Beyond the Usual Suspects: Physiological Roles of the Arabidopsis Amidase Signature (AS) Superfamily Members in Plant Growth Processes and Stress Responses
Abstract
1. Introduction
2. The AS Superfamily
2.1. The Arabidopsis AS Superfamily Members
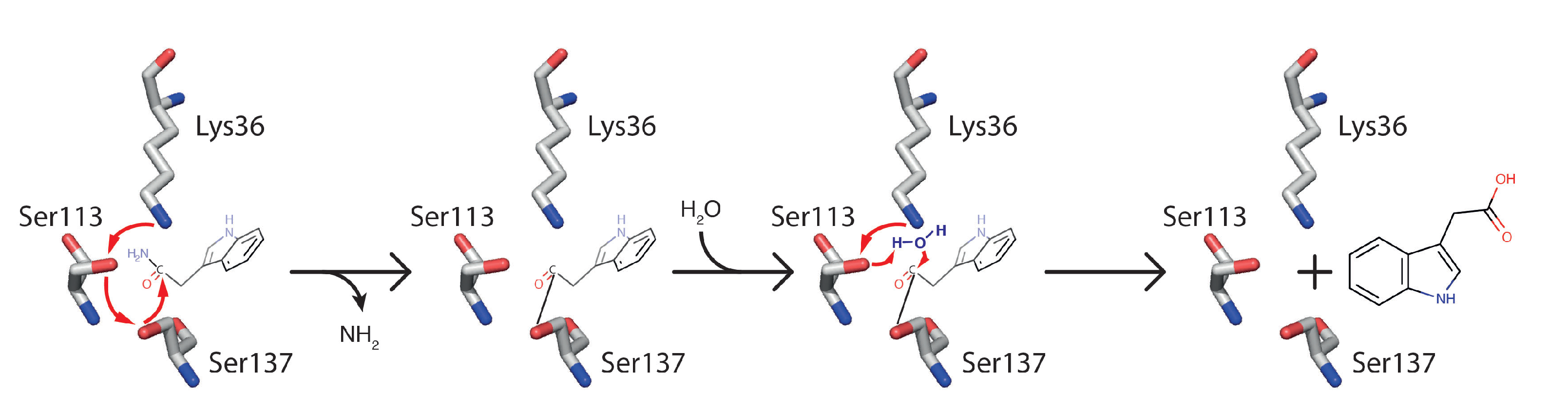
2.1.1. AMI1
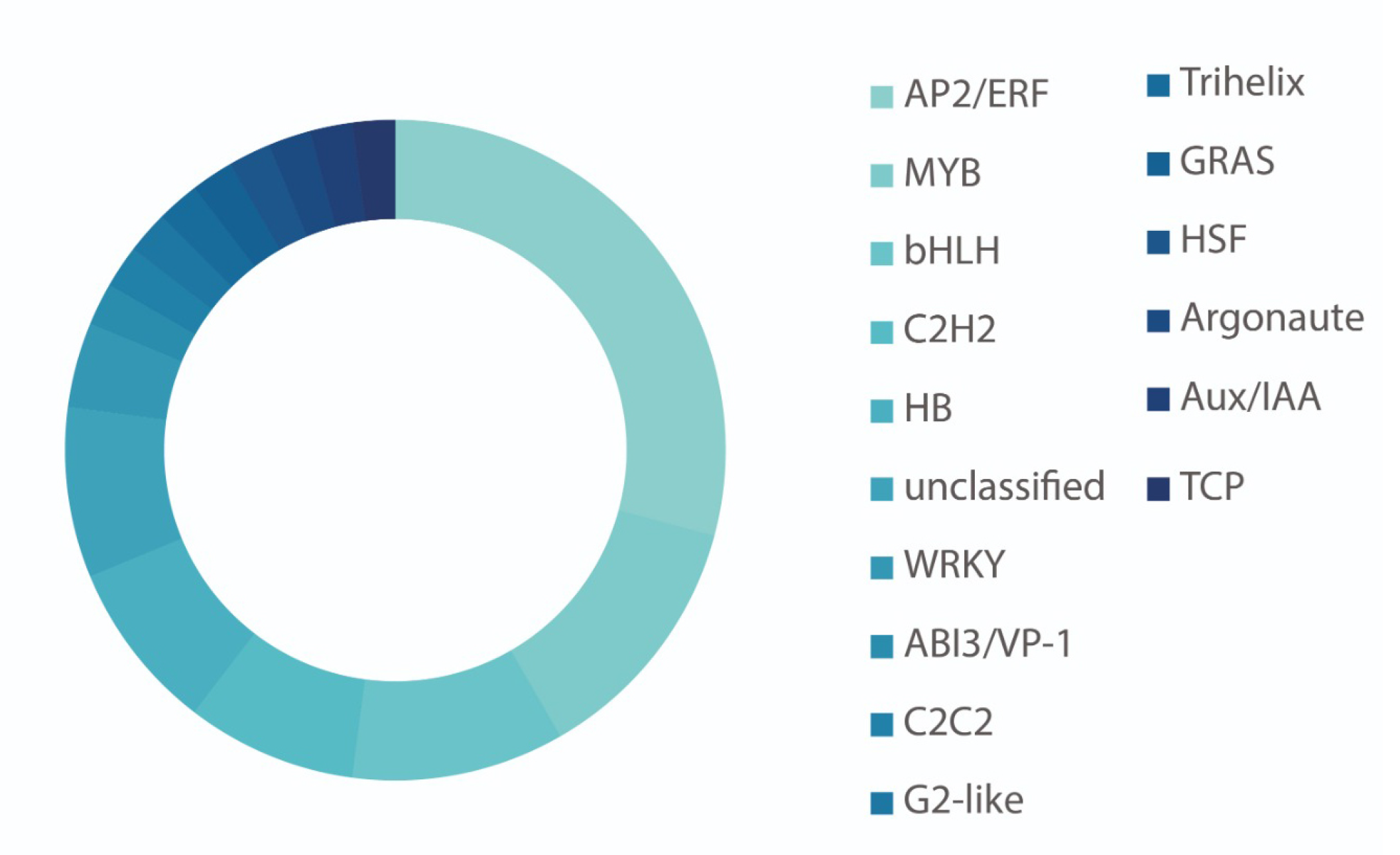
The Atypical Member of the Family
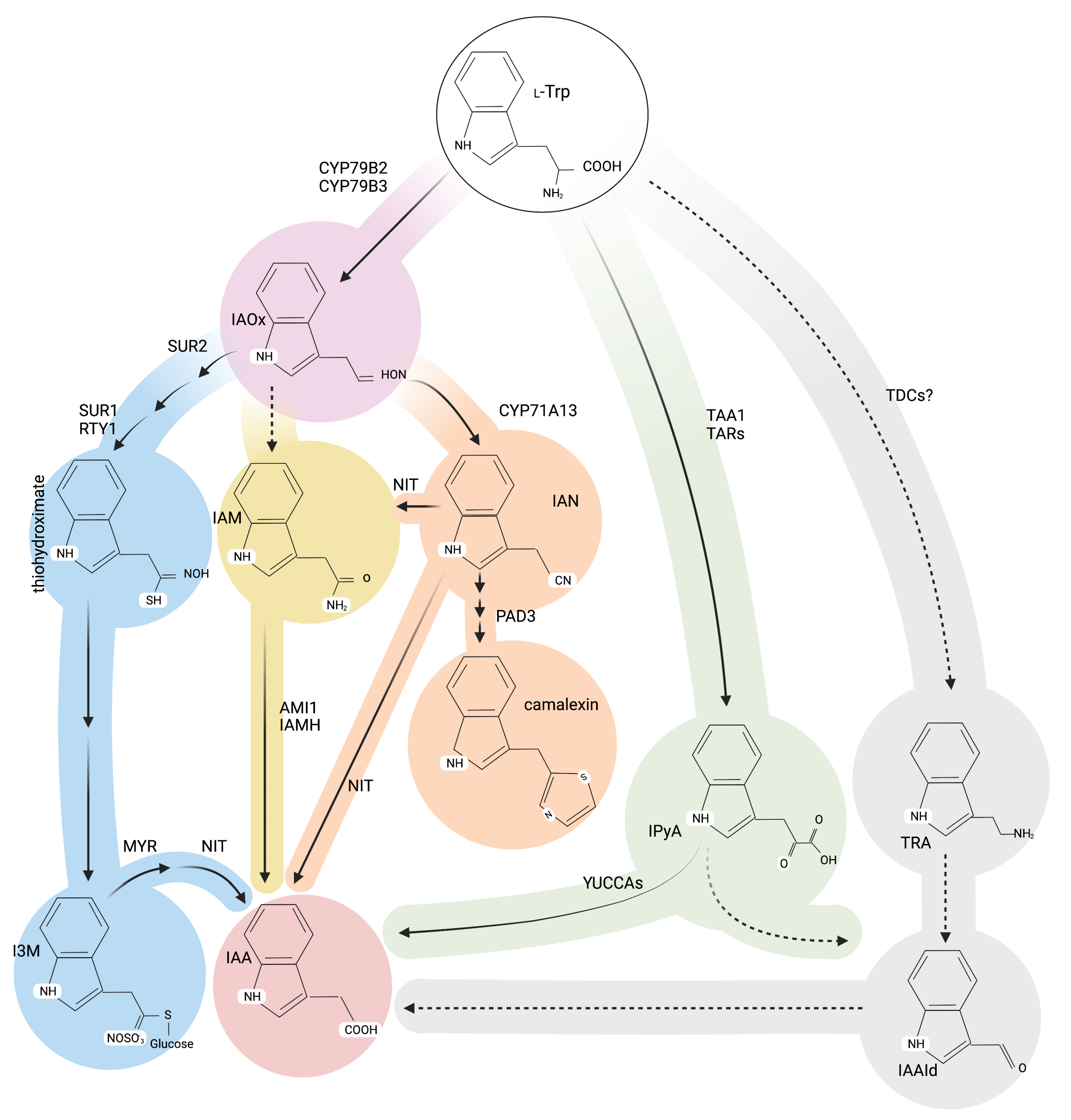
Alternative Roads to IAA Biosynthesis
The More IAM, the Less Plant Growth
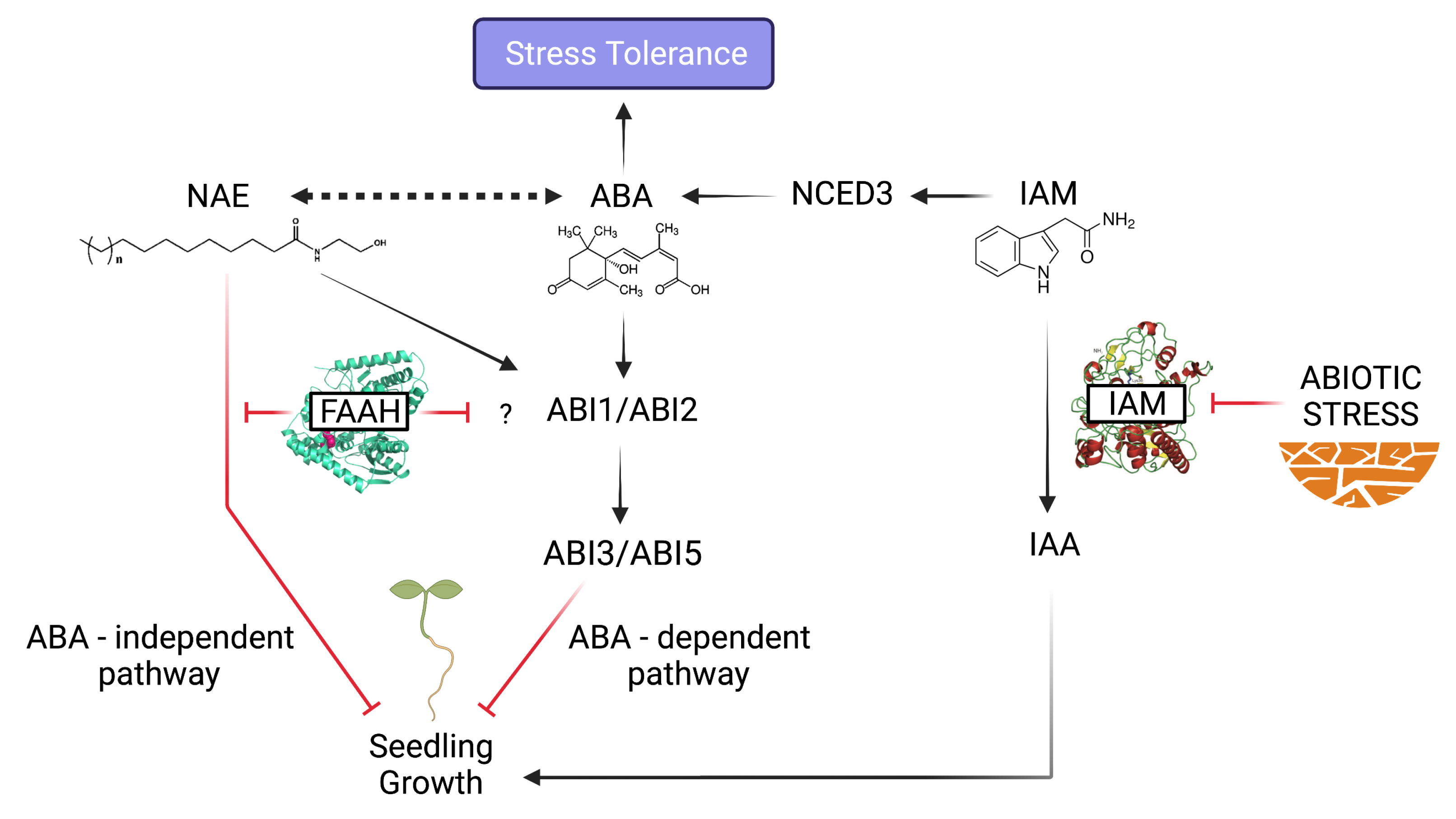
The AMI1 Connection: IAM-ABA Crosstalk in Stress Responses
2.1.2. FAAH
What Lies Beneath the Structure
One FAAH to Terminate Them All
NAE Signaling Alterations: Plant Physiological Processes in Jeopardy
Convergent and Bifurcating Pathways at the NAE-ABA Signaling Crossroads
3. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Spoel, S.H.; Dong, X. Making Sense of Hormone Crosstalk during Plant Immune Responses. Cell Host Microbe 2008, 3, 348–351. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of Hormone Signaling Networks in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.; Fukui, K.; Seto, Y.; Takaoka, Y.; Okamoto, M. Ligand-receptor interactions in plant hormone signaling. Plant J. 2021, 105, 290–306. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2017, 176, 465–479. [Google Scholar] [CrossRef]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136. [Google Scholar] [CrossRef]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 2017, 41, 33–46. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Pinon, V.; Prasad, K.; Grigg, S.P.; Sanchez-Perez, G.F.; Scheres, B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1107–1112. [Google Scholar] [CrossRef]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef]
- Gomez-Cadenas, A.; Vives, V.; Zandalinas, S.I.; Manzi, M.; Sanchez-Perez, A.M.; Perez-Clemente, R.M.; Arbona, V. Abscisic Acid: A Versatile Phytohormone in Plant Signaling and Beyond. Curr. Protein Pept. Sci. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef]
- Kumar, M.; Kesawat, M.S.; Ali, A.; Lee, S.C.; Gill, S.S.; Kim, H.U. Integration of Abscisic Acid Signaling with Other Signaling Pathways in Plant Stress Responses and Development. Plants 2019, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.A.; Gori, A.; Da-Silva, C.J.; Brunetti, C. Abscisic Acid Biosynthesis and Signaling in Plants: Key Targets to Improve Water Use Efficiency and Drought Tolerance. Appl. Sci. 2020, 10, 6322. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of reactive oxygen species and hormone signaling during abiotic stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.N.; Vieira, S.A.; Ribeiro, R.V.; Ponte, L.F.A.; Ferreira-Silva, S.L.; Silveira, J.A.G. Contrasting Physiological Responses of Jatropha curcas Plants to Single and Combined Stresses of Salinity and Heat. J. Plant Growth Regul. 2013, 32, 159–169. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Perdomo, J.A.; Conesa, M.À.; Medrano, H.; Ribas-Carbó, M.; Galmés, J. Effects of long-term individual and combined water and temperature stress on the growth of rice, wheat and maize: Relationship with morphological and physiological acclimation. Physiol. Plant. 