Abstract
Pregnane X Receptor (PXR) belongs to the nuclear receptors’ superfamily and mainly functions as a xenobiotic sensor activated by a variety of ligands. PXR is widely expressed in normal and malignant tissues. Drug metabolizing enzymes and transporters are also under PXR’s regulation. Antineoplastic agents are of particular interest since cancer patients are characterized by significant intra-variability to treatment response and severe toxicities. Various PXR polymorphisms may alter the function of the protein and are linked with significant effects on the pharmacokinetics of chemotherapeutic agents and clinical outcome variability. The purpose of this review is to summarize the roles of PXR polymorphisms in the metabolism and pharmacokinetics of chemotherapeutic drugs. It is also expected that this review will highlight the importance of PXR polymorphisms in selection of chemotherapy, prediction of adverse effects and personalized medicine.
1. Introduction
Pregnane X receptor (PXR), also known as nuclear receptor subfamily 1 group I member 2 (NR1I2) and steroid and xenobiotic receptor (SXR), is an orphan receptor of the nuclear receptor gene superfamily and plays a key role in the metabolism of xenobiotics and endobiotics [1,2,3,4,5,6]. Human PXR (hPXR), a 49.7 kDa protein of 434 amino acids, is the product of the NR1I2 gene which is located in chromosome 3 (3q12-q13.3) and consists of approximately 40 kb [7,8]. hPXR is mostly expressed in normal liver tissue, the small intestine and the kidney whereas the PXR expression in tissues like stomach, ovaries, lungs, breast and peripheral blood cells is less frequent [9,10]. PXR expression in neoplastic tissues has also been reported, which differs from the expression levels in normal tissues [6,11].
The structure of the PXR protein is presented in Figure 1. The enlarged, hydrophobic pocket of PXR enables it to accommodate a larger and more diverse number of ligands than the rest of the nuclear receptors, such as endobiotics, pharmaceutical and herbal compounds, environmental factors and dietary supplements and other xenobiotics [12,13].
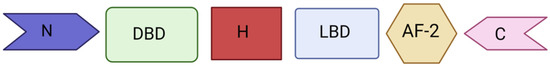
Figure 1.
Structure of PXR protein. PXR consists of a ligand-depended activation function 2 (AF-2) and a highly conserved DNA-binding domain (DBD) in the N-terminal with two zinc-fingers while the DBD is connected to the C-terminal ligand binding domain (LBD) using a hinge.
2. PXR Function
As a master regulator of xenobiotic response PXR adjusts the expression of many phase I and phase II drug metabolizing enzymes (DME), such as cytochrome P450, uridine diphosphate (UDP)-glucuronosyltransferases, sulfotransferases and carboxylesterases [13,14]. PXR also regulates drug-efflux pumps multi-drug resistance gene 1 (MDR1), MDR2, ATP-binding cassette transporter C 2 (ABCC) and anion-transporting polypeptide 2 (OATP) [14,15,16]. All the aforementioned enzymes, under the control of PXR play a significant role in the biotransformation, metabolism and clearance of therapeutic agents, including chemotherapeutic agents, that may result in drug-drug, drug-herb and drug-food interactions, toxicity, adverse effects and reduced efficacy [13,14,16,17]. The overall functions of PXR protein are presented in Figure 2.
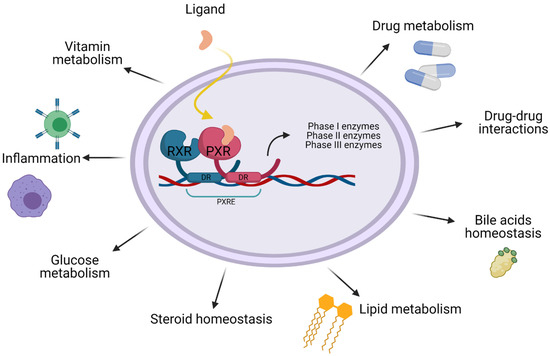
Figure 2.
Functions of PXR. Besides being a significant xenobiotic regulator PXR also seems to have an important role in lipid and glucose metabolism, bile acid detoxification, steroid homeostasis, inflammation and vitamin metabolism.
It has also been reported PXR’s participation in tumor cell proliferation and growth, apoptosis and metastasis as well as in liver regeneration and hepatic proliferation, indicating the important role of PXR in cancer [14,18].
Various nonsynonymous polymorphisms have been identified in essential domains of NR1I2, including DBD and LBD affecting either the DNA or the ligand binding process, while mutations in the 5′ UTR could affect PXR expression, resulting in modifications in drug metabolism and pharmacokinetics as well as contributing to drug resistance [3,12,19]. Polymorphisms located in gene exons can modify the LBD or the DBD domains of PXR, changing the interactions between these domains and ligands, gene promoters and co-regulators, while polymorphisms located in non-coding regions can affect the regulation of transcription and translation [3,20]. In the 3′ UTR of NR1I2 gene, specifically in the miRNA target sequence, several single nucleotide polymorphisms (SNPs) have been described to alter post-transcriptional micro-RNA (mi-RNA)-mediated regulation of PXR expression, creating or deleting regions of interaction with miRNAs, as NR1I2 rs1054190 (C > T) and NR1I2 rs1054191 (G > A), respectively [21]. Studies have also identified NR1I2 SNPs in exons, which initiate drastic changes in PXR in vivo but with very low frequencies in the population thus requiring very large cohort studies of patients [20].
3. Regulation
When PXR is not linked with an agonist its action is regulated by transcriptional co-repressors like nuclear receptor co-repressor 1 (NCoR1) and NCoR2 which inhibit PXR’s transcription via histone deacetylases (HDACs) activity [11,18]. When a ligand binds to PXR, through the LBD, the receptor heterodimerizes with retinoid X receptor (RXR), binds to xenobiotic response elements (XREs) and hormone response elements (HREs) and changes the status of coregulators (like co-repressors and co-activators, such as steroid receptor coactivator 1 and 3 [SRC-1 and SRC-3]), which remodel chromatin via histone acetyltransferase (HAT) activity and regulate transcription [11,22,23,24,25]. Once the ligand binds to the LBD, the AF-2 region at the C-terminus binds to specific amino-acid motifs of transcriptional coactivators and corepressors, resulting in the correct arrangement of the ligand in the PXR ligand-binding pocket [26,27,28]. PXR binds to different DNA response elements, including direct repeats (DRs) DR-4, DR-5 and everted repeats (ERs) ER-6 and ER-8 among others, while the receptor seems to have a higher binding preference for DNA-binding motif of DR-(5n + 4) [28,29]. Activated Nuclear Factor-kappa B (NF-κB) is reported to inhibit the activation and function of PXR, while inhibited NF-κB increases PXR activity and transcription of target genes. This mutual negative crosstalk between PXR and NF-κB indicated that PXR acts as a negative mediator of inflammation and immunity [30].
