Folding and Stability of Ankyrin Repeats Control Biological Protein Function
Abstract
1. Introduction
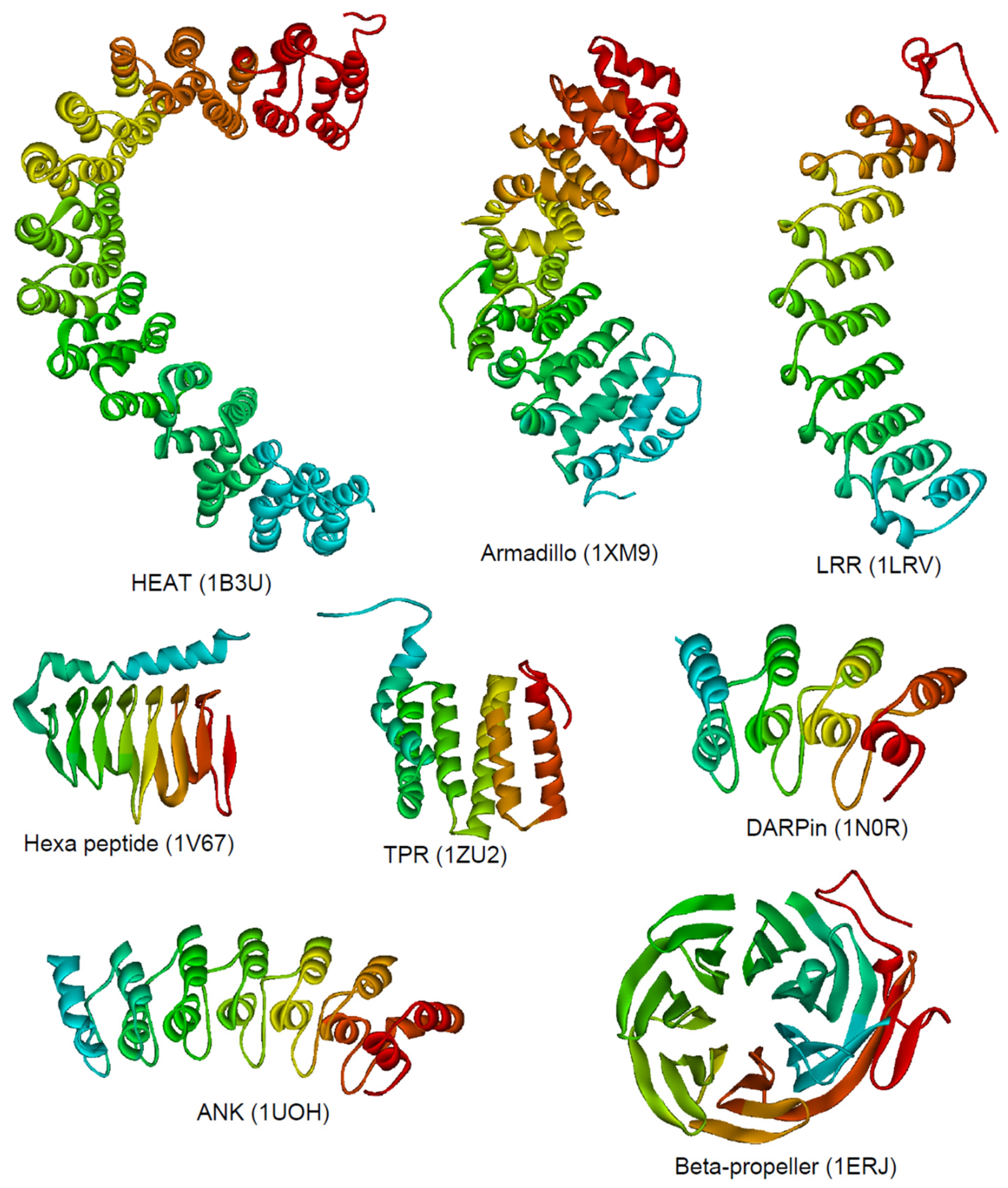
2. Structure and Classification
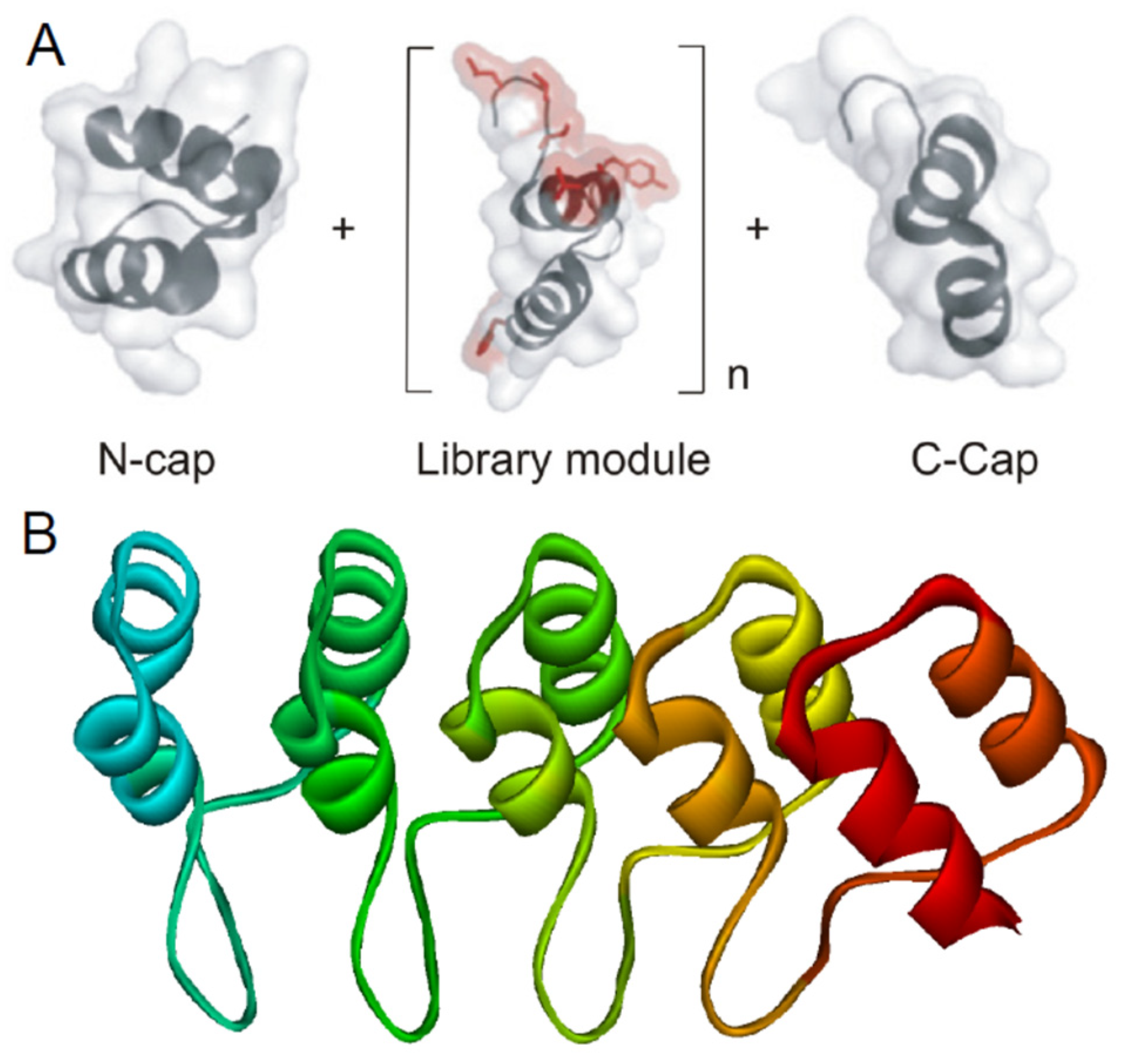
3. Ankyrin Repeats
4. General Protein Folding and Stability Aspects
5. Folding and Function of DARPins
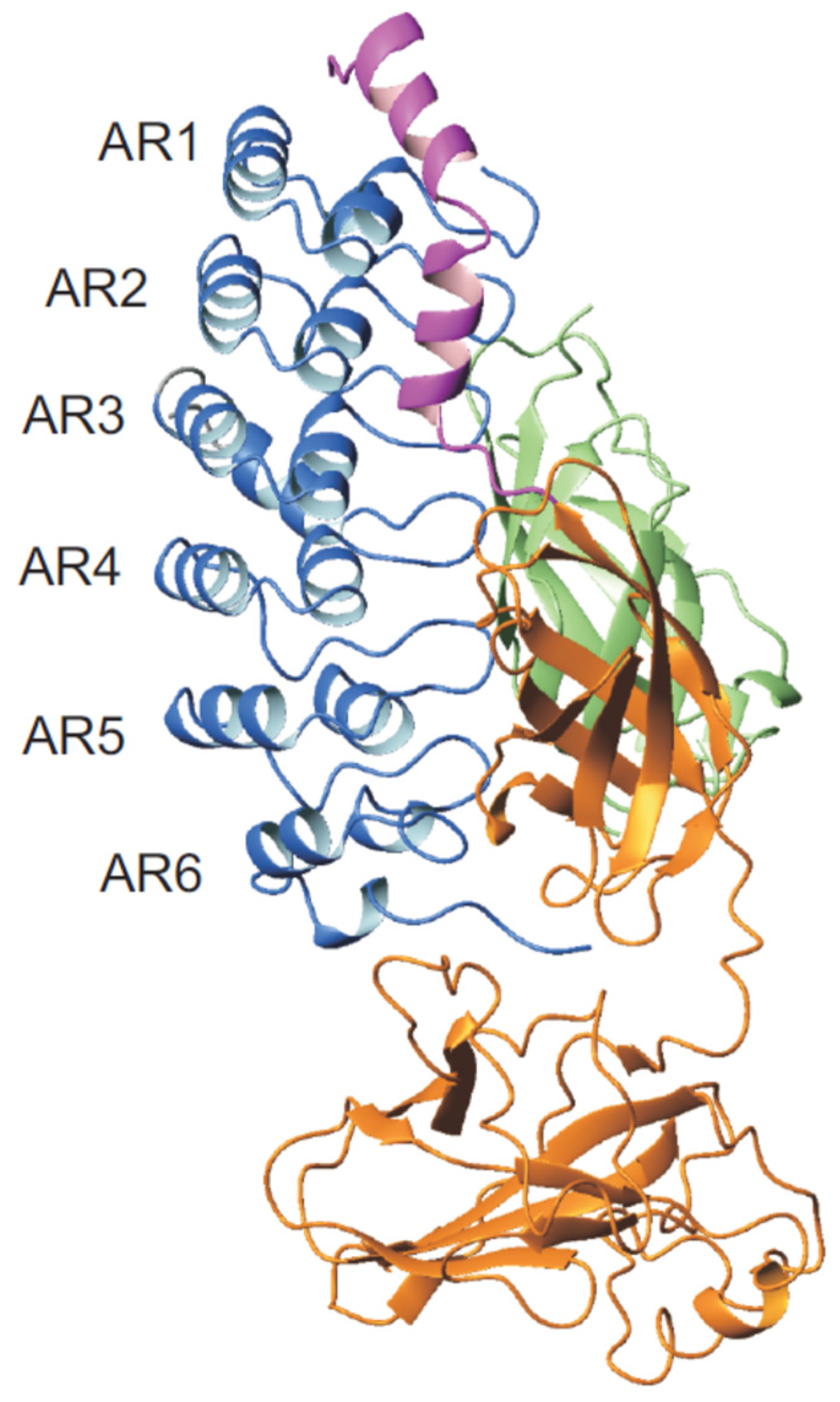
6. Notch Receptor Ankyrin Repeat Domain (Nank)
7. Ankyrin Repeat Domain of IκBα
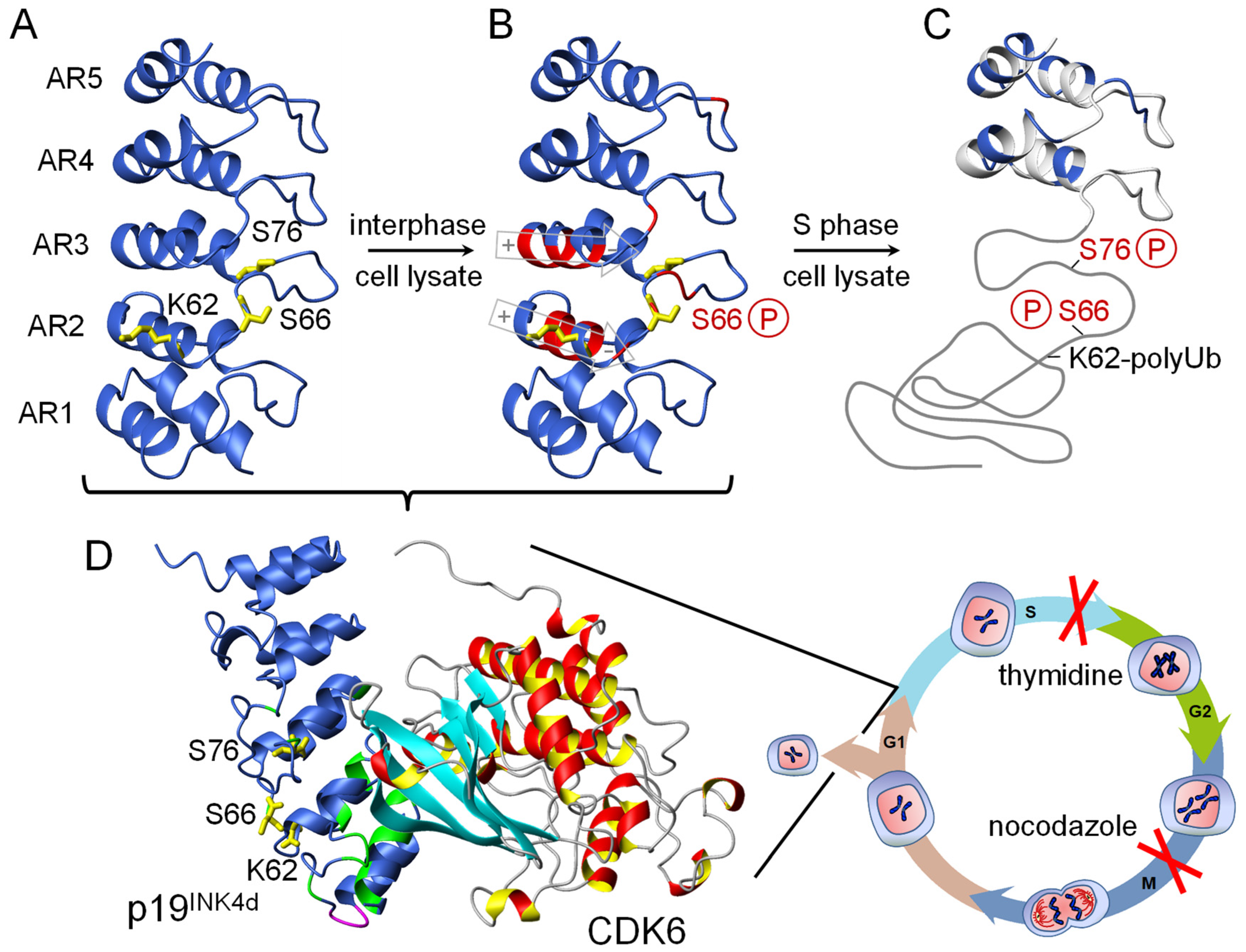
8. CDK4/6 Inhibitor p19INK4d
9. Recent Findings of Various AR Proteins
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marcotte, E.M.; Pellegrini, M.; Yeates, T.O.; Eisenberg, D. A census of protein repeats. J. Mol. Biol. 1999, 293, 151–160. [Google Scholar] [CrossRef]
- Galpern, E.A.; Freiberger, M.I.; Ferreiro, D.U. Large Ankyrin repeat proteins are formed with similar and energetically favorable units. PLoS ONE 2020, 15, e0233865. [Google Scholar] [CrossRef]
- Kajander, T.; Cortajarena, A.L.; Regan, L. Consensus design as a tool for engineering repeat proteins. Methods Mol. Biol. 2006, 340, 151–170. [Google Scholar] [CrossRef]
- Wetzel, S.K.; Settanni, G.; Kenig, M.; Binz, H.K.; Plückthun, A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J. Mol. Biol. 2008, 376, 241–257. [Google Scholar] [CrossRef]
- Binz, H.K.; Stumpp, M.T.; Forrer, P.; Amstutz, P.; Pluckthun, A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003, 332, 489–503. [Google Scholar] [CrossRef]
- Plückthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 489–511. [Google Scholar] [CrossRef]
- Barrick, D. Biological regulation via ankyrin repeat folding. ACS Chem. Biol. 2009, 4, 19–22. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barrick, D.; Ferreiro, D.U.; Komives, E.A. Folding landscapes of ankyrin repeat proteins: Experiments meet theory. Curr. Opin. Struct. Biol. 2008, 18, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Champion, E.A.; Lane, B.H.; Jackrel, M.E.; Regan, L.; Baserga, S.J. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol. Cell. Biol. 2008, 28, 6547–6556. [Google Scholar] [CrossRef]
- Ferreiro, D.U.; Komives, E.A. Molecular mechanisms of system control of NF-kappaB signaling by IkappaBalpha. Biochemistry 2010, 49, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Grove, T.Z.; Cortajarena, A.L.; Regan, L. Ligand binding by repeat proteins: Natural and designed. Curr. Opin. Struct. Biol. 2008, 18, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Javadi, Y.; Itzhaki, L.S. Tandem-repeat proteins: Regularity plus modularity equals design-ability. Curr. Opin. Struct. Biol. 2013, 23, 622–631. [Google Scholar] [CrossRef]
- Kane, E.I.; Spratt, D.E. Structural Insights into Ankyrin Repeat-Containing Proteins and Their Influence in Ubiquitylation. Int. J. Mol. Sci. 2021, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Klamt, A.; Nagarathinam, K.; Tanabe, M.; Kumar, A.; Balbach, J. Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy. J. Phys. Chem. B 2019, 123, 792–801. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Raval, M.H.; Wan, J.; Yengo, C.M.; Liu, W.; Zhang, M. Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules. Structure 2019, 27, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riba, A.; Synakewicz, M.; Itzhaki, L.S. Folding cooperativity and allosteric function in the tandem-repeat protein class. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170188. [Google Scholar] [CrossRef] [PubMed]
- Groves, M.R.; Hanlon, N.; Turowski, P.; Hemmings, B.A.; Barford, D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 1999, 96, 99–110. [Google Scholar] [CrossRef]
- Choi, H.J.; Weis, W.I. Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 2005, 346, 367–376. [Google Scholar] [CrossRef]
- Peters, J.W.; Stowell, M.H.; Rees, D.C. A leucine-rich repeat variant with a novel repetitive protein structural motif. Nat. Struct. Biol. 1996, 3, 991–994. [Google Scholar] [CrossRef]
- Jeyakanthan, J.; Rangarajan, S.; Mridula, P.; Kanaujia, S.P.; Shiro, Y.; Kuramitsu, S.; Yokoyama, S.; Sekar, K. Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 1012–1019. [Google Scholar] [CrossRef]
