Comprehensive Evaluation of PAXgene Fixation on Oral Cancer Tissues Using Routine Histology, Immunohistochemistry, and FTIR Microspectroscopy
Abstract
1. Introduction
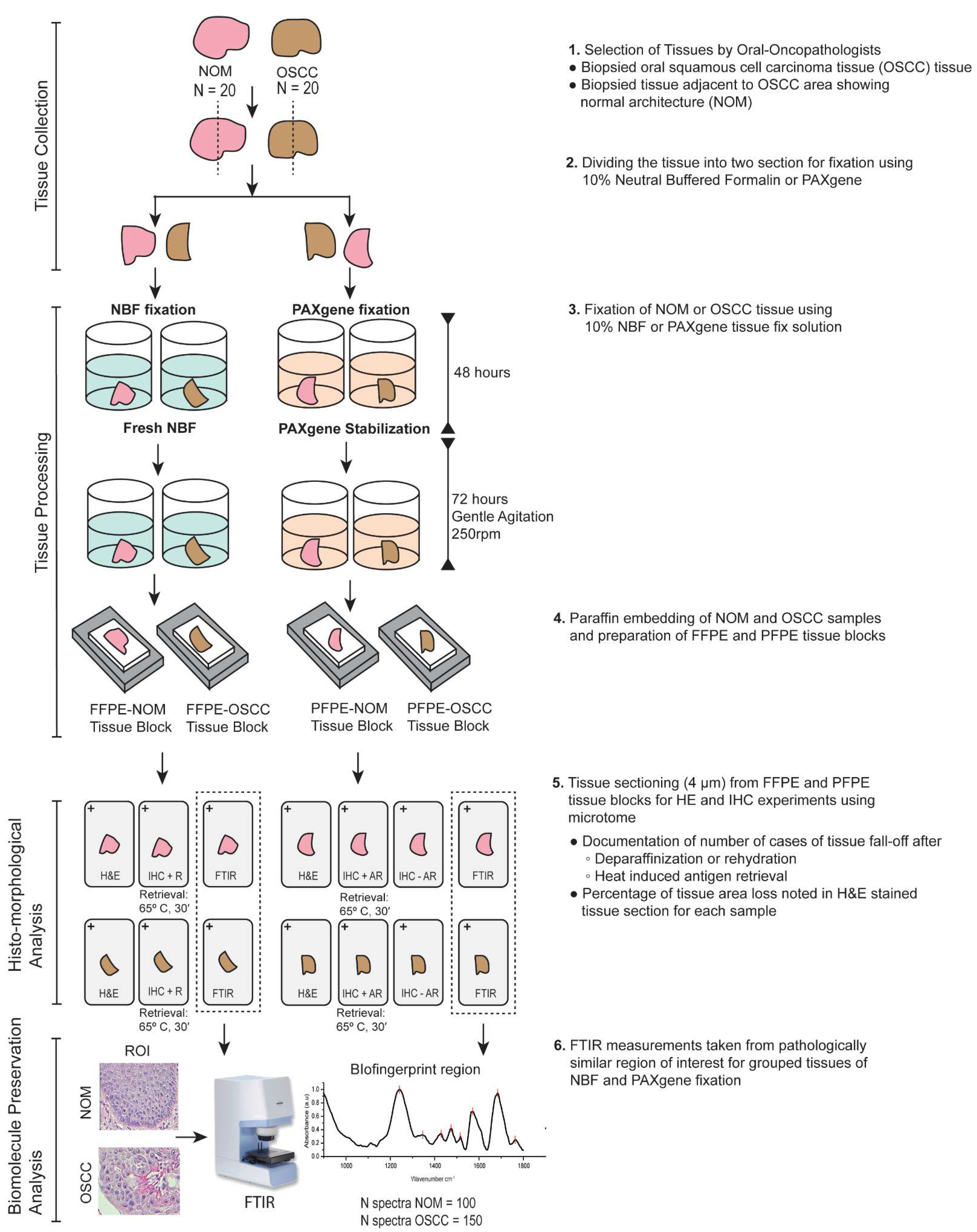
2. Materials and Methods
2.1. Collection of Tissue and Fixation
2.2. Hematoxylin and Eosin Staining
2.3. Immunohistochemistry
2.4. Microscopy
2.5. FTIR Microspectroscopy
3. Results
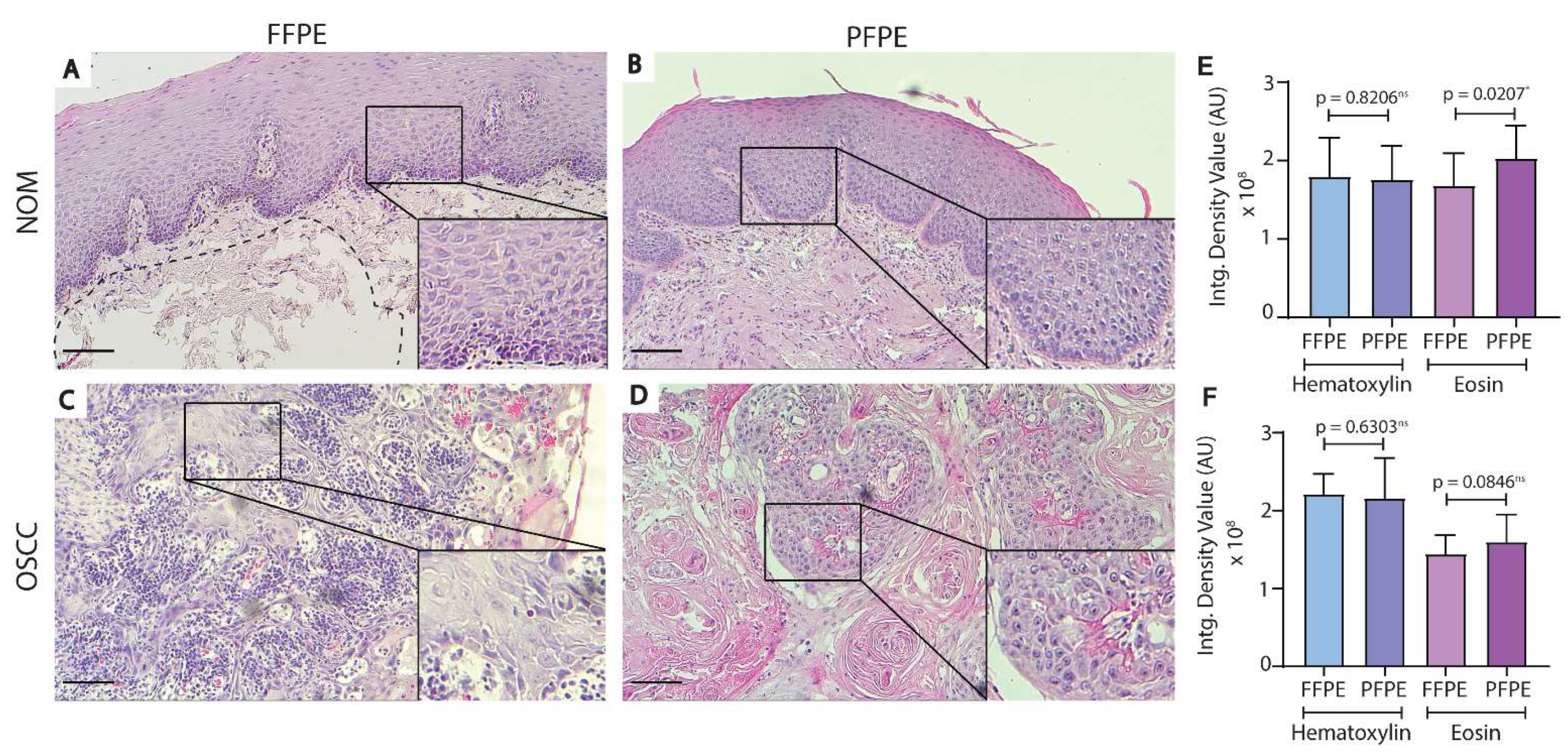
3.1. Histological Comparison of FFPE and PFPE Tissues Using H&E Staining
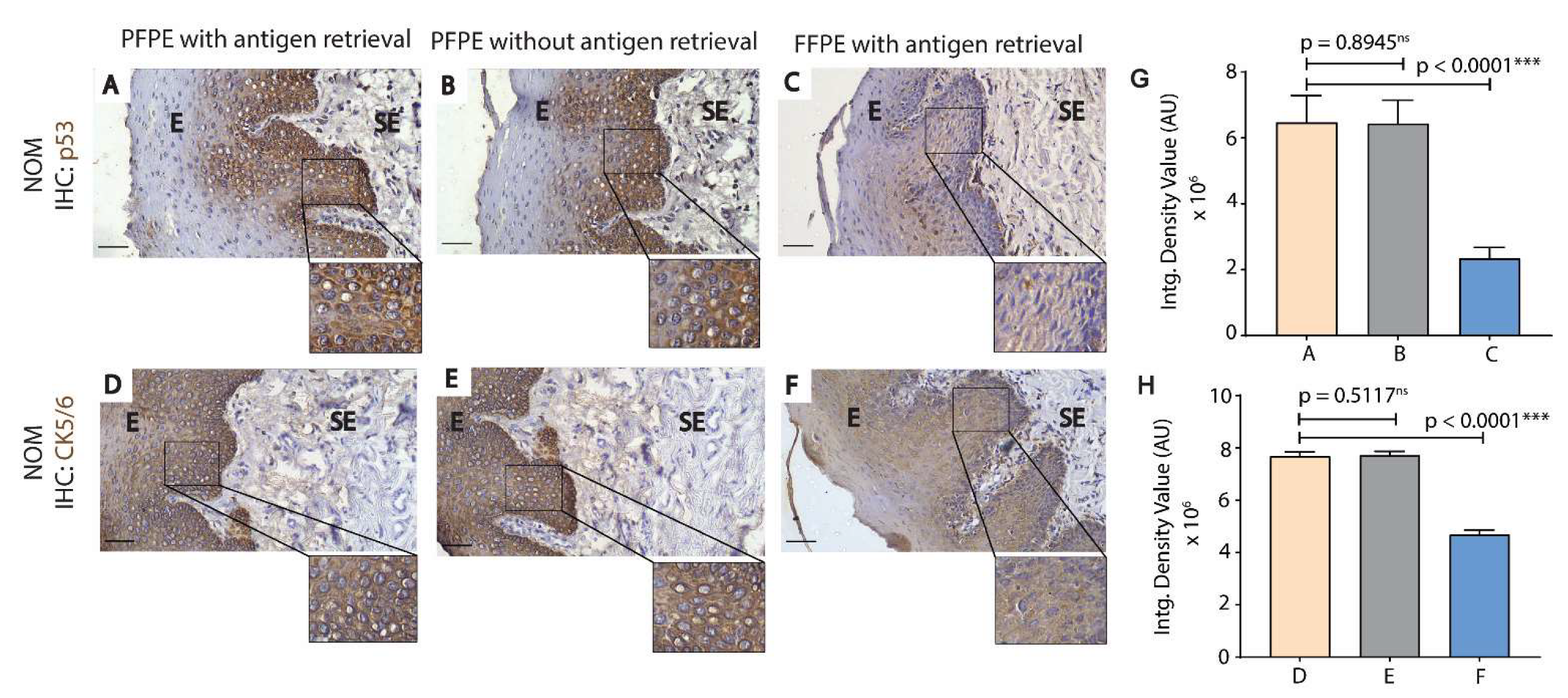
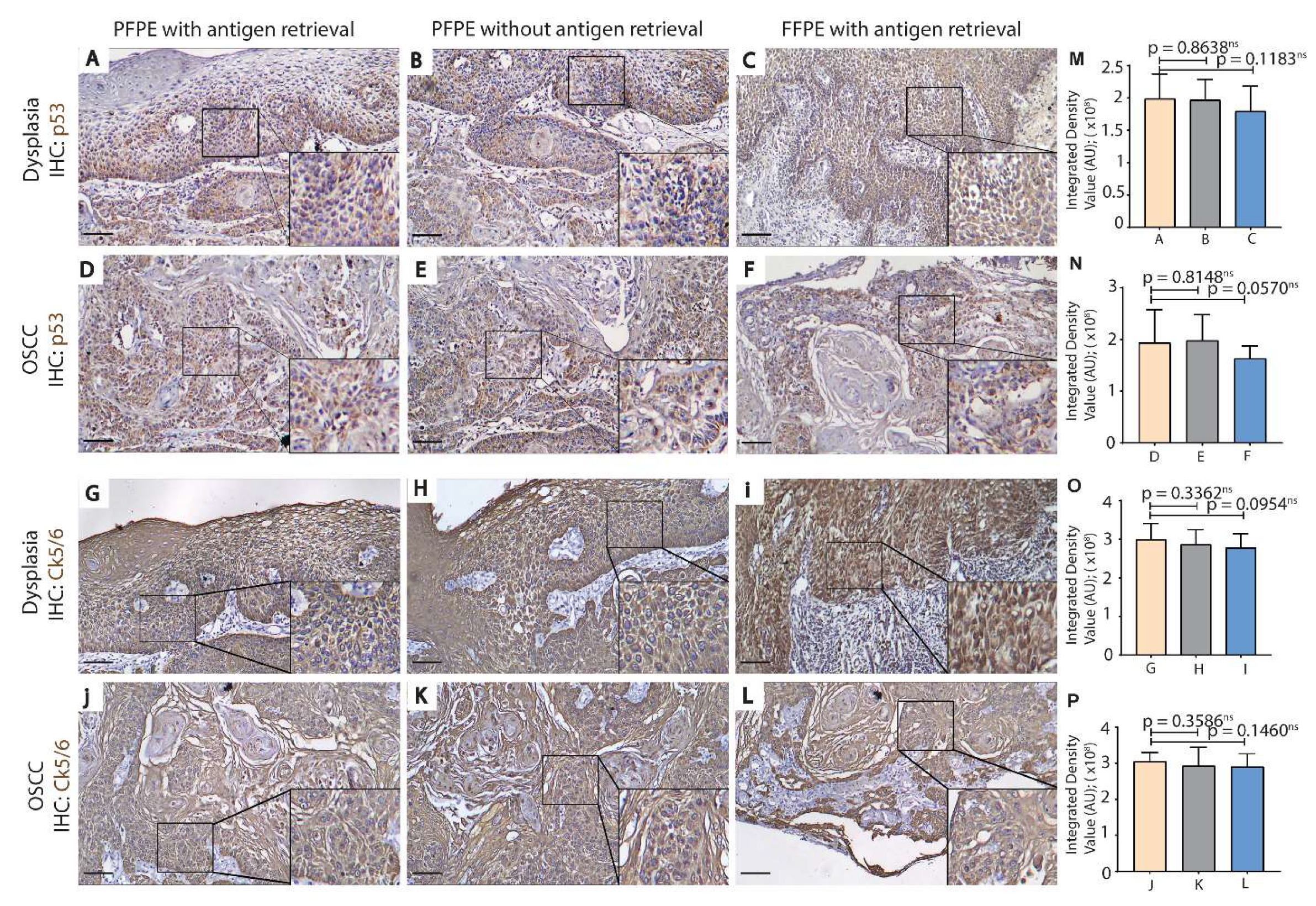
3.2. PFPE Tissues Do Not Require Heat-Induced Antigen Retrieval for Immunohistochemistry
3.3. FTIR Microspectroscopy Revealed Comparable Preservation of Bio-Components between FFPE Tissues and PFPE Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hicks, D.G.; Boyce, B.F. The challenge and importance of standardizing pre-analytical variables in surgical pathology specimens for clinical care and translational research. Biotech. Histochem. 2012, 87, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Howat, W.J.; Wilson, B.A. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods 2014, 70, 12–19. [Google Scholar] [CrossRef]
- Kap, M.; Smedts, F.; Oosterhuis, W.; Winther, R.; Christensen, N.; Reischauer, B.; Viertler, C.; Groelz, D.; Becker, K.-F.; Zatloukal, K.; et al. Histological assessment of PAXgene tissue fixation and stabilization reagents. PLoS ONE 2011, 6, e27704. [Google Scholar] [CrossRef] [PubMed]
- Gillard, M.; Tom, W.R.; Antic, T.; Paner, G.P.; Lingen, M.W.; VanderWeele, D.J. Next-gen tissue: Preservation of molecular and morphological fidelity in prostate tissue. Am. J. Transl. Res. 2015, 7, 1227–1235. [Google Scholar]