4. Post-Translational and Post-Transcriptional PXR Modifications
Apart from the direct ligand-depended activation PXR is subjected to post-translational modifications (PTMs) resulting in variations of PXR transcriptional activities which are presented in Figure 3 [31].
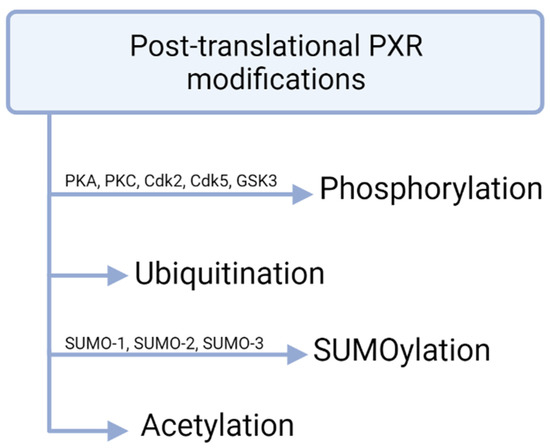
Figure 3.
PXR modifications. Phosphorylation of PXR mostly reduces transcriptional activity. Ubiquitination is mediated by the 26S proteasome and SUMOylation takes place in lysine residues. Post-translational acetylation of PXR has been shown in vivo.
There is evidence that PXR acetylation interacts with PXR SUMOylation and may be mutually excluded, but further research is needed [17,32].
Micro-RNAs (mi-RNA) are reported to alter PXR expression via post-transcriptional modifications usually targeting the 3′-untraslated region (UTR) sequence of human PXR transcripts [33]. miR-30c-1-3p is identified as a PXR silencer, thus decreases the mRNA levels of PXR and alters CYP3A4 expression [33]. Other post-translational PXR regulators are miR-18a-5p, miR-148a, miR-34a, miR-150, miR-27a and miR-140-3p, which negatively regulate PXR expression, resulting in decreased CYP3A4 expression [34,35,36,37,38]. miR-449a is described to inhibit PXR through HNF4-a in human hepatocytes [39].
5. Agonists and Antagonists
PXR can bind to a variety of agonists due to the receptor’s large cavity, as mentioned above. Some endobiotic PXR ligands are bile acids and their precursors, progesterone, pregnenolone, 17-hydroxypregnenolone, cholesterol and it’s metabolites and lithocholic acid [12,14,40]. Regarding xenobiotic PXR ligands, these may include prescription drugs and anticancer agents like ritonavir, rifampicin, clotrimazole, cyclosporin, paclitaxel, Taxol, tamoxifen, dexamethasone, troglitazone, statins, nifedipine, spironolactone, endocrine disruptors and phenobarbital [12,40]. Other xenobiotic ligands are carotenoids, vitamins like vitamin K2 and vitamin E, herbal medicines like St. John’s Wort, Kava Kava, Gugulipid, Sweet Wormwood Herb, Schisandra and environmental pollutants such as organochlorine pesticides and polybrominated diphenyl ether flame retardants [12,41].
In order to prevent PXR-mediated drug-drug interactions and restrict the variability of efficacy of therapeutics, many PXR antagonists have been identified or been developed. In 2001 the first PXR antagonist was reported, ET-743, followed by numerous other compounds like ketoconazole, fluconazole, enilconazole, camptothecin, metformin, sulforaphane, sesamin, coumestrol, allyl isothiocyanate, algal carotenoid fucoxanthin, silybin and isosilybin [42,43,44]. Environmental toxins, such as polychlorinated biphenyls (PCBs) exhibit antagonistic activity, while highly chlorinated PCBs selectively antagonizing mPXR but not human PXR [45]. However, many of these compounds bind other targets at concentrations below the range that affects PXR, resulting in incapability to inhibit PXR in vivo [43]. Some of these antagonists, ketoconazole, coumestrol and metformin, are reported to inhibit PXR’s transactivation either via interfering with PXR’s coactivators or via binding in the AF-2 domain independently of PXR LBD, while ketoconazole is reported to be able to bind to two distinct PXR binding pockets either causing allosteric or direct inhibition of coactivator binding [42,43]. Ochatoxin A, a mycotoxin, has also been shown to significantly downregulate PXR activity in human primary hepatocytes [46,47]. Other antagonists are clotrimazole, dabrafenib, SR12813, Nelfinavir and SPA70 [48,49]. SPA70 interacts with hPXR LBD, is highly specific for hPXR and has selective downregulating effects [47,50,51,52]. Although several analogs of PXR antagonists have been synthesized many function as agonists due to the flexible and large cavity of PXR, which adapts to the shape of the ligands [43].
6. PXR and Cancer
As stated, PXR regulates the expression of a variety of target genes and can participate in many physiological and pathological conditions through complex cellular circuits as PXR manipulates the expression of target genes participating in biotransformation, inflammation, cell-cycle regulation, apoptosis, tissue growth and oxidative stress [47,53]. These biological functions of PXR have an impact on cancer initiation, promotion and progression, and on the outcome of chemotherapeutic agents, as PXR and its target genes are linked with multidrug resistance, poor chemotherapy outcome as well as detoxification, defense and homeostasis maintenance, which inhibit cancer development, highlighting PXR as a central target of cancer regulation [47]. PXR is described to be associated with various cancers, including breast, esophageal, prostate, ovaries, cervix, endometrial tissues, colon, pancreas, liver, lung and hematological malignancies [18,53,54,55]. Also PXR overexpression and altered subcellular location, due to mutation, is linked with endometrial, breast and colorectal cancer [16]. Esophageal adenocarcinoma and Barrett’s epithelium were associated with increased PXR mRNA levels, while the PXR protein was not detected in normal esophageal epithelium and was detected in the nuclei of cancer cells [54].
Due to the involvement of PXR in drug transporters’ and the gene expression and regulation of drug metabolizing enzymes, such as CYP3A4, which is responsible for metabolizing more than 50% of drugs that include chemotherapeutics, PXR significantly contributes to chemotherapy resistance and variations in the chemotherapeutic outcome [16,56]. PXR contributes to the chemotherapy outcome by interfering with the metabolism, drug resistance, tumor sensitivity, apoptosis and pharmacokinetics parameters of many chemotherapeutic agents, such as tamoxifen, irinotecan, vinblastine, doxorubicin, paclitaxel, cisplatin and ixabepilone in cancer cell lines and patients [11,14]. Several studies have indicated PXR involvement in tumor sensitivity to anticancer agents. Enhanced PXR activation via miRNA-30c repression by factor that binds to the inducer of short transcripts-1 (FBI-1) was linked with chemotherapeutic resistance in triple-negative breast cancer cell lines [57]. Dabrafenib-induced hPXR activation in colon cancer cell lines was associated with enhanced expression of PXR target genes, including CYP3A4 and CYP2D6. This study indicates the potential impact of dabrafenib on its own metabolism or the metabolism of other therapeutic agents combined with dabrafenib via PXR regulation [58].