2015, 155, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Rosquete, M.; Barbez, E.; Kleine-Vehn, J. Cellular Auxin Homeostasis: Gatekeeping Is Housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local Auxin Biosynthesis Is a Key Regulator of Plant Development. Dev. Cell 2018, 47, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, D.; Geelen, D.; Verstraeten, I. Control of Endogenous Auxin Levels in Plant Root Development. Int. J. Mol. Sci. 2017, 18, 2587. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed]
- Neu, D.; Lehmann, T.; Elleuche, S.; Pollmann, S. Arabidopsis amidase 1, a member of the amidase signature family. FEBS J. 2007, 274, 3440–3451. [Google Scholar] [CrossRef]
- Aziz, M.; Wang, X.; Tripathi, A.; Bankaitis, V.A.; Chapman, K.D. Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides. J. Biol. Chem. 2019, 294, 7419–7432. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, C.; Zhang, Z.; Zheng, R.; Zheng, Y. Amidase as a versatile tool in amide-bond cleavage: From molecular features to biotechnological applications. Biotechnol. Adv. 2020, 43, 107574. [Google Scholar] [CrossRef]
- McKinney, M.K.; Cravatt, B.F. Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 2005, 74, 411–432. [Google Scholar] [CrossRef]
- Aziz, M.; Chapman, K.D. Fatty Acid Amide Hydrolases: An Expanded Capacity for Chemical Communication? Trends Plant Sci. 2020, 25, 236–249. [Google Scholar] [CrossRef]
- Sánchez-Parra, B.; Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Hentrich, M.; Pollmann, S. Accumulation of the Auxin Precursor Indole-3-Acetamide Curtails Growth through the Repression of Ribosome-Biogenesis and Development-Related Transcriptional Networks. Int. J. Mol. Sci. 2021, 22, 2040. [Google Scholar] [CrossRef]
- Echevarría, L.; Clemente, P.; Hernández-Sierra, R.; Gallardo, M.E.; Fernández-Moreno, M.A.; Garesse, R. Glutamyl-tRNAGln amidotransferase is essential for mammalian mitochondrial translation in vivo. Biochem. J. 2014, 460, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Li, Z.; Yin, H.; Xiang, S. Structure and function of allophanate hydrolase. J. Biol. Chem. 2013, 288, 21422–21432. [Google Scholar] [CrossRef] [PubMed]
- Valiña, A.L.B.; Mazumder-Shivakumar, D.; Bruice, T.C. Probing the Ser-Ser-Lys Catalytic Triad Mechanism of Peptide Amidase: Computational Studies of the Ground State, Transition State, and Intermediate. Biochemistry 2004, 43, 15657–15672. [Google Scholar] [CrossRef]
- Shin, S.; Lee, T.H.; Ha, N.C.; Koo, H.M.; Kim, S.Y.; Lee, H.S.; Kim, Y.S.; Oh, B.H. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 2002, 21, 2509–2516. [Google Scholar] [CrossRef]
- Pollmann, S.; Neu, D.; Weiler, E.W. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 2003, 62, 293–300. [Google Scholar] [CrossRef]
- Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Lehmann, T.; Sánchez-Parra, B.; Björk, R.G.; Karim, S.; Amirjani, M.R.; Aronsson, H.; Wilkinson, M.D.; et al. Endogenous indole-3-acetamide levels contribute to the crosstalk between auxin and abscisic acid, and trigger plant stress responses in Arabidopsis. J. Exp. Bot. 2021, 72, 459–475. [Google Scholar] [CrossRef]
