To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis
Abstract
1. Introduction
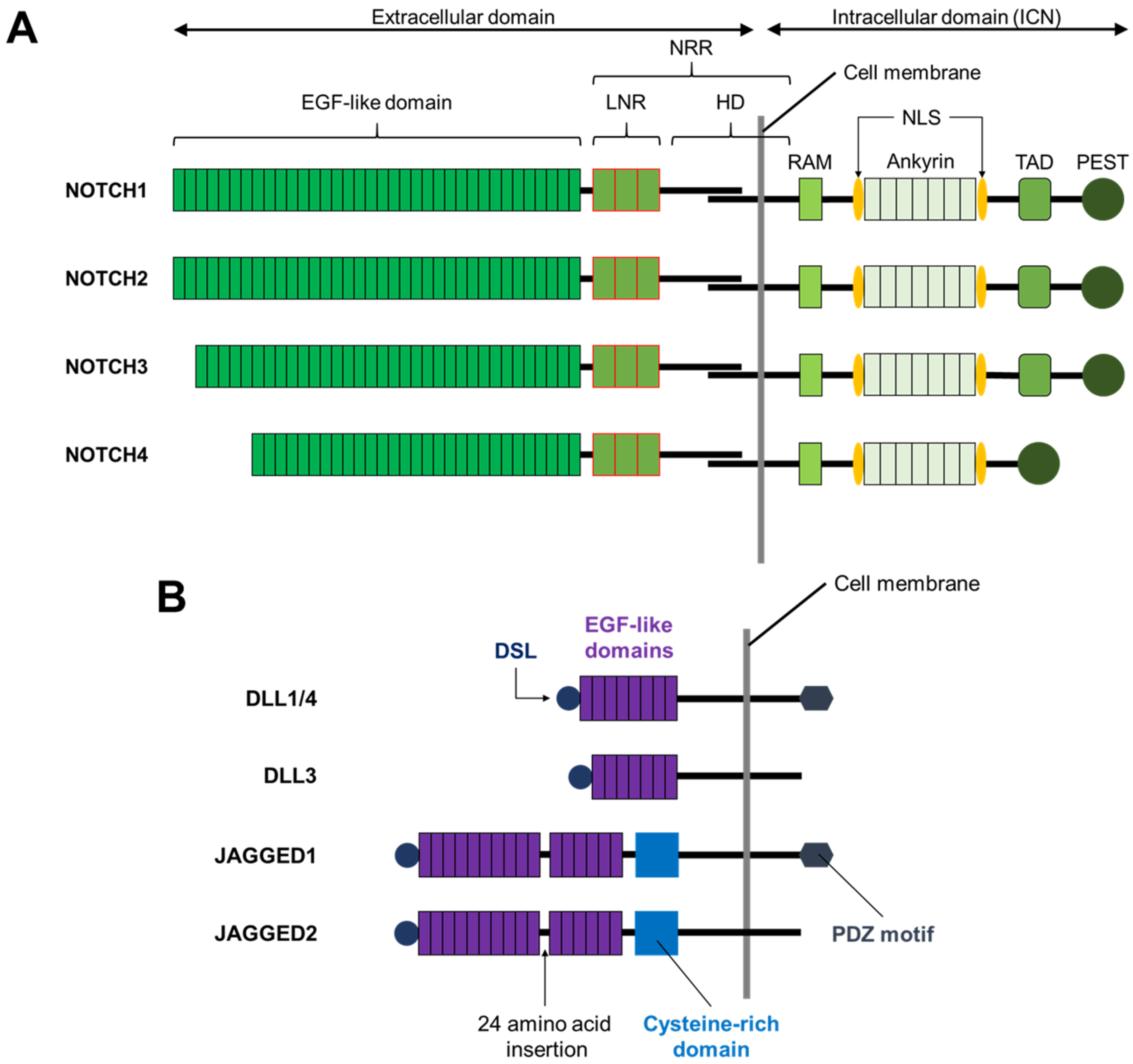
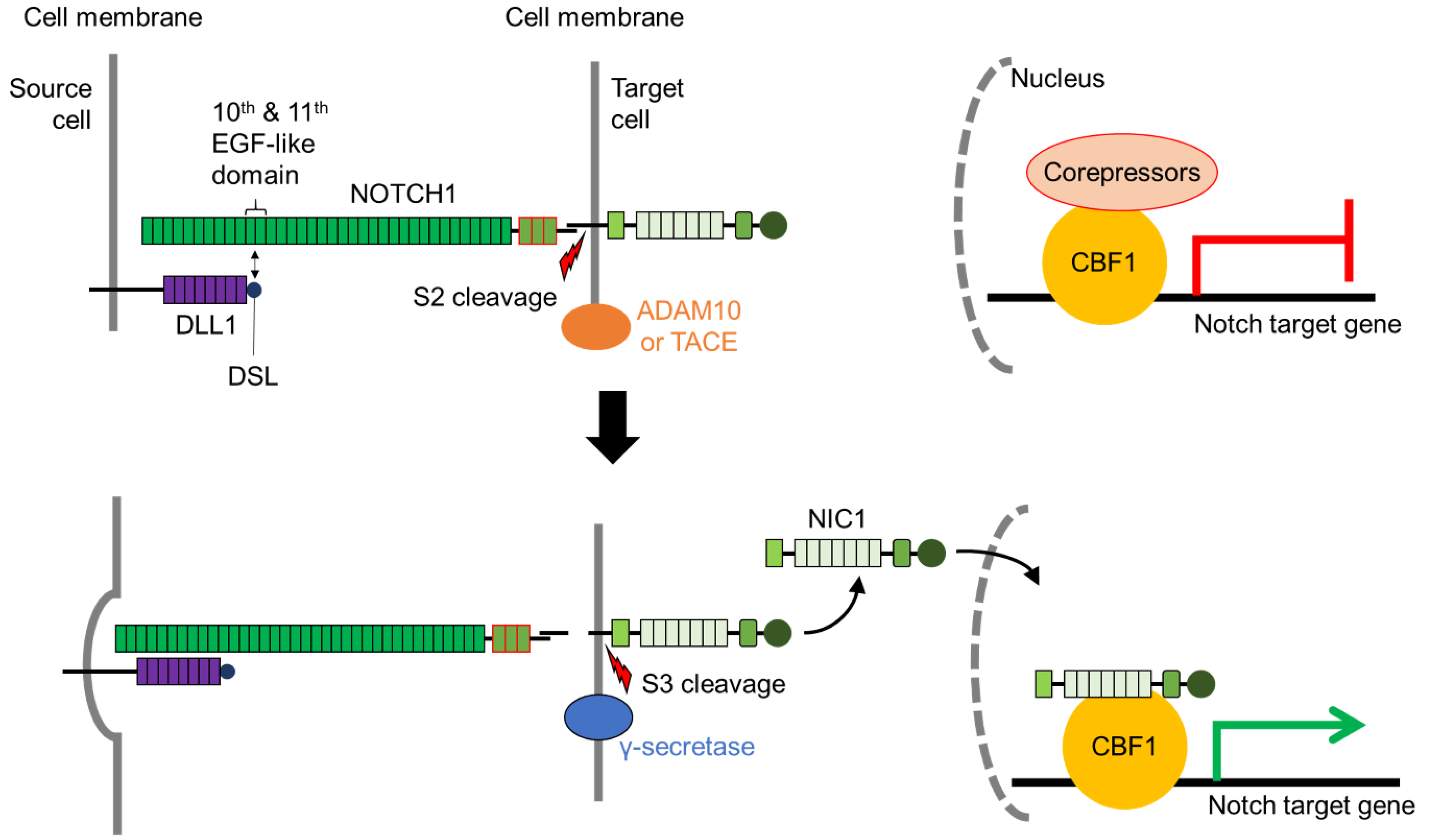
2. The Notch Signaling Pathway
3. Knockout of Notch Signaling Components Is Embryonically Lethal
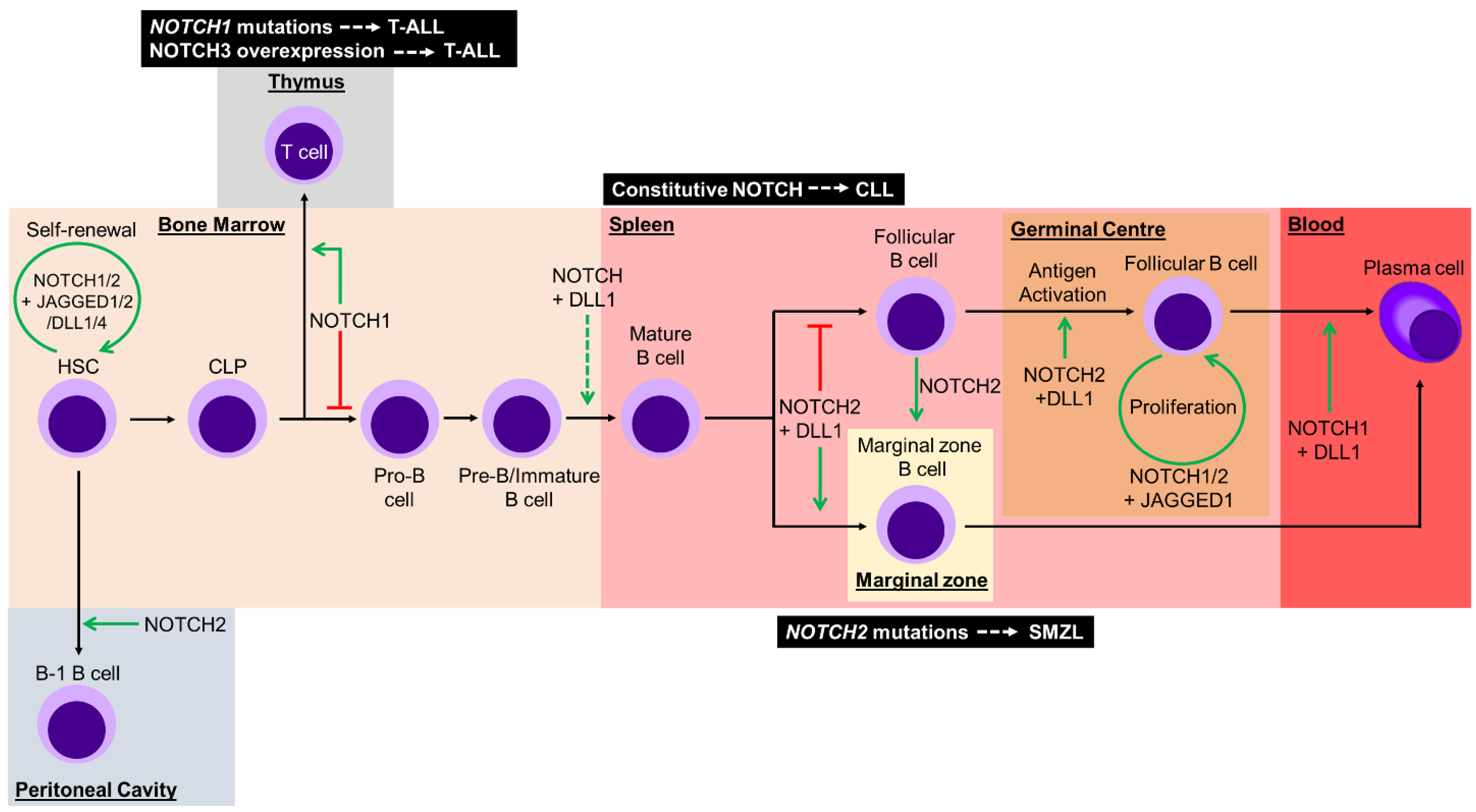
4. Notch Signal Involvement from Hematopoietic Stem Cells to B Lymphocytes
5. Notch-Related Diseases
5.1. Notch Associated Hereditary Diseases
5.2. Notch Dysregulation in Hematological Malignancies
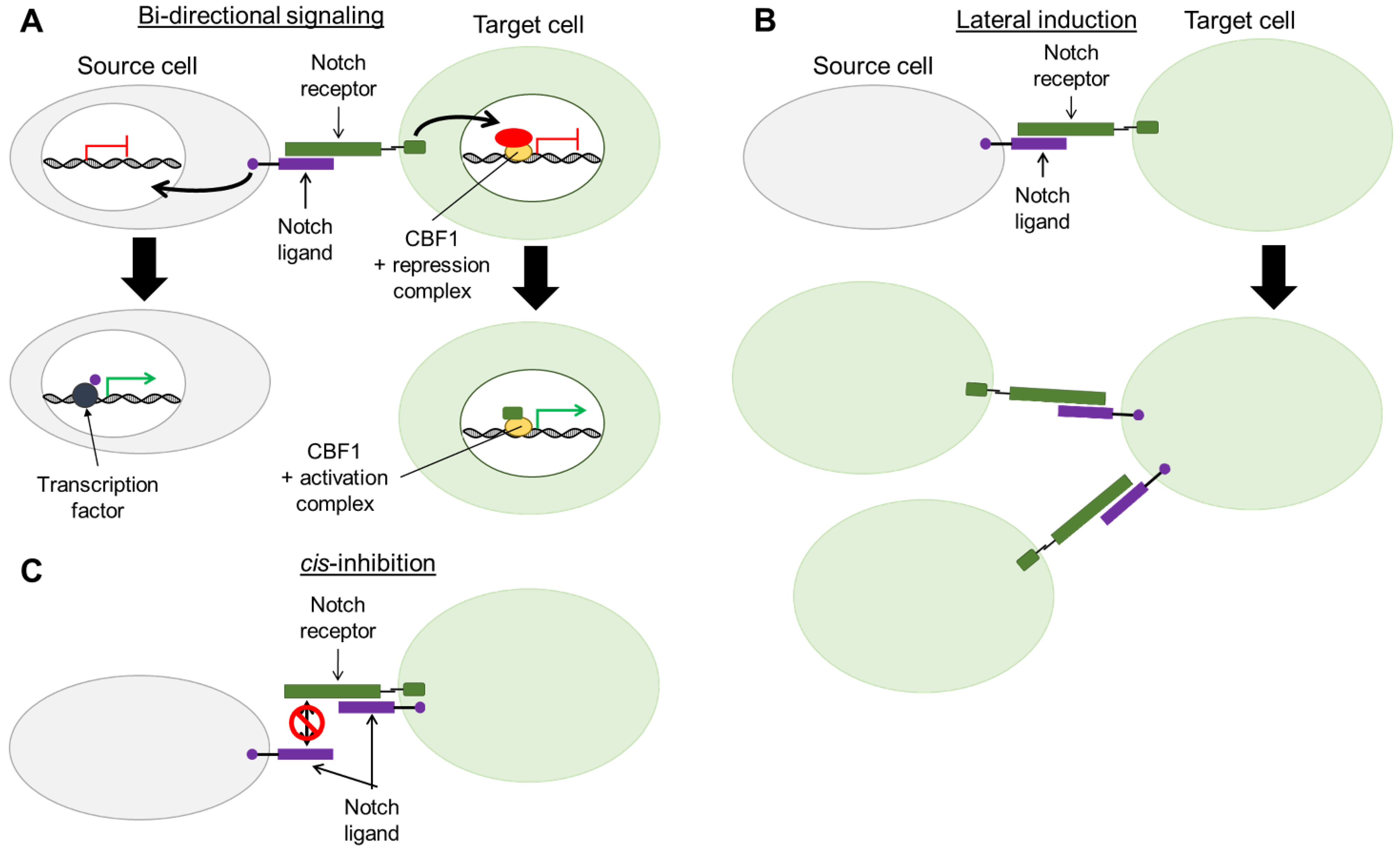
6. Bi-Directional Notch Signaling
7. Lateral Induction of Notch Signaling and Interaction with the Micro-Environment
8. Non-Canonical Cell-Autonomous Pathways
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 2017, 41, 228–241. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J.; Gomez-Lamarca, M.J. Notch after cleavage. Curr. Opin. Cell Biol. 2018, 51, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Rebay, I.; Fleming, R.J.; Fehon, R.G.; Cherbas, L.; Cherbas, P.; Artavanis-Tsakonas, S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell 1991, 67, 687–699. [Google Scholar] [CrossRef]
- Hambleton, S.; Valeyev, N.V.; Muranyi, A.; Knott, V.; Werner, J.M.; McMichael, A.J.; Handford, P.A.; Downing, A.K. Structural and Functional Properties of the Human Notch-1 Ligand Binding Region. Structure 2004, 12, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Cordle, J.; Johnson, S.; Tay, J.Z.Y.; Roversi, P.; Wilkin, M.B.; de Madrid, B.H.; Shimizu, H.; Jensen, S.; Whiteman, P.; Jin, B.; et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 2008, 15, 849–857. [Google Scholar] [CrossRef]
- Luca, V.C.; Jude, K.M.; Pierce, N.W.; Nachury, M.V.; Fischer, S.; Garcia, K.C. Structural basis for Notch1 engagement of Delta-like 4. Science 2015, 347, 847–853. [Google Scholar] [CrossRef]
- Gordon, W.R.; Vardar-Ulu, D.; Histen, G.; Sanchez-Irizarry, C.; Aster, J.C.; Blacklow, S.C. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007, 14, 295–300. [Google Scholar] [CrossRef]
- Parks, A.L.; Klueg, K.M.; Stout, J.R.; Muskavitch, M.A.T. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 2000, 127, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.T.; Miyamoto, A.; Olsen, S.L.; D’Souza, B.; Yao, C.; Weinmaster, G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 2007, 176, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.R.; Cumano, A.; Roux, P.; Black, R.A.; Israël, A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216. [Google Scholar] [CrossRef]
- Mumm, J.S.; Schroeter, E.H.; Saxena, M.T.; Griesemer, A.; Tian, X.; Pan, D.J.; Ray, W.J.; Kopan, R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 2000, 5, 197–206. [Google Scholar] [CrossRef]
- van Tetering, G.; van Diest, P.; Verlaan, I.; van der Wall, E.; Kopan, R.; Vooijs, M. Metalloprotease ADAM10 Is Required for Notch1 Site 2 Cleavage. J. Biol. Chem. 2009, 284, 31018–31027. [Google Scholar] [CrossRef] [PubMed]
- Peschon, J.J.; Slack, J.L.; Reddy, P.; Stocking, K.L.; Sunnarborg, S.W.; Lee, D.C.; Russell, W.E.; Castner, B.J.; Johnson, R.S.; Fitzner, J.N.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, D.; de Strooper, B.; Serneels, L.; Craessaerts, K.; Herreman, A.; Annaert, W.; Umans, L.; Lübke, T.; Lena Illert, A.; von Figura, K.; et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 2002, 11, 2615–2624. [Google Scholar] [CrossRef]
- De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S.; Schroeter, E.H.; Schrijvers, V.; Wolfe, M.S.; Ray, W.J.; et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 1999, 398, 518–522. [Google Scholar] [CrossRef]
- Tagami, S.; Okochi, M.; Yanagida, K.; Ikuta, A.; Fukumori, A.; Matsumoto, N.; Ishizuka-Katsura, Y.; Nakayama, T.; Itoh, N.; Jiang, J.; et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 2008, 28, 165–176. [Google Scholar] [CrossRef]
- Huenniger, K.; Krämer, A.; Soom, M.; Chang, I.; Köhler, M.; Depping, R.; Kehlenbach, R.H.; Kaether, C. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell. Mol. Life Sci. 2010, 67, 3187–3196. [Google Scholar] [CrossRef]