Considering that cancer patients are treated with multidrug regimens, PXR-mediated drug-drug interactions and drug toxicity are very important, as is PXR-mediated chemoresistance, which affects clinical outcome. So far, the exact molecular mechanisms are unclear, but SNPs within the NR1I2 gene could be a possible mechanism involved in MDR and altered clinical outcome of antineoplastic agents [56]. Up to now numerous SNPs have been reported in the NR1I2 gene, some of which may have an impact on the course of cancer treatment. In view of the above considerations, the aim of this review is to highlight the significance of researched SNPs in efficacy and toxicity of antineoplastic agents.
7. PXR Polymorphisms and Cancer Pharmacogenetics
7.1. Gastrointestinal Cancer
Pharmacogenomics have recently widely entered the personalization of CRC treatment, specifically centering on the genetic variability in metabolism-related genes, such as NR1I2. In Caucasian patients with metastatic colorectal cancer (CRC) treated with FOLFIRI (irinotecan, bolus and continuous-infusion fluorouracil, leucovorin), the T allele at NR1I2 rs1054190 (C > T) was associated with worse overall survival (OS) and progression-free survival (PFS), identifying NR1I2-rs1054190 polymorphism as a potential prognostic marker of OS [59]. The aforementioned SNP, located in 3′UTR, has been linked with downregulation of PXR and regulation of the expression of PXR via miRNA mechanisms [16,56,58]. Patients with metastatic CRC treated with FOLFIRI or FOLFIRINOX carrying the A allele of NR1I2 rs10934498 (G > A, G > C, G > T) were associated with a decreased area under the curve (AUC) of SN-38, the active metabolite of irinotecan, decreased biliary index and a decreased risk of grade 3–4 hematotoxicity. This study also highlighted the association between patients carrying the T allele at NR1I2 rs3814055 (C > T) as well as patients carrying the C allele at NR1I2 rs1523127 (C > A) and increased risk of grade 3–4 hematotoxicity, while patients with the G allele at NR1I2 rs2472677 (C > G, C > T) exhibited higher risk for all types of grade 3–4 toxicity [60]. Asian patients with gastrointestinal stromal tumors (GISTs) carrying the T allele at NR1L2 rs3814055 (C > T) exhibited lower steady-state imatinib dose-adjusted plasma concentrations than patients with the wild type (CC), while in the same study patients carrying the mutant T allele at NR1L2 rs3814055 (C > T) showed significantly lower incidence rate of continuous edema, an adverse effect of imatinib [61]. These results are in agreement with the results from Yi Qian et al. where patients with the TT genotype of NR1I2 rs3814055 (C > T) were described to have lower unbound imatinib dose-adjusted concentration [62].
7.2. Breast Cancer
Drug resistance to chemotherapeutic agents and toxicity are often noticed in breast cancer patients during treatment and worsen the chemotherapy outcome [16,63]. In Caucasian women with breast cancer treated with FAC (doxorubicin, 5′-fluorouracil and cyclophosphamide) the presence of NR1L2 rs3732359 (G > A) was an independent predictor of OS as patients carrying the AA genotype were described to have a 2 times higher risk of death compared to homozygous for the wild type allele (GG) and heterozygous (AG) [63]. Regarding the clearance of doxorubicin in Asian women with invasive breast cancer treated with adjuvant chemotherapy with doxorubicin/cyclophosphamide, patients carrying the haplotype cluster tagged by IVS6-17C > T (NR1I2 rs2276707) and 2654T > C (NR1I2 rs3814058) were characterized by reduced doxorubicin clearance [7]. SNPs NR1I2 rs3732360 (C > T, C > G), rs1054190 (C > T) and rs1054191 (G > A) are described to be linked with change in doxorubicin pharmacokinetics via altering the miRNA mediated post-transcriptional regulation of PXR in Asian (Indian) breast cancer patients [16]. Haplotype PXR*1B, which consists of NR1I2 rs2276707 (C > G, C > T) and NR1I2 rs3814058 (T > C), has been linked with decreased plasma expression of PXR in hepatic tissue, while NR1I2 rs3732359 and NR1I2 rs3732360, located in the 3′UTR, affect the effectiveness of miRNA and PXR mRNA resulting in modifications of PXR’s expression [7,16].
7.3. Renal Cell Carcinoma
Sunitinib is currently registered as first-line and second-line therapy for metastatic renal cell carcinoma (mRCC) and its efficacy may be dependent on its exposure, regulated by efflux pumps and metabolizing enzymes. Clear-cell RCC patients, treated with sunitinib, with the T allele at NR1I2 rs2276707 (C > T, C > G) and patients carrying the T allele at NR1I2 rs3814055 (C > T) were described to have a shorter PFS and a shorter OS for the T allele at rs3814055 (C > T), results confirmed by Beuselinck, B. et al. [64,65]. Another study described the link between the response rate (RR) for pazopanib in RCC patients carrying the NR1I2 rs3814055 (C > T). Patients carrying the T allele showed significantly reduced RR with a potential consequence for drug exposure and a trend to have reduced PFS compared with carriers of the wild type genotype (CC) [66,67].
7.4. Others
The presence of NR1I2 rs6785049 (G > A, G > T) or rs3814055 (C > T) was linked with inter-patient variability of temsirolimus pharmacokinetics and toxicity in patients with metastatic bladder cancer. Patients with the T allele of NR1L2 rs3814055 or the G allele of NR1L2 rs6785049 showed significantly lower frequency of adverse events, while patients homozygous for the NR1L2 rs3814055 wild type C allele (CC) and patients homozygous for the NR1L2 rs6785049 mutant A allele (AA) exhibited higher frequencies of severe temsirolimus toxicity. This study also indicated that NR1L2 rs6785049 GG genotype was correlated with increased exposure to active entities (AUCsum) and that NR1L2 rs3814055 TT genotype was linked with extended tesmirolimus T1/2 although the effect was not additive [68].
Patients with nasopharyngeal cancer treated with docetaxel, carrying the mutant allele of SNPs NR1L2 rs3732359 (G > A), rs3732360 (C > T, C > G) or rs3814058 (T > C), exhibited a decrease in nadir hemoglobin from baseline in cycle 1 but did not show any correlation with the pharmacokinetics of docetaxel [69]. Also, homozygous for the mutant allele and heterozygous (CC + TC) Asian non-small cell lung cancer (NSCLC) patients treated with platinum-based chemotherapy with NR1I2 rs3814058 (T > C) exhibited higher risk of hematological toxicity than patients homozygous for the wild type allele (TT) [70]. Asian patients with chronic myeloid leukemia (CML) treated with bosutinib, who also carried the genotypes (GG) or (TT) of NR1L2 rs6785049 (G > A, G > T) or NR1L2 rs2276707 (C > T), respectively, exhibited lower bosutinib through plasma concentration (C0) than patients carrying alleles A of rs6785049 or allele C of rs2276707, while patients carrying both (GG) and (TT) genotypes showed lower bosutinib C0 than other genotypes, indicating increased clearance of the anticancer agent [71].