- Perry, A.J.; Hulett, J.M.; Likic, V.A.; Lithgow, T.; Gooley, P.R. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 2006, 16, 221–229. [Google Scholar] [CrossRef]
- Mosavi, L.K.; Minor, D.L., Jr.; Peng, Z.Y. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. USA 2002, 99, 16029–16034. [Google Scholar] [CrossRef]
- Krzywda, S.; Brzozowski, A.M.; Higashitsuji, H.; Fujita, J.; Welchman, R.; Dawson, S.; Mayer, R.J.; Wilkinson, A.J. The crystal structure of gankyrin, an oncoprotein found in complexes with cyclin-dependent kinase 4, a 19 S proteasomal ATPase regulator, and the tumor suppressors Rb and p53. J. Biol. Chem. 2004, 279, 1541–1545. [Google Scholar] [CrossRef] [PubMed]
- Sprague, E.R.; Redd, M.J.; Johnson, A.D.; Wolberger, C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000, 19, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Aves, S.J.; Durkacz, B.W.; Carr, A.; Nurse, P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 ‘start’ gene. EMBO J. 1985, 4, 457–463. [Google Scholar] [CrossRef]
- Breeden, L.; Nasmyth, K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature 1987, 329, 651–654. [Google Scholar] [CrossRef]
- Sharma, N.; Bham, K.; Senapati, S. Human ankyrins and their contribution to disease biology: An update. J. Biosci. 2020, 45, 1–16. [Google Scholar] [CrossRef]
- Bork, P. Hundreds of ankyrin-like repeats in functionally diverse proteins: Mobile modules that cross phyla horizontally? Proteins 1993, 17, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef]
- Kohl, A.; Binz, H.K.; Forrer, P.; Stumpp, M.T.; Pluckthun, A.; Grutter, M.G. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. USA 2003, 100, 1700–1705. [Google Scholar] [CrossRef]
- Ferreiro, D.U.; Cervantes, C.F.; Truhlar, S.M.; Cho, S.S.; Wolynes, P.G.; Komives, E.A. Stabilizing IkappaBalpha by “consensus” design. J. Mol. Biol. 2007, 365, 1201–1216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Löw, C.; Weininger, U.; Neumann, P.; Klepsch, M.; Lilie, H.; Stubbs, M.T.; Balbach, J. Structural insights into an equilibrium folding intermediate of an archaeal ankyrin repeat protein. Proc. Natl. Acad. Sci. USA 2008, 105, 3779–3784. [Google Scholar] [CrossRef]
- Zhang, B.; Peng, Z. A minimum folding unit in the ankyrin repeat protein p16(INK4). J. Mol. Biol. 2000, 299, 1121–1132. [Google Scholar] [CrossRef]
- Huxford, T.; Huang, D.B.; Malek, S.; Ghosh, G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell 1998, 95, 759–770. [Google Scholar] [CrossRef]
- Jacobs, M.D.; Harrison, S.C. Structure of an IkappaBalpha/NF-kappaB complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef]
- Brotherton, D.H.; Dhanaraj, V.; Wick, S.; Brizuela, L.; Domaille, P.J.; Volyanik, E.; Xu, X.; Parisini, E.; Smith, B.O.; Archer, S.J.; et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature 1998, 395, 244–250. [Google Scholar] [CrossRef]
- Kumar, A.; Gopalswamy, M.; Wolf, A.; Brockwell, D.J.; Hatzfeld, M.; Balbach, J. Phosphorylation-induced unfolding regulates p19(INK4d) during the human cell cycle. Proc. Natl. Acad. Sci. USA 2018, 115, 3344–3349. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, A.H.; Piper, D.E.; de la Brousse, F.C.; McKnight, S.L.; Wolberger, C. The structure of GABPalpha/beta: An ETS domain- ankyrin repeat heterodimer bound to DNA. Science 1998, 279, 1037–1041. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Tompa, P. Fuzzy complexes: A more stochastic view of protein function. Adv. Exp. Med. Biol. 2012, 725, 1–14. [Google Scholar] [CrossRef]
- Kumar, A.; Balbach, J. Real-time protein NMR spectroscopy and investigation of assisted protein folding. Biochim. Biophys. Acta 2015, 1850, 1965–1972. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Sali, A.; Karplus, M. Protein Folding: A Perspective from Theory and Experiment. Angew. Chem. Int. Ed. Engl. 1998, 37, 868–893. [Google Scholar] [CrossRef]
- Dill, K.A.; Chan, H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997, 4, 10–19. [Google Scholar] [CrossRef]
