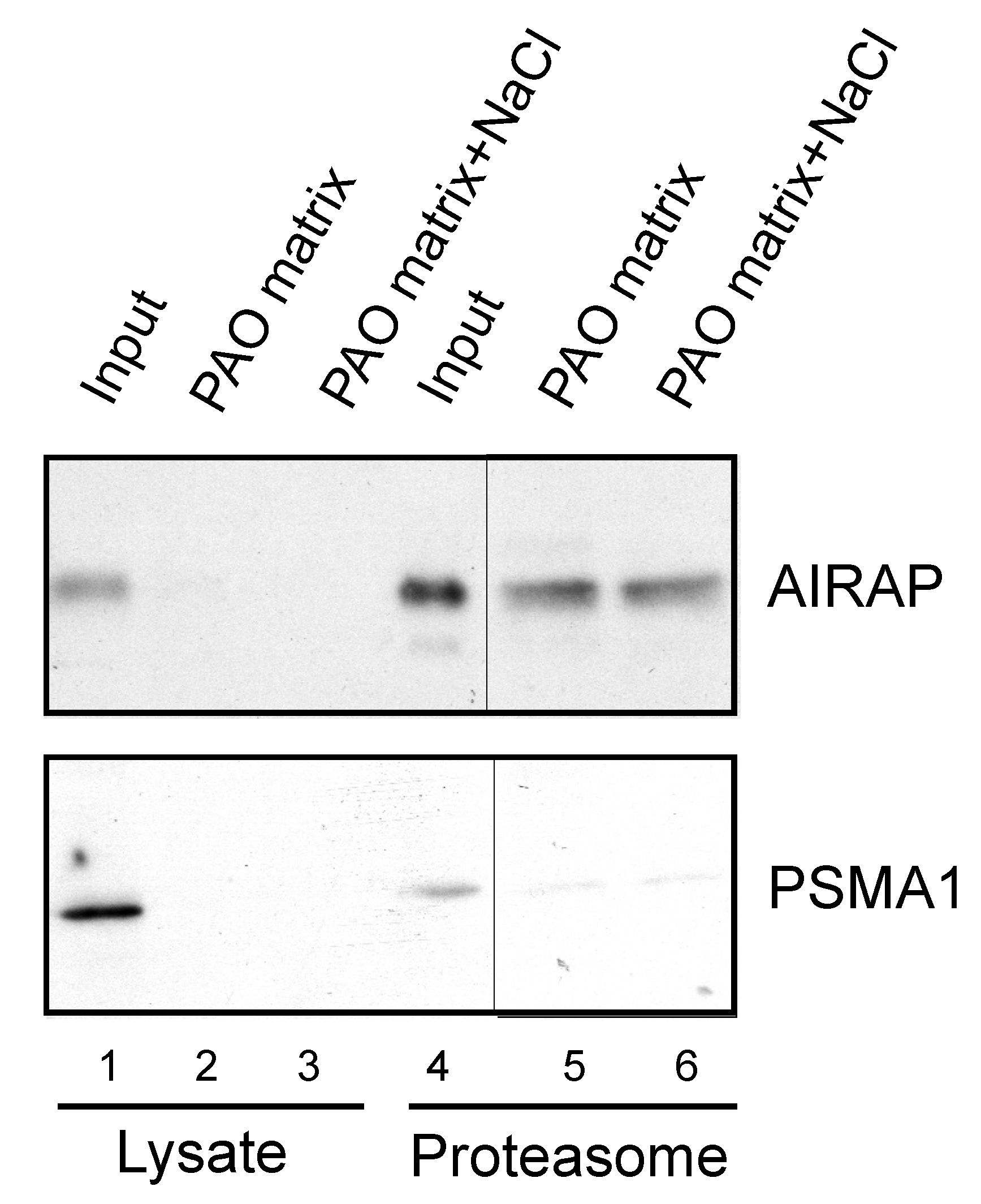
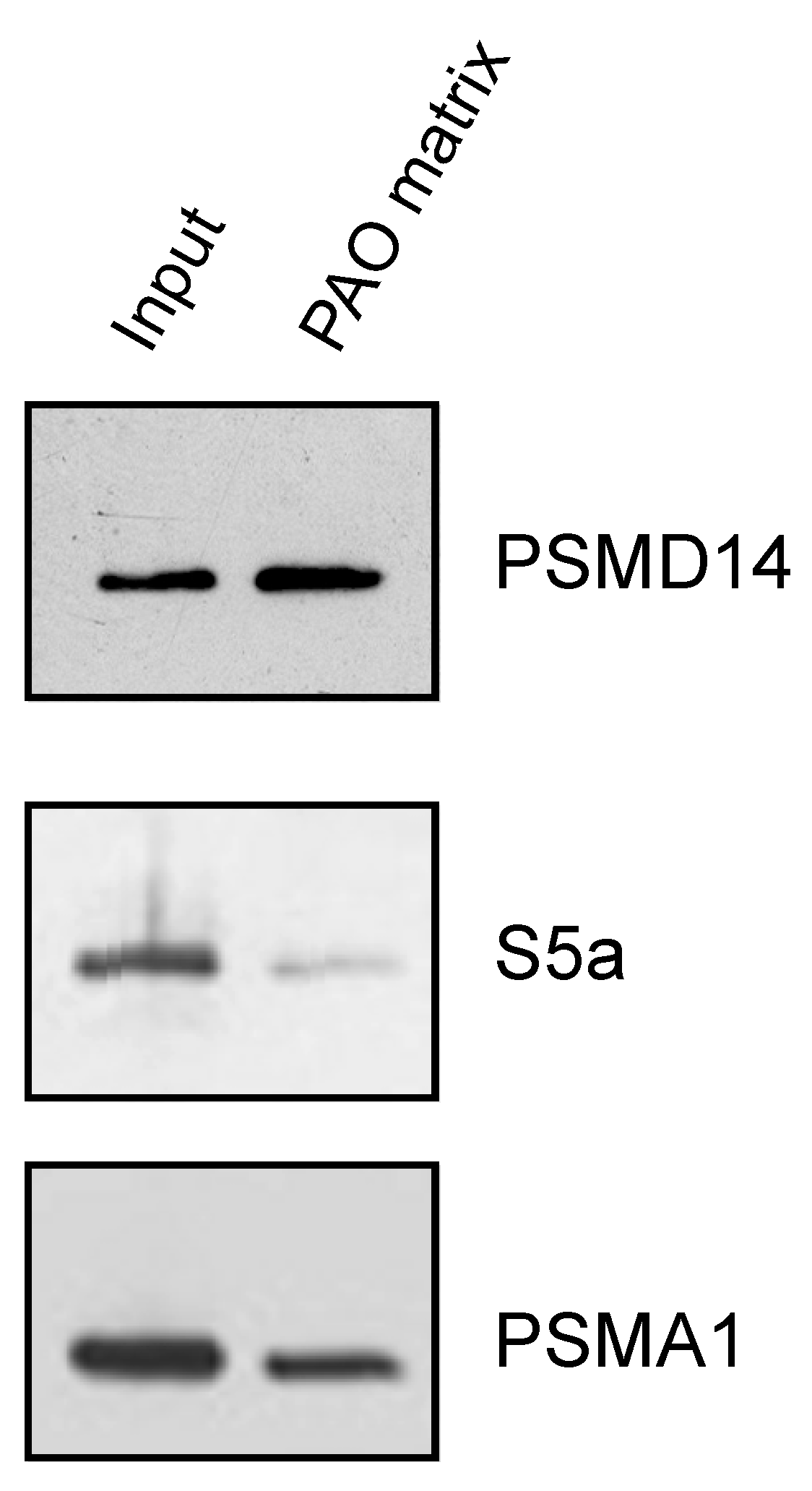
3.3. Endogenous Rpn11/PSMD14 Binds PAO Matrix More Strongly Than Other Proteasome Subunits
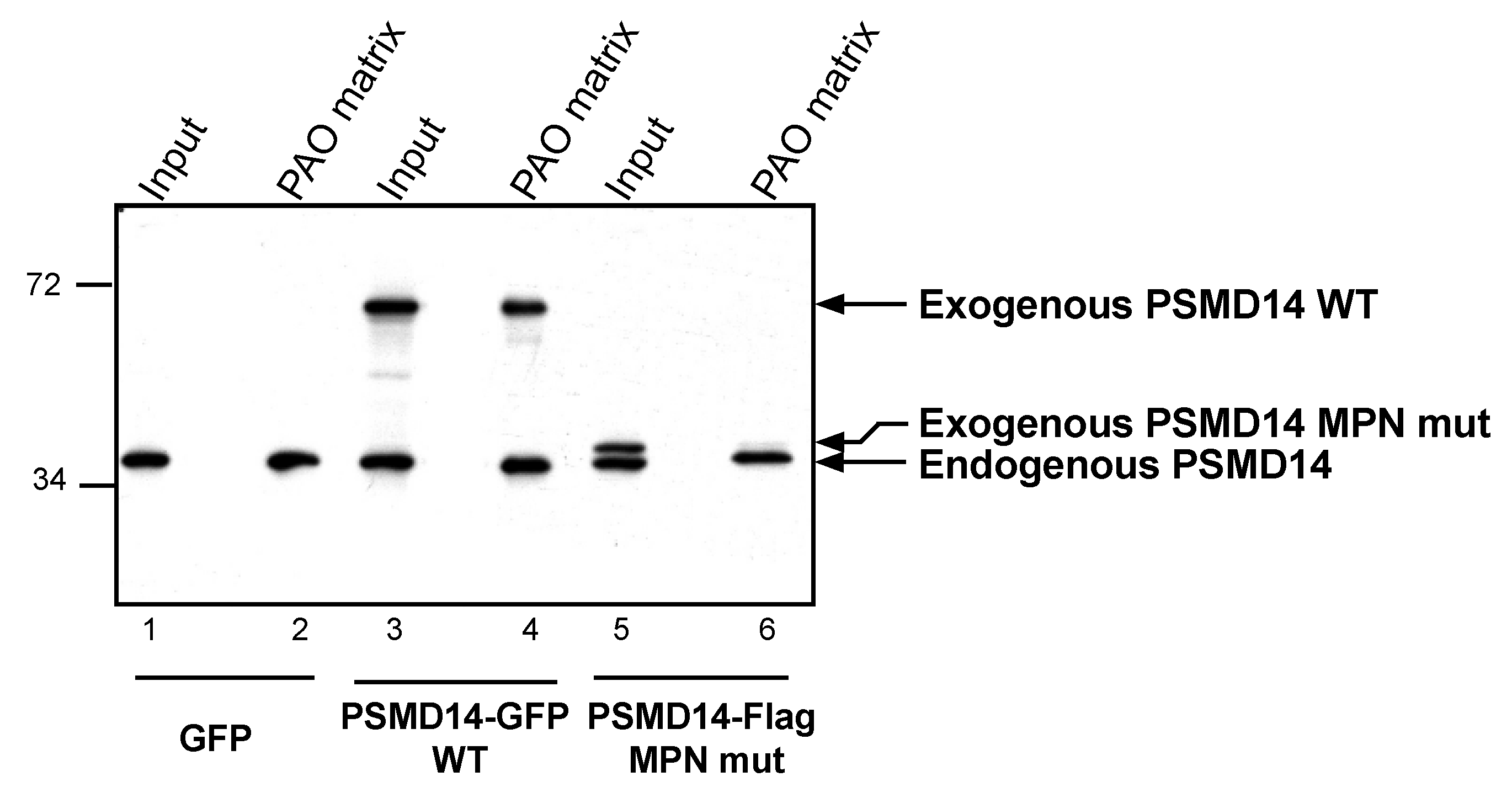
3.4. Arsenite Regulation of Psmd14 Activity via MPN/JAMM Motif
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta. 2014, 1843, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannhaupt, G.; Schnall, R.; Karpov, V.; Vetter, I.; Feldmann, H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999, 450, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, S.K.; Lee, C.S.; Young, P.; Beskow, A.; Chan, J.Y.; Deshaies, R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 2010, 38, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffen, J.; Seeger, M.; Koch, A.; Krüger, E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 2010, 40, 147–158. [Google Scholar] [CrossRef]
- Panasenko, O.O.; Somasekharan, S.P.; Villanyi, Z.; Zagatti, M.; Bezrukov, F.; Rashpa, R.; Cornut, J.; Iqbal, J.; Longis, M.; Carl, S.H.; et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct Mol. Biol. 2019, 26, 110–120. [Google Scholar] [CrossRef]
- Hirano, H.; Kimura, Y.; Kimura, A. Biological significance of co- and post-translational modifications of the yeast 26S proteasome. J. Proteom. 2016, 134, 37–46. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar] [CrossRef] [Green Version]
- Aki, M.; Shimbara, N.; Takashina, M.; Akiyama, K.; Kagawa, S.; Tamura, T.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994, 115, 257–269. [Google Scholar] [CrossRef]