In docetaxel-based treatment for patients with solid tumors the SNP NR1I2 rs3732359 (G > A) was significantly associated with docetaxel-induced myelosuppression grade ≥3, as carriers of the wild type G allele were of higher myelosuppression risk [72]. Another study indicated that, in cancer patients who received carboplatin plus paclitaxel as chemotherapy, the NR1I2 rs1523130 (T > A, T > C, T > G) and rs1523127 (T > G) were related, with altered sensitivity to thrombocytopenia; as the A allele of rs1523130 and the G allele of rs1523127 exhibited a recessive and genotypic effect, the AA genotype of rs15233130 and the GG genotype of rs1523127 were correlated with a decreased sensitivity to thrombocytopenia. This study also showed that carriers of two copies of the ATG haplotypes of NR1I2 rs1523130 (T > A, T > C, T > G), NR1I2 rs1523127 (T > G) and NR1I2 rs3814055 (C > T) were less sensitive to thrombocytopenia [73]. Osteosarcoma patients treated with methotrexate (MTX) presented differences in MTX pharmacokinetics and toxicities depending on their genotype. Patients carrying the SNPs NR1I2 rs3814055 (C > T) and rs7643038 (A > G) exhibited longer first half-life of MTX, while SNPs NR1I2 rs6785049 (G > A, G > T) and rs3732361 (G > A) were linked with higher 48 h MTX concentration. The same study indicates correlation between SNPs NR1I2 rs3732361, rs3814058 and rs6785049 and lower risk of hepatic and bone marrow toxicity [74]. All of the above are summarized in Figure 4 and Table 1.
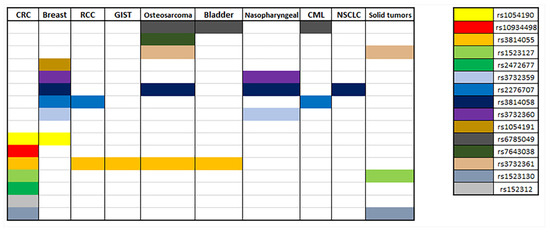
Figure 4.
Summary of correlations of NR1I2 polymorphisms with different types of cancer.

Table 1.
PXR polymorphisms and their associations with efficacy, toxicity and pharmacokinetics of chemotherapeutic agents.
8. Discussion
Numerous NR1I2 SNPs have been researched regarding their association with pharmacotherapy outcome and severe toxicities. In Chinese Han tuberculosis patients who received anti-tuberculosis treatment, with NR1I2 rs7643645 (A > G) were associated with increased risk of anti-tuberculosis drug-induced hepatotoxicity (ATDH) (GG genotype), while carriers of the NR1I2 rs2276707 (C > T) were linked with reduced risk of ATDH [75]. These results were at conflict with the results from Yu Wang et al., where the G allele of NR1I2 rs7643645 was associated with reduced risk of ATDH [76]. NR1I2 polymorphisms detected in patients who received cyclosporin (CsA) after their first renal transplantation were correlated with altered CsA C0/D and C2/D after the first month of transplantation, as the haplotype cluster PXR*1B tagged by NR1I2 rs2276707 and rs3814058 was correlated with increased CsA C2/D [77]. A study also indicated the importance of NR1I2 rs13059232 (T > C) as a biomarker for clopidogrel therapy in acute ischemic stroke (IS) patients, since patients carrying the CC genotype exhibited poorer clinical outcome than patients carrying the T allele, while comparable results were not observed in the aspirin cohort [78]. HIV-positive patients treated with atazanavir and ritonavir and carrying the G allele of NR1I2 rs1523130 (T > G) exhibited higher ritonavir intracellular concentrations [79]. Furthermore, HIV-positive patients, homozygous for the mutant allele of NR1I2 rs2472677 (C > T) and treated with atazanavir, were associated with a 17.0% higher clearance of atazanavir [80]. Alzheimer’s patients treated with memantine and carrying the T allele of NR1I2 rs1523130 (T > C) were reported to exhibit slower memantine clearance [81]. It has also been reported that the Cssmin of voriconazole, given to patients with hematological malignancies, is affected significantly by SNPs NR1I2 rs2461817 (A > C), rs7643645 (A > G), rs3732359 (G > A), rs3814057 (A > C) and rs6785049 (G > A) [82]. The studies above highlight the possible significant involvement of PXR in the pharmacokinetics of therapeutic agents, the therapeutic outcome along with adverse effects and severe toxicity. Especially in neoplastic diseases, where patients exhibit important intra-individual variations of the therapeutic outcome and severe toxicities, identification and characterization of PXR polymorphisms are deemed necessary.
Although chemotherapy is still the main strategy followed for the treatment of many solid tumors and systematic malignancies, it is characterized by significant inter-individual heterogeneity of chemotherapeutic response, toxicity and drug resistance, which alter the clinical outcome of the chemotherapeutic treatment. Evidently, better understanding of factors that determine the chemotherapeutic response will help to detect patients who are at risk of displaying severe toxicities or benefit the most from a specific therapeutic combination, providing personalized treatment [14,83]. To achieve this goal extended research which incorporate modern techniques like next generation sequencing (NGS) is required as well as conducting more clinical studies with a larger cohort of patients.
9. Conclusions
PXR is characterized as a master regulator of xenobiotic and endobiotic metabolism, since it modulates the expression of significant enzymes involved in drug distribution, metabolism and clearance, and has been linked with various functions, such as inflammation, drug-drug interactions, detoxification, and vitamin and bile acid metabolism. PXR polymorphisms in the NR1I2 gene are considered to have noteworthy consequences on the protein’s function, such as abnormal DNA binding and changes in target genes transactivation and expression; thus detection and identification of functional PXR SNPs, and their distribution in the different populations and ethnicities, are crucial for understanding and explaining the mechanisms behind the variations in drug pharmacokinetics and clinical outcome. Cancer patients receiving chemotherapeutic agents are often characterized by variability in chemotherapeutic outcome, severe adverse events, resistance to therapy and drug-drug interactions due to polypharmacy. Since PXR regulates the metabolism and pharmacokinetics of the majority of the antineoplastic agents, it is important to study the effects PXR polymorphisms have on them so we can enhance the personalized therapy of cancer therapy. The current review highlights the importance of PXR polymorphisms in cancer precision medicine. Current literature data report that gene PXR polymorphisms appear to interfere with pharmacokinetics, metabolism and toxicity of antineoplastic factors. Some of these seem to affect the response and toxicity of more than one antineoplastic therapy, for instance NR1I2 rs3814055 has been linked with variability in pharmacokinetics parameters and toxicity of FOLFIRINOX/FOLFIRI, imatinib, sunitinib, pazopanib, methotrexate and temsirolimus. The study of PXR SNPs in combination with the development of PXR antagonists with antineoplastic therapies that activate PXR receptors have the potential to decrease or eradicate adverse events, toxicity and chemoresistance, enhancing precision medicine in cancer. In summary, 15 polymorphisms were found to be associated with variations in chemotherapy clinical outcome or toxicity in cancers such as breast, gastrointestinal and renal, all of them located in non-translated regions of the NR1I2 gene. Further information is required to fully understand the role of PXR SNPs in response to treatment and clinical outcome of cancer patients, although the currently available data indicate the significance of PXR polymorphisms for the pharmacotherapy of cancer patients.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable. No new data were created or analyzed in this study.