- Ferreiro, D.U.; Walczak, A.M.; Komives, E.A.; Wolynes, P.G. The energy landscapes of repeat-containing proteins: Topology, cooperativity, and the folding funnels of one-dimensional architectures. PLoS Comput. Biol. 2008, 4, e1000070. [Google Scholar] [CrossRef] [PubMed]
- Buchner, J.; Kiefhaber, T. Protein Folding Handbook; Buchner, J., Kiefhaber, T., Eds.; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; Volume 1, p. 2623. [Google Scholar]
- Löw, C.; Weininger, U.; Zeeb, M.; Zhang, W.; Laue, E.D.; Schmid, F.X.; Balbach, J. Folding mechanism of an ankyrin repeat protein: Scaffold and active site formation of human CDK inhibitor p19(INK4d). J. Mol. Biol. 2007, 373, 219–231. [Google Scholar] [CrossRef]
- Löw, C.; Homeyer, N.; Weininger, U.; Sticht, H.; Balbach, J. Conformational switch upon phosphorylation: Human CDK inhibitor p19INK4d between the native and partially folded state. ACS Chem. Biol. 2009, 4, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Rowling, P.J.; Sivertsson, E.M.; Perez-Riba, A.; Main, E.R.; Itzhaki, L.S. Dissecting and reprogramming the folding and assembly of tandem-repeat proteins. Biochem. Soc. Trans. 2015, 43, 881–888. [Google Scholar] [CrossRef]
- Ferreiro, D.U.; Komives, E.A. The plastic landscape of repeat proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 7735–7736. [Google Scholar] [CrossRef]
- Bachmann, A.; Kiefhaber, T. Apparent two-state tendamistat folding is a sequential process along a defined route. J. Mol. Biol. 2001, 306, 375–386. [Google Scholar] [CrossRef]
- Bachmann, A.; Kiefhaber, T. Kinetic Mechanisms in Protein Folding. In Protein Folding Handbook; Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH: Weinheim, Germany, 2005; Volume 1, pp. 402–406. [Google Scholar]
- Main, E.R.; Stott, K.; Jackson, S.E.; Regan, L. Local and long-range stability in tandemly arrayed tetratricopeptide repeats. Proc. Natl. Acad. Sci. USA 2005, 102, 5721–5726. [Google Scholar] [CrossRef]
- Stumpp, M.T.; Binz, H.K.; Amstutz, P. DARPins: A new generation of protein therapeutics. Drug Discov. Today 2008, 13, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Chagula, D.B.; Rechcinski, T.; Rudnicka, K.; Chmiela, M. Ankyrins in human health and disease—An update of recent experimental findings. Arch. Med. Sci. 2020, 16, 715–726. [Google Scholar] [CrossRef]
- Zahnd, C.; Wyler, E.; Schwenk, J.M.; Steiner, D.; Lawrence, M.C.; McKern, N.M.; Pecorari, F.; Ward, C.W.; Joos, T.O.; Pluckthun, A. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J. Mol. Biol. 2007, 369, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Amstutz, P.; Kohl, A.; Stumpp, M.T.; Briand, C.; Forrer, P.; Grutter, M.G.; Pluckthun, A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004, 22, 575–582. [Google Scholar] [CrossRef]
- Kohl, A.; Amstutz, P.; Parizek, P.; Binz, H.K.; Briand, C.; Capitani, G.; Forrer, P.; Pluckthun, A.; Grutter, M.G. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure 2005, 13, 1131–1141. [Google Scholar] [CrossRef]
- Schweizer, A.; Roschitzki-Voser, H.; Amstutz, P.; Briand, C.; Gulotti-Georgieva, M.; Prenosil, E.; Binz, H.K.; Capitani, G.; Baici, A.; Pluckthun, A.; et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: Specificity, structure, and inhibition mechanism. Structure 2007, 15, 625–636. [Google Scholar] [CrossRef]
- Sennhauser, G.; Amstutz, P.; Briand, C.; Storchenegger, O.; Grutter, M.G. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007, 5, e7. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, P.; Koch, H.; Binz, H.K.; Deuber, S.A.; Pluckthun, A. Rapid selection of specific MAP kinase-binders from designed ankyrin repeat protein libraries. Protein Eng. Des. Sel. 2006, 19, 219–229. [Google Scholar] [CrossRef][Green Version]
- Siegel, P.M.; Bojti, I.; Bassler, N.; Holien, J.; Flierl, U.; Wang, X.; Waggershauser, P.; Tonnar, X.; Vedecnik, C.; Lamprecht, C.; et al. A DARPin targeting activated Mac-1 is a novel diagnostic tool and potential anti-inflammatory agent in myocarditis, sepsis and myocardial infarction. Basic. Res. Cardiol. 2021, 116, 17. [Google Scholar] [CrossRef]