- Shrestha, R.; Dixon, R.A.; Chapman, K.D. Molecular Identification of a Functional Homologue of the Mammalian Fatty Acid Amide Hydrolase in Arabidopsis thaliana *. J. Biol. Chem. 2003, 278, 34990–34997. [Google Scholar] [CrossRef]
- Chew, O.; Lister, R.; Qbadou, S.; Heazlewood, J.L.; Soll, J.; Schleiff, E.; Millar, A.H.; Whelan, J. A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 2004, 557, 109–114. [Google Scholar] [CrossRef]
- Sohrt, K.; Soll, J. Toc64, a New Component of the Protein Translocon of Chloroplasts. J. Cell Biol. 2000, 148, 1213–1222. [Google Scholar] [CrossRef]
- Prasad, B.D.; Goel, S.; Krishna, P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS ONE 2010, 5, e12761. [Google Scholar] [CrossRef]
- Wu, X.; Huang, R.; Liu, Z.; Zhang, G. Functional Characterization of cis-Elements Conferring Vascular Vein Expression of At4g34880 Amidase Family Protein Gene in Arabidopsis. PLoS ONE 2013, 8, e67562. [Google Scholar] [CrossRef]
- Aryal, U.K.; Xiong, Y.; McBride, Z.; Kihara, D.; Xie, J.; Hall, M.C.; Szymanski, D.B. A Proteomic Strategy for Global Analysis of Plant Protein Complexes. Plant Cell 2014, 26, 3867–3882. [Google Scholar] [CrossRef]
- Pollmann, S.; Neu, D.; Lehmann, T.; Berkowitz, O.; Schäfer, T.; Weiler, E.W. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 2006, 224, 1241–1253. [Google Scholar] [CrossRef]
- Cilia, E.; Fabbri, A.; Uriani, M.; Scialdone, G.G.; Ammendola, S. The signature amidase from Sulfolobus solfataricus belongs to the CX3C subgroup of enzymes cleaving both amides and nitriles. Ser195 and Cys145 are predicted to be the active site nucleophiles. FEBS J. 2005, 272, 4716–4724. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Cravatt, B.F. Clarifying the Catalytic Roles of Conserved Residues in the Amidase Signature Family *. J. Biol. Chem. 2000, 275, 19177–19184. [Google Scholar] [CrossRef]
- Labahn, J.; Neumann, S.; Büldt, G.; Kula, M.R.; Granzin, J. An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 2002, 322, 1053–1064. [Google Scholar] [CrossRef]
- Shrestha, R.; Noordermeer, M.A.; van der Stelt, M.; Veldink, G.A.; Chapman, K.D. N-acylethanolamines are metabolized by lipoxygenase and amidohydrolase in competing pathways during cottonseed imbibition. Plant Physiol. 2002, 130, 391–401. [Google Scholar] [CrossRef]
- Upadhyaya, N.M.; Zhou, X.R.; Wu, L.; Ramm, K.; Dennis, E.S. Thetms2 gene as a negative selection marker in rice. Plant Mol. Biol. Report. 2000, 18, 227–233. [Google Scholar] [CrossRef]
- Kemper, E.; Wafenschmidt, S.; Weiler, E.W.; Rausch, T.; Schröder, J. T-DNA-encoded auxin formation in crown-gall cells. Planta 1985, 163, 257–262. [Google Scholar] [CrossRef]
- Pollmann, S.; Müller, A.; Piotrowski, M.; Weiler, E.W. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 2002, 216, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Pollmann, S.; Müller, A.; Weiler, E.W. Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol. 2006, 8, 326–333. [Google Scholar] [CrossRef]