- Sasaki, K.; Takada, K.; Ohte, Y.; Kondo, H.; Sorimachi, H.; Tanaka, K.; Takahama, Y.; Murata, S. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat. Commun. 2015, 6, 7484. [Google Scholar] [CrossRef] [Green Version]
- Frantzeskakis, M.; Takahama, Y.; Ohigashi, I. The Role of Proteasomes in the Thymus. Front. Immunol. 2021, 12, 646209. [Google Scholar] [CrossRef]
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef] [Green Version]
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef] [Green Version]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28αβ reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanahashi, N.; Yokota, K.Y.; Ahn, J.Y.; Chung, C.H.; Fujiwara, T.; Takahashi, E.I.; George, N.; DeMartino, G.N.; Slaughter, C.A.; Toyonaga, T.; et al. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 1997, 2, 195–211. [Google Scholar] [CrossRef]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef]
- Chen, X.; Htet, Z.M.; López-Alfonzo, E.; Martin, A.; Walters, K.J. Proteasome interaction with ubiquitinated substrates: From mechanisms to therapies. FEBS J. 2020. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Randles, L.; Shi, K.; Tarasov, S.G.; Aihara, H.; Walters, K.J. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 2016, 24, 1257–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, G.A.; Goldberg, A.L. Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proc. Natl. Acad. Sci. USA 2020, 117, 4664–4674. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.P.; Kolaitis, R.M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castañeda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978. [Google Scholar] [CrossRef] [Green Version]