- Mittl, P.R.; Ernst, P.; Pluckthun, A. Chaperone-assisted structure elucidation with DARPins. Curr. Opin. Struct. Biol. 2020, 60, 93–100. [Google Scholar] [CrossRef]
- Ernst, P.; Honegger, A.; van der Valk, F.; Ewald, C.; Mittl, P.R.E.; Pluckthun, A. Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts. Sci. Rep. 2019, 9, 16162. [Google Scholar] [CrossRef]
- Ernst, P.; Pluckthun, A.; Mittl, P.R.E. Structural analysis of biological targets by host:guest crystal lattice engineering. Sci. Rep. 2019, 9, 15199. [Google Scholar] [CrossRef]
- Bery, N.; Legg, S.; Debreczeni, J.; Breed, J.; Embrey, K.; Stubbs, C.; Kolasinska-Zwierz, P.; Barrett, N.; Marwood, R.; Watson, J.; et al. KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe. Nat. Commun. 2019, 10, 2607. [Google Scholar] [CrossRef]
- Stumpp, M.T.; Dawson, K.M.; Binz, H.K. Beyond Antibodies: The DARPin((R)) Drug Platform. BioDrugs 2020, 34, 423–433. [Google Scholar] [CrossRef]
- Gebauer, M.; Skerra, A. Engineered Protein Scaffolds as Next-Generation Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 391–415. [Google Scholar] [CrossRef] [PubMed]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. Notch signaling in leukemia. Annu. Rev. Pathol. 2008, 3, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.L.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66, 649–661. [Google Scholar] [CrossRef]
- Koch, U.; Radtke, F. Notch and cancer: A double-edged sword. Cell. Mol. Life Sci. 2007, 64, 2746–2762. [Google Scholar] [CrossRef] [PubMed]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris, J.P.t.; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Jarriault, S.; Brou, C.; Logeat, F.; Schroeter, E.H.; Kopan, R.; Israel, A. Signalling downstream of activated mammalian Notch. Nature 1995, 377, 355–358. [Google Scholar] [CrossRef]
- Kurooka, H.; Kuroda, K.; Honjo, T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998, 26, 5448–5455. [Google Scholar] [CrossRef]
- Wettstein, D.A.; Turner, D.L.; Kintner, C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 1997, 124, 693–702. [Google Scholar] [CrossRef]
- Roehl, H.; Bosenberg, M.; Blelloch, R.; Kimble, J. Roles of the RAM and ANK domains in signaling by the C. elegans GLP-1 receptor. EMBO J. 1996, 15, 7002–7012. [Google Scholar] [CrossRef] [PubMed]
- Roehl, H.; Kimble, J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature 1993, 364, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.E.; Ilagan, M.X.; Kopan, R.; Barrick, D. Thermodynamic analysis of the CSL x Notch interaction: Distribution of binding energy of the Notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J. Biol. Chem. 2010, 285, 6681–6692. [Google Scholar] [CrossRef] [PubMed]
- Lubman, O.Y.; Ilagan, M.X.; Kopan, R.; Barrick, D. Quantitative dissection of the Notch:CSL interaction: Insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 2007, 365, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Taniguchi, Y.; Minoguchi, S.; Sakai, T.; Tun, T.; Furukawa, T.; Honjo, T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol. 1995, 5, 1416–14123. [Google Scholar] [CrossRef]
- Jarrett, S.M.; Seegar, T.C.M.; Andrews, M.; Adelmant, G.; Marto, J.A.; Aster, J.C.; Blacklow, S.C. Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Leahy, D.J.; Hughson, F.M.; Barrick, D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003, 12, 2622–2632. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Barrick, D. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 1. Solution conformational and hydrodynamic properties. Biochemistry 2001, 40, 14344–14356. [Google Scholar] [CrossRef]
- Nam, Y.; Sliz, P.; Song, L.; Aster, J.C.; Blacklow, S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 2006, 124, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.C.; Bradley, C.M.; Tripp, K.W.; Barrick, D. Experimental characterization of the folding kinetics of the notch ankyrin domain. J. Mol. Biol. 2005, 352, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.J.; Kovall, R.A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 2006, 124, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Lubman, O.Y.; Kopan, R.; Waksman, G.; Korolev, S. The crystal structure of a partial mouse Notch-1 ankyrin domain: Repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005, 14, 1274–1281. [Google Scholar] [CrossRef]