- Kasahara, H. Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 2016, 80, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Igoshi, M.; Yamaguchi, I.; Takahashi, N.; Hirose, K. Plant Growth Substances in the Young Fruit of Citrus unshiu. Agric. Biol. Chem. 1971, 35, 629–631. [Google Scholar] [CrossRef]
- Takahashi, N.; Yamaguchi, I.; Kono, T.; Igoshi, M.; Hirose, K.; Suzuki, K. Characterization of plant growth substances in Citrus unshiu and their change in fruit development. Plant Cell Physiol. 1975, 16, 1101–1111. [Google Scholar] [CrossRef]
- Saotome, M.; Shirahata, K.; Nishimura, R.; Yahaba, M.; Kawaguchi, M.; Syōno, K.; Kitsuwa, T.; Ishii, Y.; Nakamura, T. The Identification of Indole-3-Acetic Acid and Indole-3-Acetamide in the Hypocotyls of Japanese Cherry. Plant Cell Physiol. 1993, 34, 157–159. [Google Scholar] [CrossRef]
- Rajagopal, R.; Tsurusaki, K.i.; Kannangara, G.; Kuraishi, S.; Sakurai, N. Natural Occurrence of Indoleacetamide and Amidohydrolase Activity in Etiolated Aseptically-Grown Squash Seedlings. Plant Cell Physiol. 1994, 35, 329–339. [Google Scholar] [CrossRef]
- Fawcett, C.H.; Wain, R.L.; Wightman, F. The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc. R. Soc. Lond. B Biol. Sci. 1960, 152, 231–254. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Kobayashi, M.; Sakurai, A.; Syōno, K. The Presence of an Enzyme that Converts IndoIe-3-acetamide into IAA in Wild and Cultivated Rice. Plant Cell Physiol. 1991, 32, 143–149. [Google Scholar] [CrossRef]
- Arai, Y.; Kawaguchi, M.; Syono, K.; Ikuta, A. Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J. Plant Res. 2004, 117, 191–198. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Fujioka, S.; Sakurai, A.; Yamaki, Y.T.; Syōno, K. Presence of a Pathway for the Biosynthesis of Auxin via Indole-3-Acetamide in Trifoliata Orange. Plant Cell Physiol. 1993, 34, 121–128. [Google Scholar] [CrossRef]
- Nemoto, K.; Hara, M.; Suzuki, M.; Seki, H.; Muranaka, T.; Mano, Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 2009, 583, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Parra, B.; Frerigmann, H.; Alonso, M.M.P.; Loba, V.C.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants 2014, 3, 324–347. [Google Scholar] [CrossRef]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef]
- Glawischnig, E.; Hansen, B.G.; Olsen, C.E.; Halkier, B.A. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 8245–8250. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Glawischnig, E. Camalexin. Phytochemistry 2007, 68, 401–406. [Google Scholar] [CrossRef]
- Frerigmann, H.; Glawischnig, E.; Gigolashvili, T. The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 654. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dai, X.; Aoi, Y.; Takebayashi, Y.; Yang, L.; Guo, X.; Zeng, Q.; Yu, H.; Kasahara, H.; Zhao, Y. Two homologous INDOLE-3-ACETAMIDE (IAM) HYDROLASE genes are required for the auxin effects of IAM in Arabidopsis. J. Genet. Genom. 2020, 47, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Davies, P. Plant Hormones: Biosynthesis, Signal Transduction, Action; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.T.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef]