Acknowledgments
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yoshinari, K. Role of Nuclear Receptors PXR and CAR in Xenobiotic-Induced Hepatocyte Proliferation and Chemical Carcinogenesis. Biol. Pharm. Bull. 2019, 42, 1243–1252. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Goodwin, B.; Willson, T.M. The Nuclear Pregnane X Receptor: A Key Regulator of Xenobiotic Metabolism. Endocr. Rev. 2002, 23, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Patsouris, E.; Theocharis, S. Pregnane X Receptor Polymorphisms Associated with Human Diseases. Expert Opin. Ther. Targets 2013, 17, 1167–1177. [Google Scholar] [CrossRef]
- Koutsounas, I.; Patsouris, E.; Theocharis, S. Pregnane X Receptor and Human Malignancy. Histol. Histopathol. 2013, 28, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Koutsounas, I.; Theocharis, S.; Patsouris, E.; Giaginis, C. Pregnane X Receptor (PXR) at the Crossroads of Human Metabolism and Disease. Curr. Drug Metab. 2013, 14, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Koutsounas, I.; Giaginis, C.; Alexandrou, P.; Zizi-Serbetzoglou, A.; Patsouris, E.; Kouraklis, G.; Theocharis, S. Pregnane X Receptor Expression in Human Pancreatic Adenocarcinoma. Pancreas 2015, 44, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Sandanaraj, E.; Lal, S.; Selvarajan, V.; Ooi, L.L.; Zee, W.W.; Nan, S.W.; Ang, P.C.S.; Lee, E.J.D.; Chowbay, B. PXR Pharmacogenetics: Association of Haplotypes with Hepatic CYP3A4 and ABCB1 Messenger RNA Expression and Doxorubicin Clearance in Asian Breast Cancer Patients. Clin. Cancer Res. 2008, 14, 7116–7126. [Google Scholar] [CrossRef]
- Zhang, J.; Kuehl, P.; Green, E.D.; Touchman, J.W.; Watkins, P.B.; Daly, A.; Hall, S.D.; Maurel, P.; Relling, M.; Brimer, C.; et al. The Human Pregnane X Receptor: Genomic Structure and Identification and Functional Characterization of Natural Allelic Variants. Pharmacogenetics 2001, 11, 555–572. [Google Scholar] [CrossRef]
- Pavek, P.; Dvorak, Z. Xenobiotic-Induced Transcriptional Regulation of Xenobiotic Metabolizing Enzymes of the Cytochrome P450 Superfamily in Human Extrahepatic Tissues. Curr. Drug Metab. 2008, 9, 129–143. [Google Scholar] [CrossRef]
- Nishimura, M.; Naito, S.; Yokoi, T. Tissue-Specific MRNA Expression Profiles of Human Nuclear Receptor Subfamilies. Drug Metab. Pharmacokinet. 2004, 19, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Pondugula, S.R.; Mani, S. Pregnane Xenobiotic Receptor in Cancer Pathogenesis and Therapeutic Response. Cancer Lett. 2013, 328, 1–9. [Google Scholar] [CrossRef]
- Zhang, B.; Xie, W.; Krasowski, M.D. PXR: A xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics 2008, 9, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Hogle, B.C.; Guan, X.; Folan, M.M.; Xie, W. PXR as a Mediator of Herb–Drug Interaction. J. Food Drug Anal. 2018, 26, S26–S31. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, W.; Hu, L.; Lv, J.; Wang, H.; Zhou, H.; Fan, L. Role of Pregnane X Receptor in Chemotherapeutic Treatment. Cancer Chemother. Pharmacol. 2014, 74, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.E.; Davis-searles, P.R.; Lambert, M.H.; Redinbo, M.R.; Hill, C. Coactivator Binding Promotes the Specific Interaction Between Ligand and the Pregnane X Receptor. J. Mol. Biol. 2003, 2836, 815–828. [Google Scholar] [CrossRef]
- Revathidevi, S.; Sudesh, R.; Vaishnavi, V.; Kaliyanasundaram, M.; MaryHelen, K.G.; Sukanya, G.; Munirajan, A.K. Screening for the 3’ UTR Polymorphism of the PXR Gene in South Indian Breast Cancer Patients and Its Potential Role in Pharmacogenomics. Asian Pac. J. Cancer Prev. 2016, 17, 3969–3975. [Google Scholar]
- Smutny, T.; Mani, S.; Pavek, P. Post-Translational and Post-Transcriptional Modifications of Pregnane X Receptor (PXR) in Regulation of the Cytochrome P450 Superfamily. Curr. Drug Metab. 2013, 14, 1059–1069. [Google Scholar] [CrossRef]
- Pondugula, S.R.; Pavek, P.; Mani, S. Pregnane X Receptor and Cancer: Context-Specificity Is Key. Nucl. Recept. Res. 2016, 3, 101198. [Google Scholar] [CrossRef]
- Qiao, E.; Ji, M.; Wu, J.; Ma, R.; Zhang, X.; He, Y.; Zha, Q.; Song, X.; Zhu, L.W.; Tang, J. Expression of the PXR Gene in Various Types of Cancer and Drug Resistance (Review). Oncol. Lett. 2013, 5, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Mbatchi, L.C.; Brouillet, J.P.; Evrard, A. Genetic Variations of the Xenoreceptors NR1I2 and NR1I3 and Their Effect on Drug Disposition and Response Variability. Pharmacogenomics 2018, 19, 61–77. [Google Scholar] [CrossRef]
- Swart, M.; Dandara, C. Genetic Variation in the 3′-UTR of CYP1A2, CYP2B6, CYP2D6, CYP3A4, NR1I2, and UGT2B7: Potential Effects on Regulation by MicroRNA and Pharmacogenomics Relevance. Front. Genet. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dussault, I.; Lin, M.; Hollister, K.; Fan, M.; Termini, J.; Sherman, M.A.; Forman, B.M. A Structural Model of the Constitutive Androstane Receptor Defines Novel Interactions That Mediate Ligand-Independent Activity. Mol. Cell. Biol. 2002, 22, 5270–5280. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Zeng, S.; Xie, W. Nuclear Receptors PXR and CAR: Implications for Drug Metabolism Regulation, Pharmacogenomics and Beyond. Expert Opin. Drug Metab. Toxicol. 2013, 9, 253–266. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mani, S. Orphan Nuclear Receptors as Targets for Drug Development. Pharm. Res. 2010, 27, 1439–1468. [Google Scholar] [CrossRef]