- Groelz, D.; Sobin, L.; Branton, P.; Compton, C.; Wyrich, R.; Rainen, L. Non-formalin fixative versus formalin-fixed tissue: A comparison of histology and RNA quality. Exp. Mol. Pathol. 2013, 94, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Ohnishi, T.; Kawamoto, S.; Monden, M.; Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999, 27, 4436–4443. [Google Scholar] [CrossRef]
- Mc Sherry, E.A.; Mc Goldrick, A.; Kay, E.W.; Hopkins, A.M.; Gallagher, W.M.; Dervan, P.A. Formalin-fixed paraffin-embedded clinical tissues show spurious copy number changes in array-CGH profiles. Clin. Genet. 2007, 72, 441–447. [Google Scholar] [CrossRef]
- Mathieson, W.; Marcon, N.; Antunes, L.; Ashford, D.A.; Betsou, F.; Frasquilho, S.G.; Kofanova, O.A.; McKay, S.C.; Pericleous, S.; Smith, C.; et al. A Critical Evaluation of the PAXgene Tissue Fixation System: Morphology, Immunohistochemistry, Molecular Biology, and Proteomics. Am. J. Clin. Pathol. 2016, 146, 25–40. [Google Scholar] [CrossRef]
- Pegg, D.E. The relevance of ice crystal formation for the cryopreservation of tissues and organs. Cryobiology 2010, 60, S36–S44. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.J.E.; Zwirner, J.; Ramani, R.S.; Ma, S.; Hussaini, H.M.; Waddell, J.N.; Hammer, N. Mechanical properties of human oral mucosa tissues are site dependent: A combined biomechanical, histological and ultrastructural approach. Clin. Exp. Dent. Res. 2020, 6, 602–611. [Google Scholar] [CrossRef]
- Belloni, B.; Lambertini, C.; Nuciforo, P.; Phillips, J.; Bruening, E.; Wong, S.; Dummer, R. Will PAXgene substitute formalin? A morphological and molecular comparative study using a new fixative system. J. Clin. Pathol. 2013, 66, 124–135. [Google Scholar] [CrossRef]