- Sok, J.; Calfon, M.; Lu, J.; Lichtlen, P.; Clark, S.G.; Ron, D. Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones 2001, 6, 6–15. [Google Scholar] [CrossRef] [Green Version]
- Stanhill, A.; Haynes, C.M.; Zhang, Y.; Min, G.; Steele, M.C.; Kalinina, J.; Martinez, E.; Pickart, C.M.; Kong, X.P.; Ron, D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 2006, 23, 875–885. [Google Scholar] [CrossRef]
- Rossi, A.; Riccio, A.; Coccia, M.; Trotta, E.; La Frazia, S.; Santoro, M.G. The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: Role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. J. Biol. Chem. 2014, 289, 12705–12715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isakov, E.; Stanhill, A. Stalled proteasomes are directly relieved by P97 recruitment. J. Biol. Chem. 2011, 286, 30274–30283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piterman, R.; Braunstein, I.; Isakov, E.; Ziv, T.; Navon, A.; Cohen, S.; Stanhill, A. VWA domain of S5a restricts the ability to bind ubiquitin and Ubl to the 26S proteasome. Mol. Biol Cell 2014, 25, 3988–3998. [Google Scholar] [CrossRef]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar] [CrossRef]
- Saeki, Y.; Isono, E.; Toh-E, A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005, 399, 215–227. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Shen, S.; Li, X.F.; Cullen, W.R.; Weinfeld, M.; Le, X.C. Arsenic binding to proteins. Chem. Rev. 2013, 113, 7769–7792. [Google Scholar] [CrossRef]