- Bourguet, W.; Germain, P.; Gronemeyer, H. Nuclear Receptor Ligand-Binding Domains: Three-Dimensional Structures, Molecular Interactions and Pharmacological Implications. Trends Pharmacol. Sci. 2000, 21, 381–388. [Google Scholar] [CrossRef]
- Lazar, M.A. Nuclear Receptor Corepressors. Nucl. Recept. Signal. 2003, 1, nrs.01001. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.G.; Lunyak, V.V.; Glass, C.K. Sensors and Signals: A Coactivator/Corepressor/Epigenetic Code for Integrating Signal-Dependent Programs of Transcriptional Response. Genes Dev. 2006, 20, 1405–1428. [Google Scholar] [CrossRef]
- Orans, J.; Teotico, D.G.; Redinbo, M.R. The Nuclear Xenobiotic Receptor Pregnane X Receptor: Recent Insights and New Challenges. Mol. Endocrinol. 2005, 19, 2891–2900. [Google Scholar] [CrossRef]
- Cui, J.Y.; Gunewardena, S.S.; Rockwell, C.E.; Klaassen, C.D. ChIPing the Cistrome of PXR in Mouse Liver. Nucleic Acids Res. 2010, 38, 7943–7963. [Google Scholar] [CrossRef]
- Wahli, W. A Gut Feeling of the PXR, PPAR and NF-ΚB Connection. J. Intern. Med. 2008, 263, 613–619. [Google Scholar] [CrossRef]
- Staudinger, J.L.; Xu, C.; Biswas, A.; Mani, S. Post-Translational Modification of Pregnane x Receptor. Pharmacol. Res. 2011, 64, 4–10. [Google Scholar] [CrossRef]
- Cui, W.; Sun, M.; Zhang, S.; Shen, X.; Gal-, N.; Williams, T.D.; Staudinger, J.L. A SUMO-Acetyl Switch in PXR Biology. BBA Gene Regul. Mech. 2016, 1859, 1170–1182. [Google Scholar] [CrossRef]
- Vachirayonstien, T.; Yan, B. MicroRNA-30c-1-3p Is a Silencer of the Pregnane X Receptor by Targeting the 3′-Untranslated Region and Alters the Expression of Its Target Gene Cytochrome P450 3A4. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 1238–1244. [Google Scholar] [CrossRef][Green Version]
- Takagi, S.; Nakajima, M.; Mohri, T.; Yokoi, T. Post-Transcriptional Regulation of Human Pregnane X Receptor by Micro-RNA Affects the Expression Of Cytochrome P450 3A4. J. Biol. Chem. 2008, 283, 9674–9680. [Google Scholar] [CrossRef] [PubMed]
- Lamba, V.; Ghodke, Y.; Guan, W.; Tracy, T.S. MicroRNA-34a Is Associated with Expression of Key Hepatic Transcription Factors and Cytochromes P450. Biochem. Biophys. Res. Commun. 2014, 445, 404–411. [Google Scholar] [CrossRef]
- Li, J.; Zhao, J.; Wang, H.; Li, X.; Liu, A.; Qin, Q.; Li, B. MicroRNA-140-3p Enhances the Sensitivity of Hepatocellular Carcinoma Cells to Sorafenib by Targeting Pregnenolone X Receptor. OncoTargets Ther. 2018, 5885–5894. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Turkistani, A.A.; Chang, W.; Hu, C.; Xu, Z.; Chang, T.K.H. Negative Regulation of Human Pregnane X Receptor by MicroRNA-18a-5p: Evidence for Suppression of MicroRNA-18a-5p Expression by Rifampin and Rilpivirine. Mol. Pharmacol. 2017, 48–56. [Google Scholar] [CrossRef]
- Reuter, T.; Herold-Mende, C.; Dyckhoff, G.; Rigalli, J.P.; Weiss, J. Functional Role of MiR-148a in Oropharyngeal Cancer: Influence on Pregnane X Receptor and P-Glycoprotein Expression. J. Recept. Signal Transduct. 2019, 39, 451–459. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Li, L.; Gaedigk, A.; Bradford, L.D.A.; Benson, E.A.; Flockhart, D.A.; Skaar, T.C. In Silico and in Vitro Identification of MicroRNAs That Regulate Hepatic Nuclear Factor 4α Expression. Drug Metab. Dispos. 2012, 40, 726–733. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, Y.; Chen, S.; Nie, D.; Chen, Y.; Tang, Y.; Chen, S.; Nie, D. Regulation of Drug Resistance by Human Pregnane X Receptor in Breast Cancer Regulation of Drug Resistance by Human Pregnane X Receptor in Breast Cancer. Cancer Biol. Ther. 2016, 4047, 1265–1272. [Google Scholar] [CrossRef]
- Zhou, H.; Tomlinson, B. Theranostics Meets Traditional Chinese Medicine: Rational Prediction of Drug—Herb Interactions. Expert Rev. Mol. Diagn. 2012, 12, 815–830. [Google Scholar]
- Smutny, T.; Pavek, P. Resveratrol as an Inhibitor of Pregnane X Receptor ( PXR ): Another Lesson in PXR Antagonism. J. Pharmacol. Sci. 2014, 178, 177–178. [Google Scholar] [CrossRef]
- Mani, S.; Dou, W.; Redinbo, M.R. PXR Antagonists and Implication in Drug Metabolism. Drug Metab. Rev. 2013, 45, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Synold, T.W.; Dussault, I.; Forman, B.M. The Orphan Nuclear Receptor SXR Coordinately Regulates Drug Metabolism and Efflux. Nat. Med. 2001, 7, 584–590. [Google Scholar] [CrossRef]
- Chai, S.C.; Wright, W.C.; Chen, T. Strategies for Developing Pregnane X Receptor Antagonists: Implications from Metabolism to Cancer. Med. Res. Rev. 2020, 40, 1061–1083. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, Z.; Fan, J.T.; Yan, B. Dechlorination and Demethylation of Ochratoxin A Enhance Blocking Activity of PXR Activation, Suppress PXR Expression and Reduce Cytotoxicity. Toxicol. Lett. 2020, 332, 171–180. [Google Scholar] [CrossRef]
- Xing, Y.; Yan, J.; Niu, Y. PXR: A Center of Transcriptional Regulation in Cancer. Acta Pharm. Sin. B 2020, 10, 197–206. [Google Scholar] [CrossRef]
- Toporova, L.; Grimaldi, M.; Boulahtouf, A.; Balaguer, P. Assessing the Selectivity of FXR, LXRs, CAR, and RORγ Pharmaceutical Ligands with Reporter Cell Lines. Front. Pharmacol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Burk, O.; Kronenberger, T.; Keminer, O.; Lee, S.M.L.; Schiergens, T.S.; Schwab, M.; Windshügel, B. Nelfinavir and Its Active Metabolite M8 Are Partial Agonists and Competitive Antagonists of the Human Pregnane X Receptor. Mol. Pharmacol. 2021, 99, 184–196. [Google Scholar] [CrossRef]