- Kovall, R.A.; Hendrickson, W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004, 23, 3441–3451. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Oswald, F.; Borggrefe, T. Dynamic chromatin regulation at Notch target genes. Transcription 2017, 8, 61–66. [Google Scholar] [CrossRef]
- Oka, C.; Nakano, T.; Wakeham, A.; de la Pompa, J.L.; Mori, C.; Sakai, T.; Okazaki, S.; Kawaichi, M.; Shiota, K.; Mak, T.W.; et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 1995, 121, 3291–3301. [Google Scholar] [CrossRef] [PubMed]
- Swiatek, P.J.; Lindsell, C.E.; del Amo, F.F.; Weinmaster, G.; Gridley, T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994, 8, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Conlon, R.A.; Reaume, A.G.; Rossant, J. Notch1 is required for the coordinate segmentation of somites. Development 1995, 121, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Kadokawa, Y.; Okabe, M.; Ikawa, M.; Coleman, J.R.; Tsujimoto, Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 1999, 126, 3415–3424. [Google Scholar] [CrossRef]
- Domenga, V.; Fardoux, P.; Lacombe, P.; Monet, M.; Maciazek, J.; Krebs, L.T.; Klonjkowski, B.; Berrou, E.; Mericskay, M.; Li, Z.; et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004, 18, 2730–2735. [Google Scholar] [CrossRef]
- Krebs, L.T.; Xue, Y.; Norton, C.R.; Sundberg, J.P.; Beatus, P.; Lendahl, U.; Joutel, A.; Gridley, T. Characterization of Notch3-deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 2003, 37, 139–143. [Google Scholar] [CrossRef]
- Krebs, L.T.; Xue, Y.; Norton, C.R.; Shutter, J.R.; Maguire, M.; Sundberg, J.P.; Gallahan, D.; Closson, V.; Kitajewski, J.; Callahan, R.; et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000, 14, 1343–1352. [Google Scholar]
- James, A.C.; Szot, J.O.; Iyer, K.; Major, J.A.; Pursglove, S.E.; Chapman, G.; Dunwoodie, S.L. Notch4 reveals a novel mechanism regulating Notch signal transduction. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1272–1284. [Google Scholar] [CrossRef]
- de Angelis, M.H.; Mclntyre, J.; Gossler, A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature 1997, 386, 717–721. [Google Scholar] [CrossRef]
- Kusumi, K.; Sun, E.; Kerrebrock, A.W.; Bronson, R.T.; Chi, D.-C.; Bulotsky, M.; Spencer, J.B.; Birren, B.W.; Frankel, W.N.; Lander, E.S. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat. Genet. 1998, 19, 274–278. [Google Scholar] [CrossRef]
- Duarte, A.; Hirashima, M.; Benedito, R.; Trindade, A.; Diniz, P.; Bekman, E.; Costa, L.; Henrique, D.; Rossant, J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004, 18, 2474–2478. [Google Scholar] [CrossRef]
- Gale, N.W.; Dominguez, M.G.; Noguera, I.; Pan, L.; Hughes, V.; Valenzuela, D.M.; Murphy, A.J.; Adams, N.C.; Lin, H.C.; Holash, J.; et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA 2004, 101, 15949–15954. [Google Scholar] [CrossRef]
- Krebs, L.T.; Shutter, J.R.; Tanigaki, K.; Honjo, T.; Stark, K.L.; Gridley, T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004, 18, 2469–2473. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Gao, X.; Lindsell, C.E.; Norton, C.R.; Chang, B.; Hicks, C.; Gendron-Maguire, M.; Rand, E.B.; Weinmaster, G.; Gridley, T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 1999, 8, 723–730. [Google Scholar] [CrossRef]
- Sidow, A.; Bulotsky, M.S.; Kerrebrock, A.W.; Bronson, R.T.; Daly, M.J.; Reeve, M.P.; Hawkins, T.L.; Birren, B.W.; Jaenisch, R.; Lander, E.S. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature 1997, 389, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Lan, Y.; Chapman, H.D.; Shawber, C.; Norton, C.R.; Serreze, D.V.; Weinmaster, G.; Gridley, T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998, 12, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Varnum-Finney, B.; Purton, L.E.; Yu, M.; Brashem-Stein, C.; Flowers, D.; Staats, S.; Moore, K.A.; Le Roux, I.; Mann, R.S.; Gray, G.E.; et al. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood 1998, 91, 4084–4091. [Google Scholar] [CrossRef]
- Carlesso, N.; Aster, J.C.; Sklar, J.; Scadden, D.T. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood 1999, 93, 838–848. [Google Scholar] [CrossRef]