- Malek, K.; Wood, B.R.; Bambery, K.R. FTIR Imaging of Tissues: Techniques and Methods of Analysis. In Optical Spectroscopy and Computational Methods in Biology and Medicine; Baranska, M., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 419–473. [Google Scholar]
- PAXgene Tissue FIX Container Product Circular—PreAnalytiX www.preanalytix.com. Available online: https://www.preanalytix.com/storage/download/_ProductResources_/Handbooks/HB-1477-001_1074355_HB_Tissue_all_FIX_Container_0213_WW.pdf (accessed on 26 February 2021).
- Cai, S.; Singh, B.R. A Distinct Utility of the Amide III Infrared Band for Secondary Structure Estimation of Aqueous Protein Solutions Using Partial Least Squares Methods. Biochemistry 2004, 43, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1206. [Google Scholar] [CrossRef] [PubMed]
- Geetha, K.M.; Leeky, M.; Narayan, T.V.; Sadhana, S.; Saleha, J. Grading of oral epithelial dysplasia: Points to ponder. J. Oral Maxillofac. Pathol. JOMFP 2015, 19, 198–204. [Google Scholar] [CrossRef]
- Choi, P.; Jordan, C.D.; Mendez, E.; Houck, J.; Yueh, B.; Farwell, D.G.; Futran, N.; Chen, C. Examination of oral cancer biomarkers by tissue microarray analysis. Arch. Otolaryngol. Head Neck Surg. 2008, 134, 539–546. [Google Scholar] [CrossRef]
- Almangush, A.; Heikkinen, I.; Mäkitie, A.A.; Coletta, R.D.; Läärä, E.; Leivo, I.; Salo, T. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2017, 117, 856–866. [Google Scholar] [CrossRef]
- Shi, S.R.; Shi, Y.; Taylor, C.R. Antigen retrieval immunohistochemistry: Review and future prospects in research and diagnosis over two decades. J. Histochem. Cytochem. 2011, 59, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Ezaki, T. Antigen retrieval on formaldehyde-fixed paraffin sections: Its potential drawbacks and optimization for double immunostaining. Micron 2000, 31, 639–649. [Google Scholar] [CrossRef]