- Lin, W.; Wang, Y.M.; Chai, S.C.; Lv, L.; Zheng, J.; Wu, J.; Zhang, Q.; Wang, Y.D.; Griffin, P.R.; Chen, T. SPA70 Is a Potent Antagonist of Human Pregnane X Receptor. Nat. Commun. 2017, 8, 741. [Google Scholar] [CrossRef]
- Huber, A.D.; Wright, W.C.; Lin, W.; Majumder, K.; Low, J.A.; Wu, J.; Buchman, C.D.; Pintel, D.J.; Chen, T. Mutation of a Single Amino Acid of Pregnane X Receptor Switches an Antagonist to Agonist by Altering AF-2 Helix Positioning. Cell. Mol. Life Sci. 2021, 78, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, W.; Wright, W.C.; Chai, S.C.; Wu, J.; Chen, T. Building a Chemical Toolbox for Human Pregnane X Receptor Research: Discovery of Agonists, Inverse Agonists, and Antagonists Among Analogs Based on the Unique Chemical Scaffold of SPA70. J. Med. Chem. 2021, 64, 1733–1761. [Google Scholar] [CrossRef] [PubMed]
- Creamer, B.A.; Sloan, S.N.B.; Dennis, J.F.; Rogers, R.; Spencer, S.; McCuen, A.; Persaud, P.; Staudinger, J.L. Associations between Pregnane X Receptor and Breast Cancer Growth and Progression. Cells 2020, 9, 2295. [Google Scholar] [CrossRef] [PubMed]
- Van de Winkel, A.; Menke, V.; Capello, A.; Moons, L.M.G.; Pot, R.G.J.; van Dekken, H.; Siersema, P.D.; Kusters, J.G.; van der Laan, L.J.W.; Kuipers, E.J. Expression, Localization and Polymorphisms of the Nuclear Receptor PXR in Barrett’s Esophagus and Esophageal Adenocarcinoma. BMC Gastroenterol. 2011, 11, 108. [Google Scholar] [CrossRef]
- Zhang, L.; Qiu, F.; Lu, X.; Li, Y.; Fang, W.; Zhang, L.; Zhou, Y.; Yang, L.; Lu, J. A Functional Polymorphism in the 3’-UTR of PXR Interacts with Smoking to Increase Lung Cancer Risk in Southern and Eastern Chinese Smoker. Int. J. Mol. Sci. 2014, 15, 17457–17468. [Google Scholar] [CrossRef] [PubMed]
- Reuter, T.; Warta, R.; Theile, D.; Meid, A.D.; Rigalli, J.P.; Mogler, C.; Herpel, E.; Grabe, N.; Lahrmann, B.; Plinkert, P.K.; et al. Role of NR1I2 (Pregnane X Receptor) Polymorphisms in Head and Neck Squamous Cell Carcinoma. Naunyn. Schmiedebergs. Arch. Pharmacol. 2015, 388, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, L.; Wang, Y.; Bi, X.; Li, X.; Wen, M.; Zhang, Q.; Yang, Y.; Jia, Y.; Li, Y.; et al. FBI-1 Enhanced the Resistance of Triple-Negative Breast Cancer Cells to Chemotherapeutic Agents via the MiR-30c/PXR Axis. Cell Death Dis. 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Creusot, N.; Gassiot, M.; Alaterre, E.; Chiavarina, B.; Grimaldi, M.; Boulahtouf, A.; Toporova, L.; Gerbal-Chaloin, S.; Daujat-Chavanieu, M.; Matheux, A.; et al. The Anti-Cancer Drug Dabrafenib Is a Potent Activator of the Human Pregnane X Receptor. Cells 2020, 9, 1641. [Google Scholar] [CrossRef]
- De Mattia, E.; Polesel, J.; Roncato, R.; Labriet, A.; Bignucolo, A.; Dreussi, E.; Romanato, L.; Guardascione, M.; Buonadonna, A.; D’Andrea, M.; et al. Germline Polymorphisms in the Nuclear Receptors PXR and VDR as Novel Prognostic Markers in Metastatic Colorectal Cancer Patients Treated with FOLFIRI. Front. Oncol. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Mbatchi, L.C.; Robert, J.; Ychou, M.; Boyer, J.C.; Del Rio, M.; Gassiot, M.; Thomas, F.; Tubiana, N.; Evrard, A. Effect of Single Nucleotide Polymorphisms in the Xenobiotic-Sensing Receptors NR1I2 and NR1I3 on the Pharmacokinetics and Toxicity of Irinotecan in Colorectal Cancer Patients. Clin. Pharmacokinet. 2016, 55, 1145–1157. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Z.; Chen, H.; Hou, Y.; Lu, W.; He, J.; Tong, H.; Zhou, Y.; Cai, W. Genetic Polymorphisms Contribute to the Individual Variations of Imatinib Mesylate Plasma Levels and Adverse Reactions in Chinese GIST Patients. Int. J. Mol. Sci. 2017, 18, 603. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Sun, L.N.; Liu, Y.J.; Zhang, Q.; Xu, J.H.; Ma, Z.Q.; Zhang, X.H.; Xu, H.; Wang, Y.Q. Genetic Polymorphisms and Adverse Events on Unbound Imatinib and Its Active Metabolite Concentration in Patients with Gastrointestinal Stromal Tumors. Front. Pharmacol. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Pamuła-Piłat, J.; Tęcza, K.; Kalinowska-Herok, M.; Grzybowska, E. Genetic 3′UTR Variations and Clinical Factors Significantly Contribute to Survival Prediction and Clinical Response in Breast Cancer Patients. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Van Der Veldt, A.A.M.; Eechoute, K.; Gelderblom, H.; Gietema, J.; Guchelaar, H.J.; Van Erp, N.P.; Van Den Eertwegh, A.J.M.; Haanen, J.B.; Mathijssen, R.H.J.; Wessels, J.A.M. Genetic Polymorphisms Associated with a Prolonged Progression-Free Survival in Patients with Metastatic Renal Cell Cancer Treated with Sunitinib. Clin. Cancer Res. 2011, 17, 620–629. [Google Scholar] [CrossRef]
- Beuselinck, B.; Karadimou, A.; Lambrechts, D.; Claes, B.; Wolter, P.; Couchy, G.; Berkers, J.; Paridaens, R.; Schöffski, P.; Méjean, A.; et al. Single-Nucleotide Polymorphisms Associated with Outcome in Metastatic Renal Cell Carcinoma Treated with Sunitinib. Br. J. Cancer 2013, 108, 887–900. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, C.F.; Bing, N.X.; Ball, H.A.; Rajagopalan, D.; Sternberg, C.N.; Hutson, T.E.; De Souza, P.; Xue, Z.G.; McCann, L.; King, K.S.; et al. Pazopanib Efficacy in Renal Cell Carcinoma: Evidence for Predictive Genetic Markers in Angiogenesis-Related and Exposure-Related Genes. J. Clin. Oncol. 2011, 29, 2557–2564. [Google Scholar] [CrossRef]