- Lehmann, T.; Janowitz, T.; Sánchez-Parra, B.; Alonso, M.M.P.; Trompetter, I.; Piotrowski, M.; Pollmann, S. Arabidopsis NITRILASE 1 Contributes to the Regulation of Root Growth and Development through Modulation of Auxin Biosynthesis in Seedlings. Front. Plant Sci. 2017, 8, 36. [Google Scholar] [CrossRef]
- Lehmann, T.; Hoffmann, M.; Hentrich, M.; Pollmann, S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 2010, 89, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.; Horsch, R.; Hinchee, M.; Hein, M.; Hoffmann, N. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgen c petunia plants. Genes Dev. 1987, 1, 86–96. [Google Scholar] [CrossRef]
- Gielen, J.; De Beuckeleer, M.; Seurinck, J.; Deboeck, F.; De Greve, H.; Lemmers, M.; Van Montagu, M.; Schell, J. The complete nucleotide sequence of the TL-DNA of the Agrobacterium tumefaciens plasmid pTiAch5. EMBO J. 1984, 3, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Lehmann, T.; Neu, D.; Hentrich, M.; Pollmann, S. Expression of AMIDASE1 (AMI1) is suppressed during the first two days after germination. Plant Signal. Behav. 2010, 5, 1642–1644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tenorio-Berrío, R.; Pérez-Alonso, M.M.; Vicente-Carbajosa, J.; Martín-Torres, L.; Dreyer, I.; Pollmann, S. Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation. Int. J. Mol. Sci. 2018, 19, 2132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Huang, P.P.; Ji, Y.; Wang, S.; Wang, S.S.; Li, Z.; Guo, Y.; Ding, Z.; Wu, W.H.; Wang, Y. KUP9 maintains root meristem activity by regulating K(+) and auxin homeostasis in response to low K. EMBO Rep. 2020, 21, e50164. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Balsemão-Pires, E.; Andrade, L.R.; Sachetto-Martins, G. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 2013, 67, 120–125. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Truman, W.M.; Bennett, M.H.; Turnbull, C.G.N.; Grant, M.R. Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 2010, 152, 1562–1573. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An Updated Overview on the Regulation of Seed Germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef]
- Matilla, A.J. Auxin: Hormonal Signal Required for Seed Development and Dormancy. Plants 2020, 9, 705. [Google Scholar] [CrossRef] [PubMed]
- Chauffour, F.; Bailly, M.; Perreau, F.; Cueff, G.; Suzuki, H.; Collet, B.; Frey, A.; Clément, G.; Soubigou-Taconnat, L.; Balliau, T.; et al. Multi-omics Analysis Reveals Sequential Roles for ABA during Seed Maturation. Plant Physiol. 2019, 180, 1198–1218. [Google Scholar] [CrossRef]
- He, Y.; Zhao, J.; Yang, B.; Sun, S.; Peng, L.; Wang, Z. Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol. J. 2020, 18, 1933–1945. [Google Scholar] [CrossRef]
- Chapman, K.D. Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog. Lipid Res. 2004, 43, 302–327. [Google Scholar] [CrossRef]
- Bracey, M.H.; Hanson, M.A.; Masuda, K.R.; Stevens, R.C.; Cravatt, B.F. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 2002, 298, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Amborella Genome, P. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef] [PubMed]