- Coleman, M.L.; McDonough, M.A.; Hewitson, K.S.; Coles, C.; Mecinovic, J.; Edelmann, M.; Cook, K.M.; Cockman, M.E.; Lancaster, D.E.; Kessler, B.M.; et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 2007, 282, 24027–24038. [Google Scholar] [CrossRef]
- Zheng, X.; Linke, S.; Dias, J.M.; Gradin, K.; Wallis, T.P.; Hamilton, B.R.; Gustafsson, M.; Ruas, J.L.; Wilkins, S.; Bilton, R.L.; et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 3368–3373. [Google Scholar] [CrossRef]
- Kelly, L.; McDonough, M.A.; Coleman, M.L.; Ratcliffe, P.J.; Schofield, C.J. Asparagine beta-hydroxylation stabilizes the ankyrin repeat domain fold. Mol. Biosyst. 2009, 5, 52–58. [Google Scholar] [CrossRef]
- Wilkins, S.E.; Karttunen, S.; Hampton-Smith, R.J.; Murchland, I.; Chapman-Smith, A.; Peet, D.J. Factor inhibiting HIF (FIH) recognizes distinct molecular features within hypoxia-inducible factor-alpha (HIF-alpha) versus ankyrin repeat substrates. J. Biol. Chem. 2012, 287, 8769–8781. [Google Scholar] [CrossRef]
- Sen, R.; Baltimore, D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell 1986, 47, 921–928. [Google Scholar] [CrossRef]
- Courtois, G.; Gilmore, T.D. Mutations in the NF-kappaB signaling pathway: Implications for human disease. Oncogene 2006, 25, 6831–6843. [Google Scholar] [CrossRef] [PubMed]
- Basseres, D.S.; Baldwin, A.S. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene 2006, 25, 6817–6830. [Google Scholar] [CrossRef] [PubMed]
- Ceruti, J.M.; Scassa, M.E.; Flo, J.M.; Varone, C.L.; Canepa, E.T. Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene 2005, 24, 4065–4080. [Google Scholar] [CrossRef]
- Toubi, E.; Shoenfeld, Y. Toll-like receptors and their role in the development of autoimmune diseases. Autoimmunity 2004, 37, 183–188. [Google Scholar] [CrossRef]
- Hou, P.; Jia, P.; Yang, K.; Li, Z.; Tian, T.; Lin, Y.; Zeng, W.; Xing, F.; Chen, Y.; Li, C.; et al. An unconventional role of an ASB family protein in NF-kappaB activation and inflammatory response during microbial infection and colitis. Proc. Natl. Acad. Sci. USA 2021, 118, e2015416118. [Google Scholar] [CrossRef] [PubMed]
- Stancovski, I.; Baltimore, D. NF-kappaB activation: The I kappaB kinase revealed? Cell 1997, 91, 299–302. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. I kappa B: A specific inhibitor of the NF-kappa B transcription factor. Science 1988, 242, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Verma, I.M.; Stevenson, J.K.; Schwarz, E.M.; Van Antwerp, D.; Miyamoto, S. Rel/NF-kappa B/I kappa B family: Intimate tales of association and dissociation. Genes Dev. 1995, 9, 2723–2735. [Google Scholar] [CrossRef]
- Beg, A.A.; Ruben, S.M.; Scheinman, R.I.; Haskill, S.; Rosen, C.A.; Baldwin, A.S., Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: A mechanism for cytoplasmic retention. Genes Dev. 1992, 6, 1899–1913. [Google Scholar] [CrossRef]
- Ghosh, S.; Karin, M. Missing pieces in the NF-kappaB puzzle. Cell 2002, 109 (Suppl. S81), 96. [Google Scholar] [CrossRef]
- Huang, T.T.; Kudo, N.; Yoshida, M.; Miyamoto, S. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 1014–1019. [Google Scholar] [CrossRef]
- Johnson, C.; Van Antwerp, D.; Hope, T.J. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IkappaBalpha. EMBO J. 1999, 18, 6682–6693. [Google Scholar] [CrossRef]
- Malek, S.; Chen, Y.; Huxford, T.; Ghosh, G. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J. Biol. Chem. 2001, 276, 45225–45235. [Google Scholar] [CrossRef]
- Huang, T.T.; Miyamoto, S. Postrepression activation of NF-kappaB requires the amino-terminal nuclear export signal specific to IkappaBalpha. Mol. Cell. Biol. 2001, 21, 4737–4747. [Google Scholar] [CrossRef]