- Xu, C.; Ball, H.A.; Bing, N.; Sternberg, C.N.; Xue, Z.; McCann, L.; King, K.; Spraggs, C.F.; Mooser, V.E.; Pandite, L.N. Association of Genetic Markers in Angiogenesis- or Exposure-Related Genes with Overall Survival in Pazopanib (P) Treated Patients (Pts) with Advanced Renal Cell Carcinoma. J. Clin. Oncol. 2011, 29, 303. [Google Scholar] [CrossRef]
- Mbatchi, L.C.; Gassiot, M.; Pourquier, P.; Goberna, A.; Mahammedi, H.; Mourey, L.; Joly, F.; Lumbroso, S.; Evrard, A.; Houede, N. Association of NR1I2, CYP3A5 and ABCB1 Genetic Polymorphisms with Variability of Temsirolimus Pharmacokinetics and Toxicity in Patients with Metastatic Bladder Cancer. Cancer Chemother. Pharmacol. 2017, 80, 653–659. [Google Scholar] [CrossRef]
- Chew, S.C.; Lim, J.; Singh, O.; Chen, X.; Tan, E.H.; Lee, E.J.; Chowbay, B. Pharmacogenetic Effects of Regulatory Nuclear Receptors (PXR, CAR, RXRα and HNF4α) on Docetaxel Disposition in Chinese Nasopharyngeal Cancer Patients. Eur. J. Clin. Pharmacol. 2014, 70, 155–166. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Liu, Y.; Wang, L. Effect of Polymorphisms of NF-ΚB and PXR on Platinum-Based Chemotherapy for Non-Small Cell Lung Cancer. Zhong Nan Da Xue Xue Bao. Yi Xue Ban 2016, 41, 233–237. [Google Scholar] [CrossRef]
- Abumiya, M.; Mita, A.; Takahashi, S.; Yoshioka, T.; Kameoka, Y.; Takahashi, N.; Miura, M. Effects of Polymorphisms in NR1I2, CYP3A4, and ABC Transporters on the Steady-State Plasma Trough Concentrations of Bosutinib in Japanese Patient with Chronic Myeloid Leukemia. Med. Oncol. 2018, 35, 1–7. [Google Scholar] [CrossRef]
- Ren, W.; Zhou, C.; Liu, Y.; Su, K.; Jia, L.; Chen, L.; Li, M.; Ma, J.; Zhou, W.; Zhang, S.; et al. Genetic Associations of Docetaxel-Based Chemotherapy-Induced Myelosuppression in Chinese Han Population. J. Clin. Pharm. Ther. 2020, 45, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Mbatchi, L.C.; Schmitt, A.; Thomas, F.; Cazaubon, Y.; Robert, J.; Lumbroso, S.; Brouillet, J.P.; Pourquier, P.; Chatelut, E.; Boyer, J.C.; et al. Polymorphisms in SLCO1B3 and NR1I2 as Genetic Determinants of Hematotoxicity of Carboplatin and Paclitaxel Combination. Pharmacogenomics 2015, 16, 1439–1450. [Google Scholar] [CrossRef]
- Hegyi, M.; Arany, A.; Semsei, A.F.; Csordas, K.; Eipel, O.; Gezsi, A.; Kutszegi, N.; Csoka, M.; Muller, J.; Erdelyi, D.J.; et al. Pharmacogenetic Analysis of High-Dose Methotrexate Treatment in Children with Osteosarcoma. Oncotarget 2017, 8, 9388–9398. [Google Scholar] [CrossRef]
- Yang, M.; Pan, H.; Chen, H.; Liu, W.; Lu, L.; He, X.; Yi, H.; Tang, S. Association between NR1I2 Polymorphisms and Susceptibility to Anti-Tuberculosis Drug-Induced Hepatotoxicity in an Eastern Chinese Han Population: A Case-Control Study. Infect. Genet. Evol. 2020, 83, 104349. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiang, X.; Huang, W.W.; Sandford, A.J.; Wu, S.Q.; Zhang, M.M.; Wang, M.G.; Chen, G.; He, J.Q. Association of PXR and CAR Polymorphisms and Antituberculosis Drug-Induced Hepatotoxicity. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, H.; Luo, X.; Ge, W. PXR Haplotype Clusters Will Affect the Pharmacokinetics of Ciclosporin in Chinese Renal Transplant Recipients. J. Pharm. Pharmacol. 2020, 72, 271–278. [Google Scholar] [CrossRef]
- Chen, Y.B.; Zhou, Z.Y.; Li, G.M.; Xiao, C.X.; Yu, W.B.; Zhong, S.l.; Cai, Y.F.; Jin, J.; Huang, M. Influences of an NR1I2 Polymorphism on Heterogeneous Antiplatelet Reactivity Responses to Clopidogrel and Clinical Outcomes in Acute Ischemic Stroke Patients. Acta Pharmacol. Sin. 2019, 40, 762–768. [Google Scholar] [CrossRef] [PubMed]
- D’Avolio, A.; Carcieri, C.; Cusato, J.; Simiele, M.; Calcagno, A.; Allegra, S.; Sciandra, M.; Trentini, L.; Di Perri, G.; Bonora, S. Intracellular Accumulation of Atazanavir/Ritonavir According to Plasma Concentrations and OATP1B1, ABCB1 and PXR Genetic Polymorphisms. J. Antimicrob. Chemother. 2014, 69, 3061–3066. [Google Scholar] [CrossRef] [PubMed]
- Schipani, A.; Siccardi, M.; D’Avolio, A.; Baietto, L.; Simiele, M.; Bonora, S.; Novoa, S.R.; Cuenca, L.; Soriano, V.; Chierakul, N.; et al. Population Pharmacokinetic Modeling of the Association between 63396C→T Pregnane X Receptor Polymorphism and Unboosted Atazanavir Clearance. Antimicrob. Agents Chemother. 2010, 54, 5242–5250. [Google Scholar] [CrossRef]
- Noetzli, M.; Guidi, M.; Ebbing, K.; Eyer, S.; Wilhelm, L.; Michon, A.; Thomazic, V.; Alnawaqil, A.M.; Maurer, S.; Zumbach, S.; et al. Population Pharmacokinetic Study of Memantine: Effects of Clinical and Genetic Factors. Clin. Pharmacokinet. 2013, 52, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Wang, L.; Shi, L.; Li, H.; Zhu, M.; Luo, J.; Zhang, Z. Variability of Voriconazole Concentrations in Patients with Hematopoietic Stem Cell Transplantation and Hematological Malignancies: Influence of Loading Dose, Procalcitonin, and Pregnane X Receptor Polymorphisms. Eur. J. Clin. Pharmacol. 2020, 76, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Lockhart, A.C.; Kim, R.B.; Rothenberg, M.L. Cancer Pharmacogenomics: Powerful Tools in Cancer Chemotherapy and Drug Development. Oncologist 2005, 10, 104–111. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).