- Han, W.; Ye, Q.; Moore, M.A.S. A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic progenitor cells. Blood 2000, 95, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Karanu, F.N.; Murdoch, B.; Miyabayashi, T.; Ohno, M.; Koremoto, M.; Gallacher, L.; Wu, D.; Itoh, A.; Sakano, S.; Bhatia, M. Human homologues of Delta-1 and Delta-4 function as mitogenic regulators of primitive human hematopoietic cells. Blood 2001, 97, 1960–1967. [Google Scholar] [CrossRef]
- Varnum-Finney, B.; Xu, L.; Brashem-Stein, C.; Nourigat, C.; Flowers, D.; Bakkour, S.; Pear, W.S.; Bernstein, I.D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalizedby constitutive Notch1 signaling. Nat. Med. 2000, 6, 1278–1281. [Google Scholar] [CrossRef]
- Stier, S.; Cheng, T.; Dombkowski, D.; Carlesso, N.; Scadden, D.T. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood 2002, 99, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.W.; Rattis, F.M.; DiMascio, L.N.; Congdon, K.L.; Pazianos, G.; Zhao, C.; Yoon, K.; Cook, J.M.; Willert, K.; Gaiano, N.; et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005, 6, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Kumano, K.; Chiba, S.; Shimizu, K.; Yamagata, T.; Hosoya, N.; Saito, T.; Takahashi, T.; Hamada, Y.; Hirai, H. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood 2001, 98, 3283–3289. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, F.E.; Eckfeldt, C.E.; Lysholm, A.S.; LeBien, T.W. Notch-1 and Notch-2 exhibit unique patterns of expression in human B-lineage cells. Leukemia 2000, 14, 2095–2102. [Google Scholar] [CrossRef][Green Version]
- Saito, T.; Chiba, S.; Ichikawa, M.; Kunisato, A.; Asai, T.; Shimizu, K.; Yamaguchi, T.; Yamamoto, G.; Seo, S.; Kumano, K.; et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 2003, 18, 675–685. [Google Scholar] [CrossRef]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T Cell Fate Specification in Mice with an Induced Inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef]
- Feyerabend, T.B.; Terszowski, G.; Tietz, A.; Blum, C.; Luche, H.; Gossler, A.; Gale, N.W.; Radtke, F.; Fehling, H.J.; Rodewald, H.-R. Deletion of Notch1 Converts Pro-T Cells to Dendritic Cells and Promotes Thymic B Cells by Cell-Extrinsic and Cell-Intrinsic Mechanisms. Immunity 2009, 30, 67–79. [Google Scholar] [CrossRef]
- Wilson, A.; MacDonald, H.R.; Radtke, F. Notch 1–deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001, 194, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Pui, J.C.; Allman, D.; Xu, L.; DeRocco, S.; Karnell, F.G.; Bakkour, S.; Lee, J.Y.; Kadesch, T.; Hardy, R.R.; Aster, J.C.; et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 1999, 11, 299–308. [Google Scholar] [CrossRef]
- Vercauteren, S.M.; Sutherland, H.J. Constitutively active Notch4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood 2004, 104, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Tanigaki, K.; Yamamoto, N.; Kuroda, K.; Yoshimoto, M.; Nakahata, T.; Ikuta, K.; Honjo, T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 2002, 14, 637–645. [Google Scholar] [CrossRef]
- Kawamata, S.; Du, C.; Li, K.; Lavau, C. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene 2002, 21, 3855–3863. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.L.; Taylor, R.L.; Cheng, J.; Abraham, L.J.; Quail, E.; Cruickshank, M.N.; Ulgiati, D. Notch signaling induces a transcriptionally permissive state at the Complement C3d Receptor 2 (CR2) promoter in a pre-B cell model. Mol. Immunol. 2020, 128, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Negishi, N.; Suzuki, D.; Abe, N.; Sotomaru, Y.; Tamaoki, N.; Mailhos, C.; Ish-Horowicz, D.; Habu, S.; Owen, M.J. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 2004, 5, 638–644. [Google Scholar] [CrossRef]
- Sheng, Y.; Yahata, T.; Negishi, N.; Nakano, Y.; Habu, S.; Hozumi, K.; Ando, K. Expression of Delta-like 1 in the splenic non-hematopoietic cells is essential for marginal zone B cell development. Immunol. Lett. 2008, 121, 33–37. [Google Scholar] [CrossRef]