- Croy, C.H.; Bergqvist, S.; Huxford, T.; Ghosh, G.; Komives, E.A. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein. Sci. 2004, 13, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Truhlar, S.M.; Torpey, J.W.; Komives, E.A. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB.DNA interaction fold upon binding to NF-kappaB. Proc. Natl. Acad. Sci. USA 2006, 103, 18951–18956. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, C.F.; Handley, L.D.; Sue, S.C.; Dyson, H.J.; Komives, E.A. Long-range effects and functional consequences of stabilizing mutations in the ankyrin repeat domain of IkappaBalpha. J. Mol. Biol. 2013, 425, 902–913. [Google Scholar] [CrossRef][Green Version]
- DeVries, I.; Ferreiro, D.U.; Sanchez, I.E.; Komives, E.A. Folding kinetics of the cooperatively folded subdomain of the IkappaBalpha ankyrin repeat domain. J. Mol. Biol. 2011, 408, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.E.; Kiefhaber, T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J. Mol. Biol. 2003, 325, 367–376. [Google Scholar] [CrossRef]
- Huxford, T.; Ghosh, G. A structural guide to proteins of the NF-kappaB signaling module. Cold Spring Harb. Perspect. Biol. 2009, 1, a000075. [Google Scholar] [CrossRef]
- Malek, S.; Huang, D.B.; Huxford, T.; Ghosh, S.; Ghosh, G. X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex. J. Biol. Chem. 2003, 278, 23094–23100. [Google Scholar] [CrossRef]
- Chen, Z.J.; Parent, L.; Maniatis, T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell 1996, 84, 853–862. [Google Scholar] [CrossRef]
- Mukherjee, S.P.; Quintas, P.O.; McNulty, R.; Komives, E.A.; Dyson, H.J. Structural characterization of the ternary complex that mediates termination of NF-kappaB signaling by IkappaBalpha. Proc. Natl. Acad. Sci. USA 2016, 113, 6212–6217. [Google Scholar] [CrossRef]
- Napetschnig, J.; Wu, H. Molecular basis of NF-kappaB signaling. Annu. Rev. Biophys. 2013, 42, 443–468. [Google Scholar] [CrossRef]
- Sue, S.C.; Cervantes, C.; Komives, E.A.; Dyson, H.J. Transfer of flexibility between ankyrin repeats in IkappaB* upon formation of the NF-kappaB complex. J. Mol. Biol. 2008, 380, 917–931. [Google Scholar] [CrossRef]
- Bergqvist, S.; Alverdi, V.; Mengel, B.; Hoffmann, A.; Ghosh, G.; Komives, E.A. Kinetic enhancement of NF-kappaBxDNA dissociation by IkappaBalpha. Proc. Natl. Acad. Sci. USA 2009, 106, 19328–19333. [Google Scholar] [CrossRef]
- Truhlar, S.M.; Mathes, E.; Cervantes, C.F.; Ghosh, G.; Komives, E.A. Pre-folding IkappaBalpha alters control of NF-kappaB signaling. J. Mol. Biol. 2008, 380, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.O. Principles of CDK regulation. Nature 1995, 374, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Pines, J. Cell cycle: Reaching for a role for the Cks proteins. Curr. Biol. 1996, 6, 1399–1402. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Cancer cell cycles. Science 1996, 274, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Bartkova, J.; Lukas, J. The retinoblastoma protein pathway and the restriction point. Curr. Opin. Cell. Biol. 1996, 8, 805–814. [Google Scholar] [CrossRef]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995, 9, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W. Checkpoint Control and Cancer; Kastan, M.B., Ed.; CSHL Press: New York, NY, USA, 1997; Volume 29, pp. 91–107. [Google Scholar]
- Chan, F.K.; Zhang, J.; Cheng, L.; Shapiro, D.N.; Winoto, A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol. Cell. Biol. 1995, 15, 2682–2688. [Google Scholar] [CrossRef]
- Guan, K.L.; Jenkins, C.W.; Li, Y.; O’Keefe, C.L.; Noh, S.; Wu, X.; Zariwala, M.; Matera, A.G.; Xiong, Y. Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK6 and CDK4. Mol. Biol. Cell. 1996, 7, 57–70. [Google Scholar] [CrossRef][Green Version]
- Hannon, G.J.; Beach, D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 1994, 371, 257–261. [Google Scholar] [CrossRef]
- Hirai, H.; Roussel, M.F.; Kato, J.Y.; Ashmun, R.A.; Sherr, C.J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol. Cell. Biol. 1995, 15, 2672–2681. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef]
- Bartkova, J.; Thullberg, M.; Rajpert-De Meyts, E.; Skakkebaek, N.E.; Bartek, J. Lack of p19INK4d in human testicular germ-cell tumours contrasts with high expression during normal spermatogenesis. Oncogene 2000, 19, 4146–4150. [Google Scholar] [CrossRef][Green Version]