- Mascitti, M.; Rubini, C.; De Michele, F.; Balercia, P.; Girotto, R.; Troiano, G.; Lo Muzio, L.; Santarelli, A. American Joint Committee on Cancer staging system 7th edition versus 8th edition: Any improvement for patients with squamous cell carcinoma of the tongue? Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 126, 415–423. [Google Scholar] [CrossRef]
- Mascitti, M.; Zhurakivska, K.; Togni, L.; Caponio, V.C.A.; Almangush, A.; Balercia, P.; Balercia, A.; Rubini, C.; Lo Muzio, L.; Santarelli, A.; et al. Addition of the tumour–stroma ratio to the 8th edition American Joint Committee on Cancer staging system improves survival prediction for patients with oral tongue squamous cell carcinoma. Histopathology 2020, 77, 810–822. [Google Scholar] [CrossRef]
- Isacsson, G.; Shear, M. Content and distribution of glycogen in oral epithelial dysplasia. Scand. J. Dent. Res. 1981, 89, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Sabbatini, S.; Conti, C.; Rubini, C.; Librando, V.; Tosi, G.; Giorgini, E. Infrared microspectroscopy of Oral Squamous Cell Carcinoma: Spectral signatures of cancer grading. Vib. Spectrosc. 2013, 68, 196–203. [Google Scholar] [CrossRef]
- Southwood, M.; Krenz, T.; Cant, N.; Maurya, M.; Gazdova, J.; Maxwell, P.; McGready, C.; Moseley, E.; Hughes, S.; Stewart, P.; et al. Systematic evaluation of PAXgene® tissue fixation for the histopathological and molecular study of lung cancer. J. Pathol. Clin. Res. 2020, 6, 40–54. [Google Scholar] [CrossRef]
- Viertler, C.; Groelz, D.; Gündisch, S.; Kashofer, K.; Reischauer, B.; Riegman, P.H.; Winther, R.; Wyrich, R.; Becker, K.F.; Oelmüller, U.; et al. A new technology for stabilization of biomolecules in tissues for combined histological and molecular analyses. J. Mol. Diagn. 2012, 14, 458–466. [Google Scholar] [CrossRef]