- Tanigaki, K.; Han, H.; Yamamoto, N.; Tashiro, K.; Ikegawa, M.; Kuroda, K.; Suzuki, A.; Nakano, T.; Honjo, T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002, 3, 443–450. [Google Scholar] [CrossRef]
- Lechner, M.; Engleitner, T.; Babushku, T.; Schmidt-Supprian, M.; Rad, R.; Strobl, L.J.; Zimber-Strobl, U. Notch2-mediated plasticity between marginal zone and follicular B cells. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Petcherski, A.G.; Kimble, J. Mastermind is a putative activator for Notch. Curr. Biol. 2000, 10, R471–R473. [Google Scholar] [CrossRef]
- Wu, L.; Aster, J.C.; Blacklow, S.C.; Lake, R.J.; Artavanis-Tsakonas, S.; Griffin, J.D. MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000, 26, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Maillard, I.; Nakamura, M.; Pear, W.S.; Griffin, J.D. The transcriptional coactivator Maml1 is required for Notch2-mediated marginal zone B-cell development. Blood 2007, 110, 3618–3623. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Harigaya, K.; Muradil, A.; Hozumi, K.; Habu, S.; Oguro, H.; Iwama, A.; Matsuno, K.; Sakamoto, R.; Sato, M.; et al. Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc. Natl. Acad. Sci. USA 2007, 104, 9764–9769. [Google Scholar] [CrossRef]
- Yoon, S.-O.; Zhang, X.; Berner, P.; Blom, B.; Choi, Y.S. Notch ligands expressed by follicular dendritic cells protect germinal center B cells from apoptosis. J. Immunol. 2009, 183, 352–358. [Google Scholar] [CrossRef]
- Thomas, M.; Calamito, M.; Srivastava, B.; Maillard, I.; Pear, W.S.; Allman, D. Notch activity synergizes with B-cell–receptor and CD40 signaling to enhance B-cell activation. Blood 2006, 109, 3342–3350. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wang, X.; Xiao, H.; Liu, X.; Fang, Y.; Zhai, B.; Xu, R.; Han, G.; Chen, G.; Hou, C.; et al. Both Notch1 and its ligands in B cells promote antibody production. Mol. Immunol. 2017, 91, 17–23. [Google Scholar] [CrossRef]
- Santos, M.A.; Sarmento, L.M.; Rebelo, M.; Doce, A.A.; Maillard, I.; Dumortier, A.; Neves, H.; Radtke, F.; Pear, W.S.; Parreira, L.; et al. Notch1 engagement by Delta-Like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc. Natl. Acad. Sci. USA 2007, 104, 15454–15459. [Google Scholar] [CrossRef]
- Witt, C.M.; Won, W.-J.; Hurez, V.; Klug, C.A. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J. Immunol. 2003, 171, 2783–2788. [Google Scholar] [CrossRef] [PubMed]
- Witt, C.M.; Hurez, V.; Swindle, C.S.; Hamada, Y.; Klug, C.A. Activated Notch2 potentiates CD8 lineage maturation and promotes the selective development of B1 B cells. Mol. Cell. Biol. 2003, 23, 8637–8650. [Google Scholar] [CrossRef]
- Mizuno, T.; Mizuta, I.; Watanabe-Hosomi, A.; Mukai, M.; Koizumi, T. Clinical and Genetic Aspects of CADASIL. Front. Aging Neurosci. 2020, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cécillion, M.; Maréchal, E.; et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar] [CrossRef]
- Chabriat, H.; Joutel, A.; Dichgans, M.; Tournier-Lasserve, E.; Bousser, M.-G. CADASIL. Lancet Neurol. 2009, 8, 643–653. [Google Scholar] [CrossRef]
- Manini, A.; Pantoni, L. CADASIL from Bench to Bedside: Disease Models and Novel Therapeutic Approaches. Mol. Neurobiol. 2021, 1–16. [Google Scholar] [CrossRef]
- Kamath, B.M.; Baker, A.; Houwen, R.; Todorova, L.; Kerkar, N. Systematic Review: The Epidemiology, Natural History, and Burden of Alagille Syndrome. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 148–156. [Google Scholar] [CrossRef]
- Li, L.; Krantz, I.D.; Deng, Y.; Genin, A.; Banta, A.B.; Collins, C.C.; Qi, M.; Trask, B.J.; Kuo, W.L.; Cochran, J.; et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997, 16, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Elkahloun, A.G.; Pike, B.L.; Okajima, K.; Krantz, I.D.; Genin, A.; Piccoli, D.A.; Meltzer, P.S.; Spinner, N.B.; Collins, F.S.; et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997, 16, 235–242. [Google Scholar] [CrossRef]
- Kamath, B.M.; Bauer, R.C.; Loomes, K.M.; Chao, G.; Gerfen, J.; Hutchinson, A.; Hardikar, W.; Hirschfield, G.; Jara, P.; Krantz, I.D.; et al. NOTCH2 mutations in Alagille syndrome. J. Med. Genet. 2012, 49, 138–144. [Google Scholar] [CrossRef]