- Faure, L.; Cavazos, R.; Khan, B.R.; Petros, R.A.; Koulen, P.; Blancaflor, E.B.; Chapman, K.D. Effects of synthetic alkamides on Arabidopsis fatty acid amide hydrolase activity and plant development. Phytochemistry 2015, 110, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G.; Senechal, A.C.; Mukherjee, A.; Ané, J.M.; Blackwell, H.E. Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem. Biol. 2014, 9, 1834–1845. [Google Scholar] [CrossRef]
- Schmid, H.H.; Schmid, P.C.; Natarajan, V. The N-acylation-phosphodiesterase pathway and cell signalling. Chem. Phys. Lipids 1996, 80, 133–142. [Google Scholar] [CrossRef]
- Okamoto, Y.; Morishita, J.; Tsuboi, K.; Tonai, T.; Ueda, N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 2004, 279, 5298–5305. [Google Scholar] [CrossRef]
- Pappan, K.; Austin-Brown, S.; Chapman, K.D.; Wang, X. Substrate Selectivities and Lipid Modulation of Plant Phospholipase Dα, -β, and -γ. Arch. Biochem. Biophys. 1998, 353, 131–140. [Google Scholar] [CrossRef]
- Simon, G.M.; Cravatt, B.F. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J. Biol. Chem. 2006, 281, 26465–26472. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Harvey-White, J.; Osei-Hyiaman, D.; Razdan, R.; Gong, Q.; Chan, A.C.; Zhou, Z.; Huang, B.X.; Kim, H.Y.; et al. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. USA 2006, 103, 13345–13350. [Google Scholar] [CrossRef]
- Howlett, A.C.; Breivogel, C.S.; Childers, S.R.; Deadwyler, S.A.; Hampson, R.E.; Porrino, L.J. Cannabinoid physiology and pharmacology: 30 years of progress. Neuropharmacology 2004, 47 (Suppl. 1), 345–358. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, F.A.; Di Marzo, V.; Petrosino, S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog. Lipid Res. 2016, 62, 107–128. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Fride, E.; Di Marzo, V. Endocannabinoids. Eur. J. Pharmacol. 1998, 359, 1–18. [Google Scholar] [CrossRef]
- Venables, B.J.; Waggoner, C.A.; Chapman, K.D. N-acylethanolamines in seeds of selected legumes. Phytochemistry 2005, 66, 1913–1918. [Google Scholar] [CrossRef] [PubMed]
- Blancaflor, E.B.; Chapman, K.D. Similarities Between Endocannabinoid Signaling in Animal Systems and N-Acylethanolamine Metabolism in Plants. In Communication in Plants: Neuronal Aspects of Plant Life; Baluška, F., Mancuso, S., Volkmann, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 205–219. [Google Scholar] [CrossRef]
- Keereetaweep, J.; Blancaflor, E.B.; Hornung, E.; Feussner, I.; Chapman, K.D. Ethanolamide oxylipins of linolenic acid can negatively regulate Arabidopsis seedling development. Plant Cell 2013, 25, 3824–3840. [Google Scholar] [CrossRef] [PubMed]
- Keereetaweep, J.; Blancaflor, E.B.; Hornung, E.; Feussner, I.; Chapman, K.D. Lipoxygenase-derived 9-hydro(pero)xides of linoleoylethanolamide interact with ABA signaling to arrest root development during Arabidopsis seedling establishment. Plant J. 2015, 82, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Blancaflor, E.B.; Kilaru, A.; Keereetaweep, J.; Khan, B.R.; Faure, L.; Chapman, K.D. N-Acylethanolamines: Lipid metabolites with functions in plant growth and development. Plant J. 2014, 79, 568–583. [Google Scholar] [CrossRef] [PubMed]