- Singh, B.; DeOliveira, D.; Fu, F.-N.; Fuller, M. Fourier transform infrared analysis of amide III bands of proteins for the secondary structure estimation; SPIE: Bellingham, WA, USA, 1993; Volume 1890. [Google Scholar]
- Khan, S.; Vihinen, M. Spectrum of disease-causing mutations in protein secondary structures. BMC Struct. Biol. 2007, 7, 56. [Google Scholar] [CrossRef]
- Tikhonov, D.; Kulikova, L.; Kopylov, A.; Malsagova, K.; Stepanov, A.; Rudnev, V.; Kaysheva, A. Super Secondary Structures of Proteins with Post-Translational Modifications in Colon Cancer. Molecules 2020, 25, 3144. [Google Scholar] [CrossRef]
- Johnson, L.N.; Barford, D. The effects of phosphorylation on the structure and function of proteins. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 199–232. [Google Scholar] [CrossRef]
- Winck, F.V.; Belloni, M.; Pauletti, B.A.; de Lima Zanella, J.; Domingues, R.R.; Sherman, N.E.; Paes Leme, A.F. Phosphoproteome analysis reveals differences in phosphosite profiles between tumorigenic and non-tumorigenic epithelial cells. J. Proteom. 2014, 96, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Gündisch, S.; Schott, C.; Wolff, C.; Tran, K.; Beese, C.; Viertler, C.; Zatloukal, K.; Becker, K.-F. The PAXgene(®) tissue system preserves phosphoproteins in human tissue specimens and enables comprehensive protein biomarker research. PLoS ONE 2013, 8, e60638. [Google Scholar] [CrossRef] [PubMed]

| Total Number of FFPE Tissue a | Cases of Tissue Fall-Off * | Percentage | |
|---|---|---|---|
| Number of NOM samples | 35 | 10 | 28.57 |
| Number of OSCC samples | 73 | 8 | 5.48 |
| Total Number of FFPE Tissues a | Cases of Tissue Fall-Off * | Percentage | Total Number of PFPE Tissues a | Cases of Tissue Fall-Off * | Percentage | |