- McDaniell, R.; Warthen, D.M.; Sanchez-Lara, P.A.; Pai, A.; Krantz, I.D.; Piccoli, D.A.; Spinner, N.B. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 2006, 79, 169–173. [Google Scholar] [CrossRef]
- Isidor, B.; Lindenbaum, P.; Pichon, O.; Bézieau, S.; Dina, C.; Jacquemont, S.; Martin-Coignard, D.; Thauvin-Robinet, C.; Le Merrer, M.; Mandel, J.-L.; et al. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 2011, 43, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.A.; Irving, M.D.; Asilmaz, E.; Gray, M.J.; Dafou, D.; Elmslie, F.V.; Mansour, S.; Holder, S.E.; Brain, C.E.; Burton, B.K.; et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 2011, 43, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Canalis, E.; Zanotti, S. Hajdu-Cheney syndrome: A review. Orphanet J. Rare Dis. 2014, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Isidor, B.; Le Merrer, M.; Exner, G.U.; Pichon, O.; Thierry, G.; Guiochon-Mantel, A.; David, A.; Cormier-Daire, V.; Le Caignec, C. Serpentine fibula-polycystic kidney syndrome caused by truncating mutations in NOTCH2. Hum. Mutat. 2011, 32, 1239–1242. [Google Scholar] [CrossRef]
- Bulman, M.P.; Kusumi, K.; Frayling, T.M.; McKeown, C.; Garrett, C.; Lander, E.S.; Krumlauf, R.; Hattersley, A.T.; Ellard, S.; Turnpenny, P.D. Mutations in the human Delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet. 2000, 24, 438–441. [Google Scholar] [CrossRef]
- Turnpenny, P.D.; Whittock, N.; Duncan, J.; Dunwoodie, S.; Kusumi, K.; Ellard, S. Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J. Med. Genet. 2003, 40, 333–339. [Google Scholar] [CrossRef]
- Whittock, N.V.; Sparrow, D.B.; Wouters, M.A.; Sillence, D.; Ellard, S.; Dunwoodie, S.L.; Turnpenny, P.D. Mutated/MESP2 causes spondylocostal dysostosis in humans. Am. J. Hum. Genet. 2004, 74, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, D.B.; Guillén-Navarro, E.; Fatkin, D.; Dunwoodie, S.L. Mutation of HAIRY-AND-ENHANCER-OF-SPLIT-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet. 2008, 17, 3761–3766. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, D.B.; Chapman, G.; Wouters, M.A.; Whittock, N.V.; Ellard, S.; Fatkin, D.; Turnpenny, P.D.; Kusumi, K.; Sillence, D.; Dunwoodie, S.L. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 2006, 78, 28–37. [Google Scholar] [CrossRef]
- Mašek, J.; Andersson, E.R. The developmental biology of genetic Notch disorders. Development 2017, 144, 1743–1763. [Google Scholar] [CrossRef] [PubMed]
- Stittrich, A.-B.; Lehman, A.; Bodian, D.L.; Ashworth, J.; Zong, Z.; Li, H.; Lam, P.; Khromykh, A.; Iyer, R.K.; Vockley, J.G.; et al. Mutations in NOTCH1 Cause Adams-Oliver Syndrome. Am. J. Hum. Genet. 2014, 95, 275–284. [Google Scholar] [CrossRef]
- Southgate, L.; Sukalo, M.; Karountzos, A.S.V.; Taylor, E.J.; Collinson, C.S.; Ruddy, D.; Snape, K.M.; Dallapiccola, B.; Tolmie, J.L.; Joss, S.; et al. Haploinsufficiency of the NOTCH1 Receptor as a Cause of Adams–Oliver Syndrome with Variable Cardiac Anomalies. Circ. Cardiovasc. Genet. 2015, 8, 572–581. [Google Scholar] [CrossRef]
- Hassed, S.J.; Wiley, G.B.; Wang, S.; Lee, J.-Y.; Li, S.; Xu, W.; Zhao, Z.J.; Mulvihill, J.J.; Robertson, J.; Warner, J.; et al. RBPJ Mutations Identified in Two Families Affected by Adams-Oliver Syndrome. Am. J. Hum. Genet. 2012, 91, 391–395. [Google Scholar] [CrossRef]
- Meester, J.A.N.; Southgate, L.; Stittrich, A.-B.; Venselaar, H.; Beekmans, S.J.A.; den Hollander, N.; Bijlsma, E.K.; Helderman-van den Enden, A.; Verheij, J.B.G.M.; Glusman, G.; et al. Heterozygous Loss-of-Function Mutations in DLL4 Cause Adams-Oliver Syndrome. Am. J. Hum. Genet. 2015, 97, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.L.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66, 649–661. [Google Scholar] [CrossRef]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris, J.P., IV; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Yashiro-Ohtani, Y.; Wang, H.; Zang, C.; Arnett, K.L.; Bailis, W.; Ho, Y.; Knoechel, B.; Lanauze, C.; Louis, L.; Forsyth, K.S.; et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc. Natl. Acad. Sci. USA 2014, 111, E4946–E4953. [Google Scholar] [CrossRef] [PubMed]