- He, X.; Ma, Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 2009, 76, 1265–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R.; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Fuchs, A.C.D.; Maldoner, L.; Wojtynek, M.; Hartmann, M.D.; Martin, J. Rpn11-mediated ubiquitin processing in an ancestral archaeal ubiquitination system. Nat. Commun. 2018, 9, 2696. [Google Scholar] [CrossRef] [PubMed]
- Gallery, M.; Blank, J.L.; Lin, Y.; Gutierrez, J.A.; Pulido, J.C.; Rappoli, D.; Badola, S.; Rolfe, M.; Macbeth, K.J. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol. Cancer Ther. 2007, 6, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worden, E.J.; Padovani, C.; Martin, A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 2014, 21, 220–227. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukenik, S.; Braunstein, I.; Stanhill, A. An Arsenite Relay between PSMD14 and AIRAP Enables Revival of Proteasomal DUB Activity. Biomolecules 2021, 11, 1317. https://doi.org/10.3390/biom11091317
Sukenik S, Braunstein I, Stanhill A. An Arsenite Relay between PSMD14 and AIRAP Enables Revival of Proteasomal DUB Activity. Biomolecules. 2021; 11(9):1317. https://doi.org/10.3390/biom11091317
Chicago/Turabian StyleSukenik, Sigalit, Ilana Braunstein, and Ariel Stanhill. 2021. "An Arsenite Relay between PSMD14 and AIRAP Enables Revival of Proteasomal DUB Activity" Biomolecules 11, no. 9: 1317. https://doi.org/10.3390/biom11091317
APA StyleSukenik, S., Braunstein, I., & Stanhill, A. (2021). An Arsenite Relay between PSMD14 and AIRAP Enables Revival of Proteasomal DUB Activity. Biomolecules, 11(9), 1317. https://doi.org/10.3390/biom11091317