|---|---|---|---|---|---|---|
| Number of NOM samples | 20 | 7 | 35.00 | 20 | 3 | 15.00 |
| Number of OSCC samples | 20 | 1 | 5.00 | 20 | 0 | 0.00 |
| Total Number of FFPE Tissues * | Cases of Tissue Fall-Off after D or R Steps | Cases of Tissue Fall-Off after AR | Total Cases of Tissue Fall-Off | Number of Cases Left for IHC Expts. | |
|---|---|---|---|---|---|
| Number of NOM samples | 35 | 12 | 18 | 30 | 5 |
| Number of OSCC samples | 73 | 3 | 43 | 46 | 30 |
| Number of NOM Samples | Number of OSCC Samples | |
|---|---|---|
| Total number of FFPE tissues | 20 | 20 |
| Cases of tissue fall-off after D or R steps | 6 | 2 |
| Cases of tissue fall-off after AR | 12 | 9 |
| Total cases of tissue fall-off | 18 | 11 |
| Number of cases left for IHC experiments | 2 | 9 |
| Percentage of tissues left for IHC experiments | 10 | 45 |
| Total number of PFPE tissues | 20 | 20 |
| Cases of tissue fall-off after D or R steps | 3 | 1 |
| Cases of tissue fall-off after AR | 5 | 3 |
| Total cases of tissue fall-off | 7 | 4 |
| Number of cases left for IHC experiments | 13 | 16 |
| Percentage of tissues left for IHC experiments | 65 | 80 |
| Band Positions (cm−1) | ||||||
|---|---|---|---|---|---|---|
| Vibration | PFPE NOM | FFPE NOM | PFPE OSCC | FFPE OSCC | Assignment [12] (Malek et al. 2014; Wang and Wang 2021) | |
| COH deformation | 1010 | 1021 | 1010 | 1002 | Glycogen/carbohydrates |  |
| PO2- asymmetric stretch | 1085 | 1082 | 1082 | 1080 | Nucleic acids and phospholipids |  |
| C–O stretch of the ribose ring | 1113 | 1111 | 1113 | 1115 | RNA |  |
| C–O stretch | 1157 | × | 1155 | × | Glycogen |  |
| Amide III | 1245 | 1249 | 1247 | 1245 | Protein |  |
| 1324 | × | 1326 | 1330 | Protein |  | |
| COO- symmetric stretch | 1419 | × | 1419 | × | Fatty acids, amino acids |  |