- Tottone, L.; Zhdanovskaya, N.; Pestaña, Á.C.; Zampieri, M.; Simeoni, F.; Lazzari, S.; Ruocco, V.; Pelullo, M.; Caiafa, P.; Pia Felli, M.; et al. Histone modifications drive aberrant notch3 expression/activity and growth in T-ALL. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Giuli, M.V.; Diluvio, G.; Giuliani, E.; Franciosa, G.; Di Magno, L.; Pignataro, M.G.; Tottone, L.; Nicoletti, C.; Besharat, Z.M.; Peruzzi, G.; et al. Notch3 contributes to T-cell leukemia growth via regulation of the unfolded protein response. Oncogenesis 2020, 9, 1–16. [Google Scholar] [CrossRef]
- Pinazza, M.; Ghisi, M.; Minuzzo, S.; Agnusdei, V.; Fossati, G.; Ciminale, V.; Pezzè, L.; Ciribilli, Y.; Pilotto, G.; Venturoli, C.; et al. Histone deacetylase 6 controls Notch3 trafficking and degradation in T-cell acute lymphoblastic leukemia cells. Oncogene 2018, 37, 3839–3851. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Sabatini, R.; Rampino, G.; Tabilio, A.; Di Ianni, M.; Fettucciari, K.; Bartoli, A.; Coaccioli, S.; Screpanti, I.; Marconi, P. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood 2009, 113, 856–865. [Google Scholar] [CrossRef] [PubMed]
- De Falco, F.; Sabatini, R.; Del Papa, B.; Falzetti, F.; Di Ianni, M.; Sportoletti, P.; Baldoni, S.; Screpanti, I.; Marconi, P.; Rosati, E. Notch signaling sustains the expression of Mcl-1 and the activity of eIF4E to promote cell survival in CLL. Oncotarget 2015, 6, 16559–16572. [Google Scholar] [CrossRef] [PubMed]
- De Falco, F.; Del Papa, B.; Baldoni, S.; Sabatini, R.; Falzetti, F.; Di Ianni, M.; Martelli, M.P.; Mezzasoma, F.; Pelullo, M.; Marconi, P.; et al. IL-4-dependent Jagged1 expression/processing is associated with survival of chronic lymphocytic leukemia cells but not with Notch activation. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Trifonov, V.; Fangazio, M.; Bruscaggin, A.; Rasi, S.; Spina, V.; Monti, S.; Vaisitti, T.; Arruga, F.; Famà, R.; et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 2012, 209, 1537–1551. [Google Scholar] [CrossRef]
- Kiel, M.J.; Velusamy, T.; Betz, B.L.; Zhao, L.; Weigelin, H.G.; Chiang, M.Y.; Huebner-Chan, D.R.; Bailey, N.G.; Yang, D.T.; Bhagat, G.; et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 2012, 209, 1553–1565. [Google Scholar] [CrossRef]
- Campos-Martín, Y.; Martínez, N.; Martínez-López, A.; Cereceda, L.; Casado, F.; Algara, P.; Oscier, D.; Menarguez, F.J.; García, J.F.; Piris, M.A.; et al. Clinical and diagnostic relevance of NOTCH2 and KLF2 mutations in splenic marginal zone lymphoma. Haematologica 2017, 102, e310–e312. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, V.; Craig, J.W.; Hilton, L.K.; Nguyen, M.H.; Rushton, C.K.; Fahimdanesh, K.; Lovitch, S.; Ferland, B.; Scott, D.W.; Aster, J.C. Notch activation is pervasive in SMZL and uncommon in DLBCL: Implications for Notch signaling in B-cell tumors. Blood Adv. 2021, 5, 71–83. [Google Scholar] [CrossRef]
- Ryan, R.J.H.; Petrovic, J.; Rausch, D.M.; Zhou, Y.; Lareau, C.A.; Kluk, M.J.; Christie, A.L.; Lee, W.Y.; Tarjan, D.R.; Guo, B.; et al. A B Cell Regulome Links Notch to Downstream Oncogenic Pathways in Small B Cell Lymphomas. Cell Rep. 2017, 21, 784–797. [Google Scholar] [CrossRef]
- Petrovic, J.; Zhou, Y.; Fasolino, M.; Goldman, N.; Schwartz, G.W.; Mumbach, M.R.; Nguyen, S.C.; Rome, K.S.; Sela, Y.; Zapataro, Z.; et al. Oncogenic Notch Promotes Long-Range Regulatory Interactions within Hyperconnected 3D Cliques. Mol. Cell 2019, 73, 1174–1190. [Google Scholar] [CrossRef] [PubMed]
- Arcaini, L.; Rossi, D.; Lucioni, M.; Nicola, M.; Bruscaggin, A.; Fiaccadori, V.; Riboni, R.; Ramponi, A.; Ferretti, V.V.; Cresta, S.; et al. The notch pathway is recurrently mutated in diffuse large B-Cell lymphoma associated with hepatitis c virus infection. Haematologica 2015, 100, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Giuli, M.V.; Giuliani, E.; Screpanti, I.; Bellavia, D.; Checquolo, S. Notch Signaling Activation as a Hallmark for Triple-Negative Breast Cancer Subtype. J. Oncol. 2019, 2019. [Google Scholar] [CrossRef]