- Gachet, M.S.; Schubert, A.; Calarco, S.; Boccard, J.; Gertsch, J. Targeted metabolomics shows plasticity in the evolution of signaling lipids and uncovers old and new endocannabinoids in the plant kingdom. Sci. Rep. 2017, 7, 41177. [Google Scholar] [CrossRef]
- Kim, S.C.; Chapman, K.D.; Blancaflor, E.B. Fatty acid amide lipid mediators in plants. Plant Sci. 2010, 178, 411–419. [Google Scholar] [CrossRef]
- Chapman, K.D.; Venables, B.; Markovic, R.; Blair, R.W., Jr.; Bettinger, C. N-Acylethanolamines in Seeds. Quantification of Molecular Species and Their Degradation upon Imbibition1. Plant Physiol. 1999, 120, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Shrestha, R.; Kilaru, A.; Wiant, W.; Venables, B.J.; Chapman, K.D.; Blancaflor, E.B. Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc. Natl. Acad. Sci. USA 2006, 103, 12197–12202. [Google Scholar] [CrossRef]
- Blancaflor, E.B.; Hou, G.; Chapman, K.D. Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta 2003, 217, 206–217. [Google Scholar] [CrossRef]
- Motes, C.M.; Pechter, P.; Yoo, C.M.; Wang, Y.S.; Chapman, K.D.; Blancaflor, E.B. Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma 2005, 226, 109–123. [Google Scholar] [CrossRef]
- Teaster, N.D.; Motes, C.M.; Tang, Y.; Wiant, W.C.; Cotter, M.Q.; Wang, Y.S.; Kilaru, A.; Venables, B.J.; Hasenstein, K.H.; Gonzalez, G.; et al. N-Acylethanolamine metabolism interacts with abscisic acid signaling in Arabidopsis thaliana seedlings. Plant Cell 2007, 19, 2454–2469. [Google Scholar] [CrossRef]
- Cotter, M.Q.; Teaster, N.D.; Blancaflor, E.B.; Chapman, K.D. N-acylethanolamine (NAE) inhibits growth in Arabidopsis thaliana seedlings via ABI3-dependent and -independent pathways. Plant Signal. Behav. 2011, 6, 671–679. [Google Scholar] [CrossRef]
- Teaster, N.D.; Keereetaweep, J.; Kilaru, A.; Wang, Y.S.; Tang, Y.; Tran, C.N.Q.; Ayre, B.G.; Chapman, K.D.; Blancaflor, E.B. Overexpression of Fatty Acid Amide Hydrolase Induces Early Flowering in Arabidopsis thaliana. Front. Plant Sci. 2012, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Wang, Y.S.; Uppalapati, S.R.; Wang, K.; Tang, Y.; Vadapalli, V.; Venables, B.J.; Chapman, K.D.; Blancaflor, E.B.; Mysore, K.S. Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 2008, 56, 336–349. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Seo, E.; Yu, J.; Ryu, K.H.; Lee, M.M.; Lee, I. WEREWOLF, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 2011, 156, 1867–1877. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. ABA action and interactions in seeds. Trends Plant Sci. 2003, 8, 213–217. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Coursol, S.; Fan, L.M.; Stunff, H.; Spiegel, S.; Gilroy, S.; Assmann, S.M. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 2003, 423, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.F.; Wei, P.C.; Chen, Q.J.; An, R.; Chen, J.; Yang, S.; Wang, X.C. NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J. 2006, 47, 665–674. [Google Scholar] [CrossRef]