| 1427 | 1427 | 1427 | 1431 | Fatty acids, amino acids |  | |
| CH3 and CH2 deformations | 1472 | 1474 | 1474 | 1472 | Lipid Proteins |  |
| Ring CC stretch of tyrosine residues | 1517 | 1515 | 1515 | 1515 | Tyrosine Proteins |  |
| NH bend + C–N stretch Amide II | 1565 | 1571 | 1571 | 1573 | Proteins |  |
| C = O + NH bend Amide I | 1684 | 1689 | 1687 | 1684 | Proteins |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lahiri, P.; Mukherjee, S.; Ghosh, B.; Das, D.; Lahiri, B.; Varshney, S.K.; Pal, M.; Paul, R.R.; Chatterjee, J. Comprehensive Evaluation of PAXgene Fixation on Oral Cancer Tissues Using Routine Histology, Immunohistochemistry, and FTIR Microspectroscopy. Biomolecules 2021, 11, 889. https://doi.org/10.3390/biom11060889
Lahiri P, Mukherjee S, Ghosh B, Das D, Lahiri B, Varshney SK, Pal M, Paul RR, Chatterjee J. Comprehensive Evaluation of PAXgene Fixation on Oral Cancer Tissues Using Routine Histology, Immunohistochemistry, and FTIR Microspectroscopy. Biomolecules. 2021; 11(6):889. https://doi.org/10.3390/biom11060889
Chicago/Turabian StyleLahiri, Pooja, Suranjana Mukherjee, Biswajoy Ghosh, Debnath Das, Basudev Lahiri, Shailendra Kumar Varshney, Mousumi Pal, Ranjan Rashmi Paul, and Jyotirmoy Chatterjee. 2021. "Comprehensive Evaluation of PAXgene Fixation on Oral Cancer Tissues Using Routine Histology, Immunohistochemistry, and FTIR Microspectroscopy" Biomolecules 11, no. 6: 889. https://doi.org/10.3390/biom11060889
APA StyleLahiri, P., Mukherjee, S., Ghosh, B., Das, D., Lahiri, B., Varshney, S. K., Pal, M., Paul, R. R., & Chatterjee, J. (2021). Comprehensive Evaluation of PAXgene Fixation on Oral Cancer Tissues Using Routine Histology, Immunohistochemistry, and FTIR Microspectroscopy. Biomolecules, 11(6), 889. https://doi.org/10.3390/biom11060889