- Nandi, A.; Chakrabarti, R. The many facets of Notch signaling in breast cancer: Toward overcoming therapeutic resistance. Genes Dev. 2020, 34, 1422–1438. [Google Scholar] [CrossRef]
- Teodorczyk, M.; Schmidt, M.H.H. Notching on Cancer’s Door: Notch Signaling in Brain Tumors. Front. Oncol. 2015, 4, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Engler, A.; Taylor, V. Notch: An interactive player in neurogenesis and disease. Cell Tissue Res. 2018, 371, 73–89. [Google Scholar] [CrossRef]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 245–275. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, V.S.; Giuranno, L.; Dubois, L.J.; Theys, J.; Vooijs, M. Drug resistance in non-small cell lung cancer: A potential for NOTCH targeting? Front. Oncol. 2018, 8. [Google Scholar] [CrossRef]
- Vinson, K.E.; George, D.C.; Fender, A.W.; Bertrand, F.E.; Sigounas, G. The Notch pathway in colorectal cancer. Int. J. Cancer 2016, 138, 1835–1842. [Google Scholar] [CrossRef]
- LaVoie, M.J.; Selkoe, D.J. The Notch Ligands, Jagged and Delta, Are Sequentially Processed by α-Secretase and Presenilin/γ-Secretase and Release Signaling Fragments. J. Biol. Chem. 2003, 278, 34427–34437. [Google Scholar] [CrossRef] [PubMed]
- Six, E.; Ndiaye, D.; Laâbi, Y.; Brou, C.; Gupta-Rossi, N.; Israël, A.; Logeat, F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and γ-secretase. Proc. Natl. Acad. Sci. USA 2003, 100, 7638–7643. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, T.; Sisodia, S.S. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “γ-secretase” cleavage. J. Biol. Chem. 2003, 278, 7751–7754. [Google Scholar] [CrossRef]
- Hiratochi, M.; Nagase, H.; Kuramochi, Y.; Koh, C.S.; Ohkawara, T.; Nakayama, K. The Delta intracellular domain mediates TGF-β/Activin signaling through binding to Smads and has an important bi-directional function in the Notch-Delta signaling pathway. Nucleic Acids Res. 2007, 35, 912–922. [Google Scholar] [CrossRef]
- Furukawa, T.; Ishifune, C.; Tsukumo, S.I.; Hozumi, K.; Maekawa, Y.; Matsui, N.; Kaji, R.; Yasutomo, K. Transmission of survival signals through Delta-like 1 on activated CD4 + T cells. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Metrich, M.; Bezdek Pomey, A.; Berthonneche, C.; Sarre, A.; Nemir, M.; Pedrazzini, T. Jagged1 intracellular domain-mediated inhibition of Notch1 signalling regulates cardiac homeostasis in the postnatal heart. Cardiovasc. Res. 2015, 108, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Hoare, M.; Ito, Y.; Kang, T.-W.; Weekes, M.P.; Matheson, N.J.; Patten, D.A.; Shetty, S.; Parry, A.J.; Menon, S.; Salama, R.; et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol. 2016, 18, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Parry, A.J.; Hoare, M.; Bihary, D.; Hänsel-Hertsch, R.; Smith, S.; Tomimatsu, K.; Mannion, E.; Smith, A.; D’Santos, P.; Russell, I.A.; et al. NOTCH-mediated non-cell autonomous regulation of chromatin structure during senescence. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Kamdje, A.H.N.; Bassi, G.; Pacelli, L.; Malpeli, G.; Amati, E.; Nichele, I.; Pizzolo, G.; Krampera, M. Role of stromal cell-mediated Notch signaling in CLL resistance to chemotherapy. Blood Cancer J. 2012, 2, e73. [Google Scholar] [CrossRef]
- Colombo, M.; Galletti, S.; Bulfamante, G.; Falleni, M.; Tosi, D.; Todoerti, K.; Lazzari, E.; Crews, L.A.; Jamieson, C.H.M.; Ravaioli, S.; et al. Multiple myeloma-derived Jagged ligands increases autocrine and paracrine interleukin-6 expression in bone marrow niche. Oncotarget 2016, 7, 56013–56029. [Google Scholar] [CrossRef]
- Nefedova, Y.; Sullivan, D.M.; Bolick, S.C.; Dalton, W.S.; Gabrilovich, D.I. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood 2008, 111, 2220–2229. [Google Scholar] [CrossRef]
- Majumder, S.; Crabtree, J.S.; Golde, T.E.; Minter, L.M.; Osborne, B.A.; Miele, L. Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 2021, 20, 125–144. [Google Scholar] [CrossRef]
- Sprinzak, D.; Lakhanpal, A.; LeBon, L.; Santat, L.A.; Fontes, M.E.; Anderson, G.A.; Garcia-Ojalvo, J.; Elowitz, M.B. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 2010, 465, 86–90. [Google Scholar] [CrossRef]