- Rock, C.D. Tansley Review No. 120: Pathways to abscisic acid-regulated gene expression. New Phytol. 2000, 148, 357–396. [Google Scholar] [CrossRef]
- Kang, J.; Mehta, S.; Turano, F.J. The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004, 45, 1380–1389. [Google Scholar] [CrossRef]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006, 60, 51–68. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.G.; Wang, S.; Jones, A.M. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002, 129, 897–907. [Google Scholar] [CrossRef]
- Pandey, S.; Assmann, S.M. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 2004, 16, 1616–1632. [Google Scholar] [CrossRef]
- Pandey, S.; Chen, J.G.; Jones, A.M.; Assmann, S.M. G-Protein Complex Mutants Are Hypersensitive to Abscisic Acid Regulation of Germination and Postgermination Development. Plant Physiol. 2006, 141, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yue, Y.; Li, B.; Nie, Y.; Li, W.; Wu, W.H.; Ma, L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 2007, 315, 1712–1716. [Google Scholar] [CrossRef] [PubMed]
- Jacob, T.; Ritchie, S.; Assmann, S.M.; Gilroy, S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 1999, 96, 12192–12197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513. [Google Scholar] [CrossRef]
- Mishra, G.; Zhang, W.; Deng, F.; Zhao, J.; Wang, X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 2006, 312, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Ishiyama, K.; Kato, T.; Tabata, S.; Kobayashi, M.; Shinozaki, K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J. 2005, 43, 107–117. [Google Scholar] [CrossRef]
- Austin-Brown, S.L.; Chapman, K.D. Inhibition of phospholipase D alpha by N-acylethanolamines. Plant Physiol. 2002, 129, 1892–1898. [Google Scholar] [CrossRef]
- Jacobsen, J.V.; Pearce, D.W.; Poole, A.T.; Pharis, R.P.; Mander, L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant 2002, 115, 428–441. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moya-Cuevas, J.; Pérez-Alonso, M.-M.; Ortiz-García, P.; Pollmann, S. Beyond the Usual Suspects: Physiological Roles of the Arabidopsis Amidase Signature (AS) Superfamily Members in Plant Growth Processes and Stress Responses. Biomolecules 2021, 11, 1207. https://doi.org/10.3390/biom11081207
Moya-Cuevas J, Pérez-Alonso M-M, Ortiz-García P, Pollmann S. Beyond the Usual Suspects: Physiological Roles of the Arabidopsis Amidase Signature (AS) Superfamily Members in Plant Growth Processes and Stress Responses. Biomolecules. 2021; 11(8):1207. https://doi.org/10.3390/biom11081207
Chicago/Turabian StyleMoya-Cuevas, José, Marta-Marina Pérez-Alonso, Paloma Ortiz-García, and Stephan Pollmann. 2021. "Beyond the Usual Suspects: Physiological Roles of the Arabidopsis Amidase Signature (AS) Superfamily Members in Plant Growth Processes and Stress Responses" Biomolecules 11, no. 8: 1207. https://doi.org/10.3390/biom11081207
APA StyleMoya-Cuevas, J., Pérez-Alonso, M.-M., Ortiz-García, P., & Pollmann, S. (2021). Beyond the Usual Suspects: Physiological Roles of the Arabidopsis Amidase Signature (AS) Superfamily Members in Plant Growth Processes and Stress Responses. Biomolecules, 11(8), 1207. https://doi.org/10.3390/biom11081207