- Drexler, H.G. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia 1998, 12, 845–859. [Google Scholar] [CrossRef]
- Ruas, M.; Peters, G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1998, 1378, F115–F177. [Google Scholar] [CrossRef]
- Serrano, M. The tumor suppressor protein p16INK4a. Exp. Cell Res. 1997, 237, 7–13. [Google Scholar] [CrossRef]
- Baumgartner, R.; Fernandez-Catalan, C.; Winoto, A.; Huber, R.; Engh, R.A.; Holak, T.A. Structure of human cyclin-dependent kinase inhibitor p19INK4d: Comparison to known ankyrin-repeat-containing structures and implications for the dysfunction of tumor suppressor p16INK4a. Structure 1998, 6, 1279–1290. [Google Scholar] [CrossRef]
- Byeon, I.J.; Li, J.; Ericson, K.; Selby, T.L.; Tevelev, A.; Kim, H.J.; O’Maille, P.; Tsai, M.D. Tumor suppressor p16INK4A: Determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol. Cell. 1998, 1, 421–431. [Google Scholar] [CrossRef]
- Luh, F.Y.; Archer, S.J.; Domaille, P.J.; Smith, B.O.; Owen, D.; Brotherton, D.H.; Raine, A.R.; Xu, X.; Brizuela, L.; Brenner, S.L.; et al. Structure of the cyclin-dependent kinase inhibitor p19Ink4d. Nature 1997, 389, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Selby, T.L.; Li, J.; Byeon, I.J.; Tsai, M.D. Tumor suppressor INK4: Refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data. Protein Sci. 2000, 9, 1120–1128. [Google Scholar] [CrossRef]
- Scassa, M.E.; Marazita, M.C.; Ceruti, J.M.; Carcagno, A.L.; Sirkin, P.F.; Gonzalez-Cid, M.; Pignataro, O.P.; Canepa, E.T. Cell cycle inhibitor, p19INK4d, promotes cell survival and decreases chromosomal aberrations after genotoxic insult due to enhanced DNA repair. DNA Repair (Amst) 2007, 6, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Thullberg, M.; Bartek, J.; Lukas, J. Ubiquitin/proteasome-mediated degradation of p19INK4d determines its periodic expression during the cell cycle. Oncogene 2000, 19, 2870–2876. [Google Scholar] [CrossRef] [PubMed]
- Thullberg, M.; Bartkova, J.; Khan, S.; Hansen, K.; Ronnstrand, L.; Lukas, J.; Strauss, M.; Bartek, J. Distinct versus redundant properties among members of the INK4 family of cyclin-dependent kinase inhibitors. FEBS Lett. 2000, 470, 161–166. [Google Scholar] [CrossRef]
- Zeeb, M.; Rösner, H.; Zeslawski, W.; Canet, D.; Holak, T.A.; Balbach, J. Protein folding and stability of human CDK inhibitor p19INK4d. J. Mol. Biol. 2002, 315, 447–457. [Google Scholar] [CrossRef][Green Version]
- Kumar, A.; Kuhn, L.T.; Balbach, J. In-Cell NMR: Analysis of Protein-Small Molecule Interactions, Metabolic Processes, and Protein Phosphorylation. Int. J. Mol. Sci. 2019, 20, 378. [Google Scholar] [CrossRef] [PubMed]
- Carcagno, A.L.; Marazita, M.C.; Ogara, M.F.; Ceruti, J.M.; Sonzogni, S.V.; Scassa, M.E.; Giono, L.E.; Canepa, E.T. E2F1-mediated upregulation of p19INK4d determines its periodic expression during cell cycle and regulates cellular proliferation. PLoS ONE 2011, 6, e21938. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, R.; Swaminathan, K.; Marmorstein, R. Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations. Nat. Struct. Biol. 1998, 5, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Balbach, J.; Schmid, F.X. Proline isomerization and its catalysis in protein folding. In Mechanisms of Protein Folding, 2nd ed.; Pain, R.H., Ed.; University Press: Oxford, UK, 2000; pp. 212–237. [Google Scholar]
- Scholz, C.; Eckert, B.; Hagn, F.; Schaarschmidt, P.; Balbach, J.; Schmid, F.X. SlyD proteins from different species exhibit high prolyl isomerase and chaperone activities. Biochemistry 2006, 45, 20–33. [Google Scholar] [CrossRef]
- Kumar, A.; Balbach, J. Targeting the molecular chaperone SlyD to inhibit bacterial growth with a small molecule. Sci. Rep. 2017, 7, 42141. [Google Scholar] [CrossRef]
- Kumar, A.; Gopalswamy, M.; Wishart, C.; Henze, M.; Eschen-Lippold, L.; Donnelly, D.; Balbach, J. N-terminal phosphorylation of parathyroid hormone (PTH) abolishes its receptor activity. ACS Chem. Biol. 2014, 9, 2465–2470. [Google Scholar] [CrossRef]