- Mitra, A.; Shanthalingam, S.; Sherman, H.L.; Singh, K.; Canakci, M.; Torres, J.A.; Lawlor, R.; Ran, Y.; Golde, T.E.; Miele, L.; et al. CD28 Signaling Drives Notch Ligand Expression on CD4 T Cells. Front. Immunol. 2020, 11, 735. [Google Scholar] [CrossRef] [PubMed]
- Pelullo, M.; Quaranta, R.; Talora, C.; Checquolo, S.; Cialfi, S.; Felli, M.P.; Kronnie, G.; Borga, C.; Besharat, Z.M.; Palermo, R.; et al. Notch3/Jagged1 circuitry reinforces notch signaling and sustains T-ALL. Neoplasia 2014, 16, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Negri, V.A.; Logtenberg, M.E.W.; Renz, L.M.; Oules, B.; Walko, G.; Watt, F.M. Delta-like 1-mediated cis-inhibition of Jagged1/2 signalling inhibits differentiation of human epidermal cells in culture. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Kamińska, A.; Marek, S.; Pardyak, L.; Brzoskwinia, M.; Pawlicki, P.; Bilińska, B.; Hejmej, A. Disruption of androgen signaling during puberty affects Notch pathway in rat seminiferous epithelium. Reprod. Biol. Endocrinol. 2020, 18. [Google Scholar] [CrossRef]
- Marchetto, N.M.; Begum, S.; Wu, T.; O’besso, V.; Yarborough, C.C.; Valero-Pacheco, N.; Beaulieu, A.M.; Kitajewski, J.K.; Shawber, C.J.; Douglas, N.C. Endothelial jagged1 antagonizes dll4/notch signaling in decidual angiogenesis during early mouse pregnancy. Int. J. Mol. Sci. 2020, 21, 6477. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, N.; Santat, L.A.; Elowitz, M.B. Cis-activation in the Notch signaling pathway. eLife 2019, 8. [Google Scholar] [CrossRef]
- Del Álamo, D.; Rouault, H.; Schweisguth, F. Mechanism and significance of cis-inhibition in notch signalling. Curr. Biol. 2011, 21, R40–R47. [Google Scholar] [CrossRef] [PubMed]
- Sjöqvist, M.; Andersson, E.R. Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev. Biol. 2017, 447, 58–70. [Google Scholar] [CrossRef]
| Knockout | Genotype | Embryonic Lethality | Developmental Impairment | References |
|---|---|---|---|---|
| Notch1 | −/− | Yes (E9.5) | Somitogenesis Neurogenesis Developmental impairment | [24,25] |
| Notch2 | −/− | Yes (E11.5) | Wide-spread pycnosis and apoptosis | [26] |
| Notch3 | −/− | No | Vascular and arterial development | [27] |
| Notch1 & Notch3 | Notch1: −/− Notch3: −/− | Yes (E9.5) | Somitogenesis Neurogenesis Developmental impairment | [28] |
| Notch4 | −/− | No | Vasculature of embryonic head | [29,30] |
| Notch1 & Notch4 | Notch1: −/− Notch4: −/− | Yes (E10.5) | Vascular development Developmental impairment (more severe than Notch1 knockout) | [29] |
| Dll1 | −/− | Yes (E10–E12) | Somitogenesis | [31] |
| Dll3 | −/− | Yes (~20%) | Somitogenesis Skeletal deformities | [32] |
| Dll4 | −/− | Yes (E8.5) | Vascular and arterial development | [33] |
| Dll4 | +/− | Yes (E9.5–E10.5) | Vascular and arterial development | [33,34,35] |
| Jagged1 | −/− | Yes (E10.5) | Vascular development | [36] |
| Jagged2 | −/− | Yes (E10.5) | Craniofacial and skeletal deformities | [37,38] |
| Cbf1 | −/− | Yes (E8.5) | Somitogenesis Placenta defects | [23] |
| Adam10 | −/− | Yes (E9.5) | Somitogenesis Heart and neural defects | [17] |
| Adam17/Tace | −/− | Yes (E17.5) | Eyes and lungs defects | [16] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, H.L.; Quail, E.; Cruickshank, M.N.; Ulgiati, D. To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis. Biomolecules 2021, 11, 849. https://doi.org/10.3390/biom11060849
Ng HL, Quail E, Cruickshank MN, Ulgiati D. To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis. Biomolecules. 2021; 11(6):849. https://doi.org/10.3390/biom11060849
Chicago/Turabian StyleNg, Han Leng, Elizabeth Quail, Mark N. Cruickshank, and Daniela Ulgiati. 2021. "To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis" Biomolecules 11, no. 6: 849. https://doi.org/10.3390/biom11060849
APA StyleNg, H. L., Quail, E., Cruickshank, M. N., & Ulgiati, D. (2021). To Be, or Notch to Be: Mediating Cell Fate from Embryogenesis to Lymphopoiesis. Biomolecules, 11(6), 849. https://doi.org/10.3390/biom11060849