- Shacter, E.; Chock, P.B.; Stadtman, E.R. Regulation through phosphorylation/dephosphorylation cascade systems. J. Biol. Chem. 1984, 259, 12252–12259. [Google Scholar] [CrossRef]
- Msallam, M.; Sun, H.; Meledin, R.; Franz, P.; Brik, A. Examining the role of phosphorylation of p19(INK4d) in its stability and ubiquitination using chemical protein synthesis. Chem. Sci. 2020, 11, 5526–5531. [Google Scholar] [CrossRef]
- Han, X.; Kuang, Y.; Chen, H.; Liu, T.; Zhang, J.; Liu, J. p19INK4d: More than Just a Cyclin-Dependent Kinase Inhibitor. Curr. Drug Targets 2020, 21, 96–102. [Google Scholar] [CrossRef]
- Manjasetty, B.A.; Quedenau, C.; Sievert, V.; Bussow, K.; Niesen, F.; Delbruck, H.; Heinemann, U. X-ray structure of human gankyrin, the product of a gene linked to hepatocellular carcinoma. Proteins 2004, 55, 214–217. [Google Scholar] [CrossRef]
- Yuan, C.; Li, J.; Mahajan, A.; Poi, M.J.; Byeon, I.J.; Tsai, M.D. Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship. Biochemistry 2004, 43, 12152–12161. [Google Scholar] [CrossRef]
- Nakamura, Y.; Nakano, K.; Umehara, T.; Kimura, M.; Hayashizaki, Y.; Tanaka, A.; Horikoshi, M.; Padmanabhan, B.; Yokoyama, S. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure 2007, 15, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Guo, Y.; Yuan, C.; Weghorst, C.M.; Tsai, M.D.; Li, J. Dissection of protein-protein interaction and CDK4 inhibition in the oncogenic versus tumor suppressing functions of gankyrin and P16. J. Mol. Biol. 2007, 373, 990–1005. [Google Scholar] [CrossRef] [PubMed]
- Hutton, R.D.; Wilkinson, J.; Faccin, M.; Sivertsson, E.M.; Pelizzola, A.; Lowe, A.R.; Bruscolini, P.; Itzhaki, L.S. Mapping the Topography of a Protein Energy Landscape. J. Am. Chem. Soc. 2015, 137, 14610–14625. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, Y.; Zhou, H.; Ye, Y.; Talukdar, M.; Fu, Z.; Liu, Z.; Li, J.; Neculai, D.; Gao, J.; et al. Sunitinib inhibits RNase L by destabilizing its active dimer conformation. Biochem. J. 2020, 477, 3387–3399. [Google Scholar] [CrossRef]
- Cai, Q.; Hosokawa, T.; Zeng, M.; Hayashi, Y.; Zhang, M. Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms. Structure 2020, 28, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Kao, T.H.; Chang, S.W. The structural changes of the mutated ankyrin repeat domain of the human TRPV4 channel alter its ATP binding ability. J. Mech. Behav. Biomed. Mater. 2020, 101, 103407. [Google Scholar] [CrossRef] [PubMed]
- Siegel, A.; McAvoy, C.Z.; Lam, V.; Liang, F.C.; Kroon, G.; Miaou, E.; Griffin, P.; Wright, P.E.; Shan, S.O. A Disorder-to-Order Transition Activates an ATP-Independent Membrane Protein Chaperone. J. Mol. Biol. 2020, 432, 166708. [Google Scholar] [CrossRef]
- Ivankov, D.N.; Garbuzynskiy, S.O.; Alm, E.; Plaxco, K.W.; Baker, D.; Finkelstein, A.V. Contact order revisited: Influence of protein size on the folding rate. Protein Sci. 2003, 12, 2057–2062. [Google Scholar] [CrossRef]
- Espada, R.; Parra, R.G.; Sippl, M.J.; Mora, T.; Walczak, A.M.; Ferreiro, D.U. Repeat proteins challenge the concept of structural domains. Biochem. Soc. Trans. 2015, 43, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Nagampalli, R.S.K.; Fatima, M.T.; Ashraf, G.M. New paradigm in ankyrin repeats: Beyond protein-protein interaction module. Int. J. Biol. Macromol. 2018, 109, 1164–1173. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, A.; Balbach, J. Folding and Stability of Ankyrin Repeats Control Biological Protein Function. Biomolecules 2021, 11, 840. https://doi.org/10.3390/biom11060840
Kumar A, Balbach J. Folding and Stability of Ankyrin Repeats Control Biological Protein Function. Biomolecules. 2021; 11(6):840. https://doi.org/10.3390/biom11060840
Chicago/Turabian StyleKumar, Amit, and Jochen Balbach. 2021. "Folding and Stability of Ankyrin Repeats Control Biological Protein Function" Biomolecules 11, no. 6: 840. https://doi.org/10.3390/biom11060840
APA StyleKumar, A., & Balbach, J. (2021). Folding and Stability of Ankyrin Repeats Control Biological Protein Function. Biomolecules, 11(6), 840. https://doi.org/10.3390/biom11060840





