Innate Immune Response to Fasting and Refeeding in the Zebrafish Kidney
Abstract
1. Introduction
2. Materials and Methods
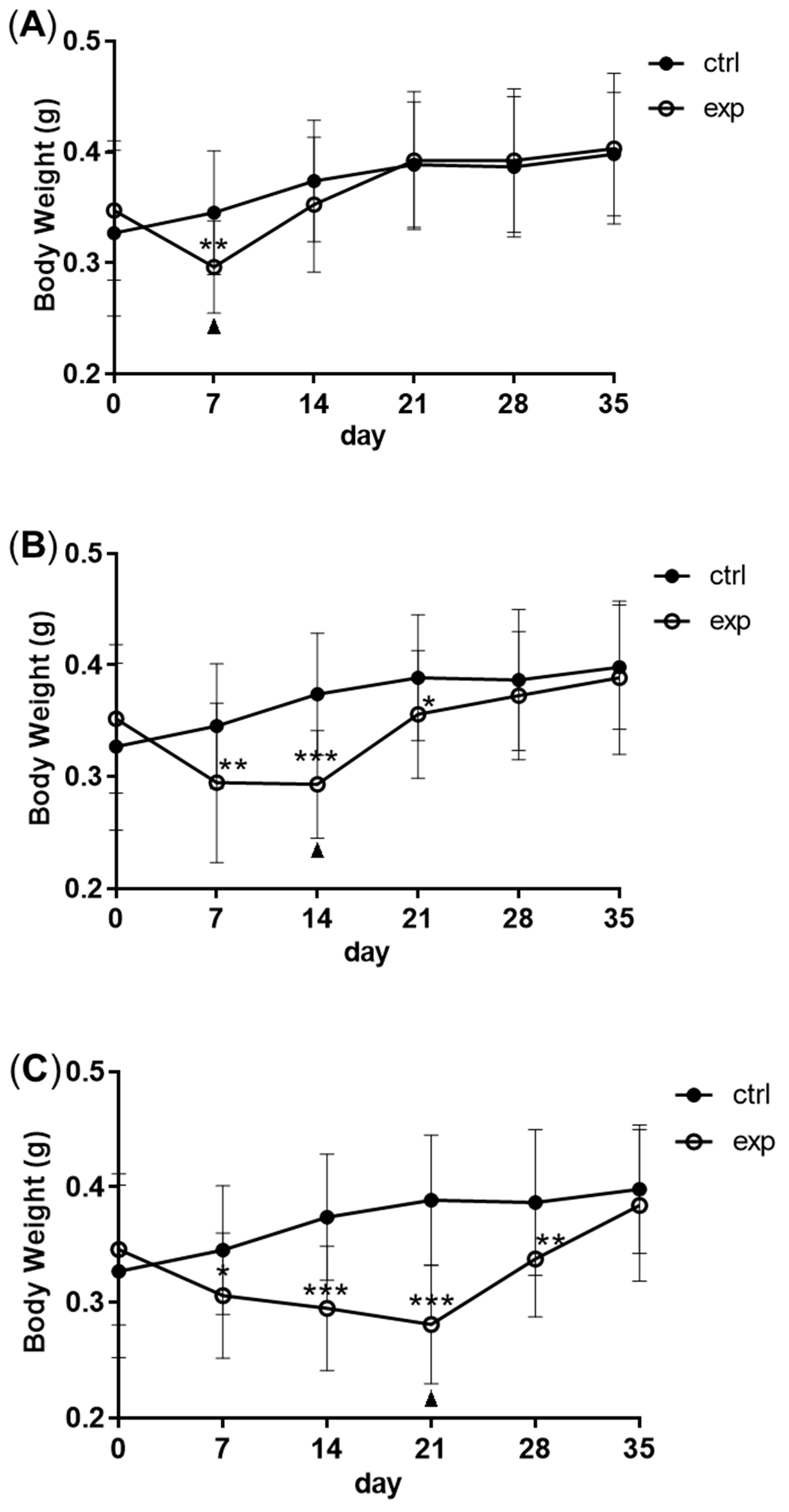
2.1. Zebrafish, Feeding Regimes and Body Weight
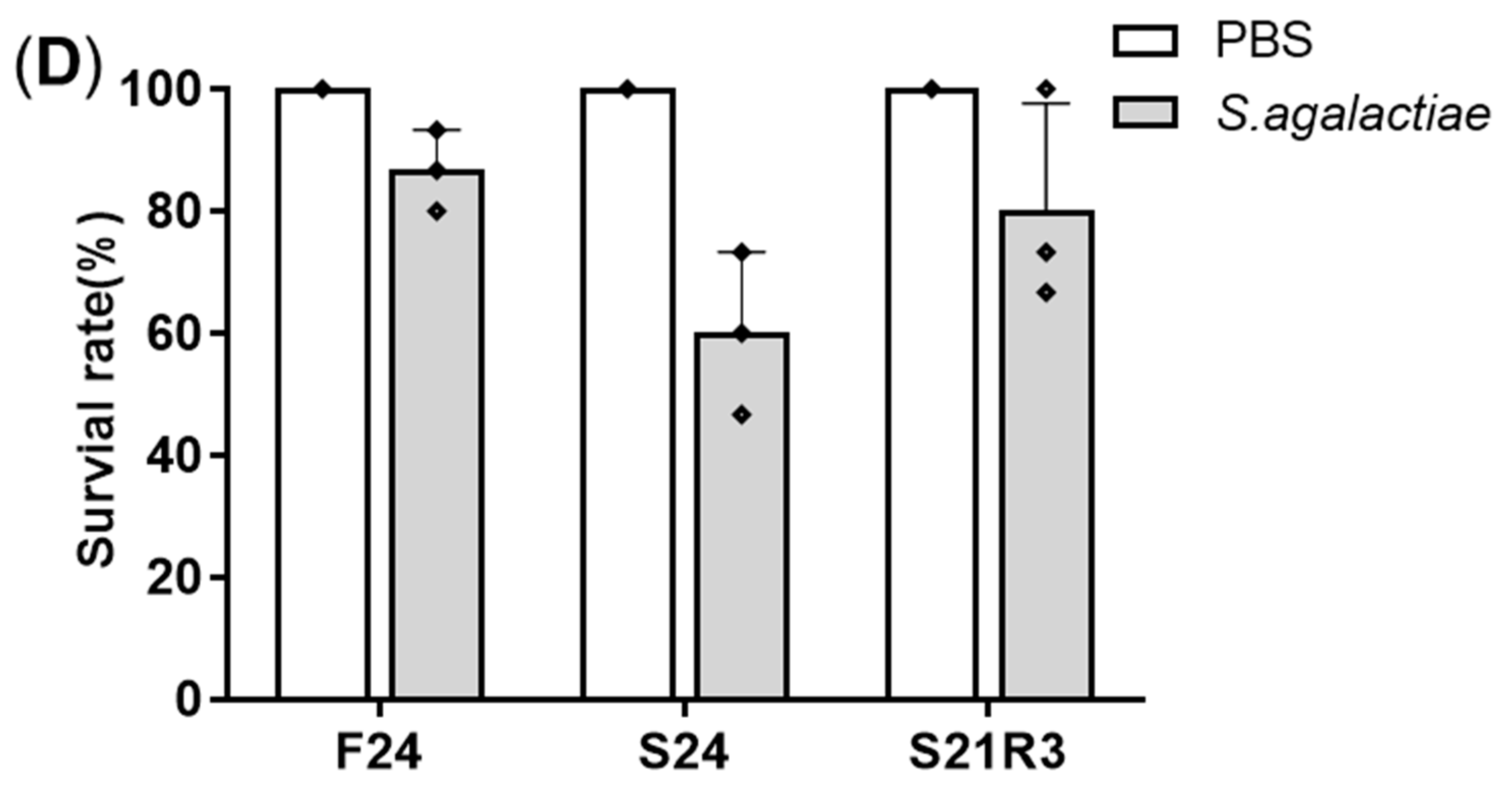
2.2. Streptococcus agalactiae Challenge
2.3. LPS Injection
2.4. Histopathology
2.5. Related Biochemical Parameter Assay
2.6. ROS Production
2.7. Respiratory Burst Assay
2.8. RNA Extraction and Real-Time PCR
2.9. Statistical Analysis
3. Results
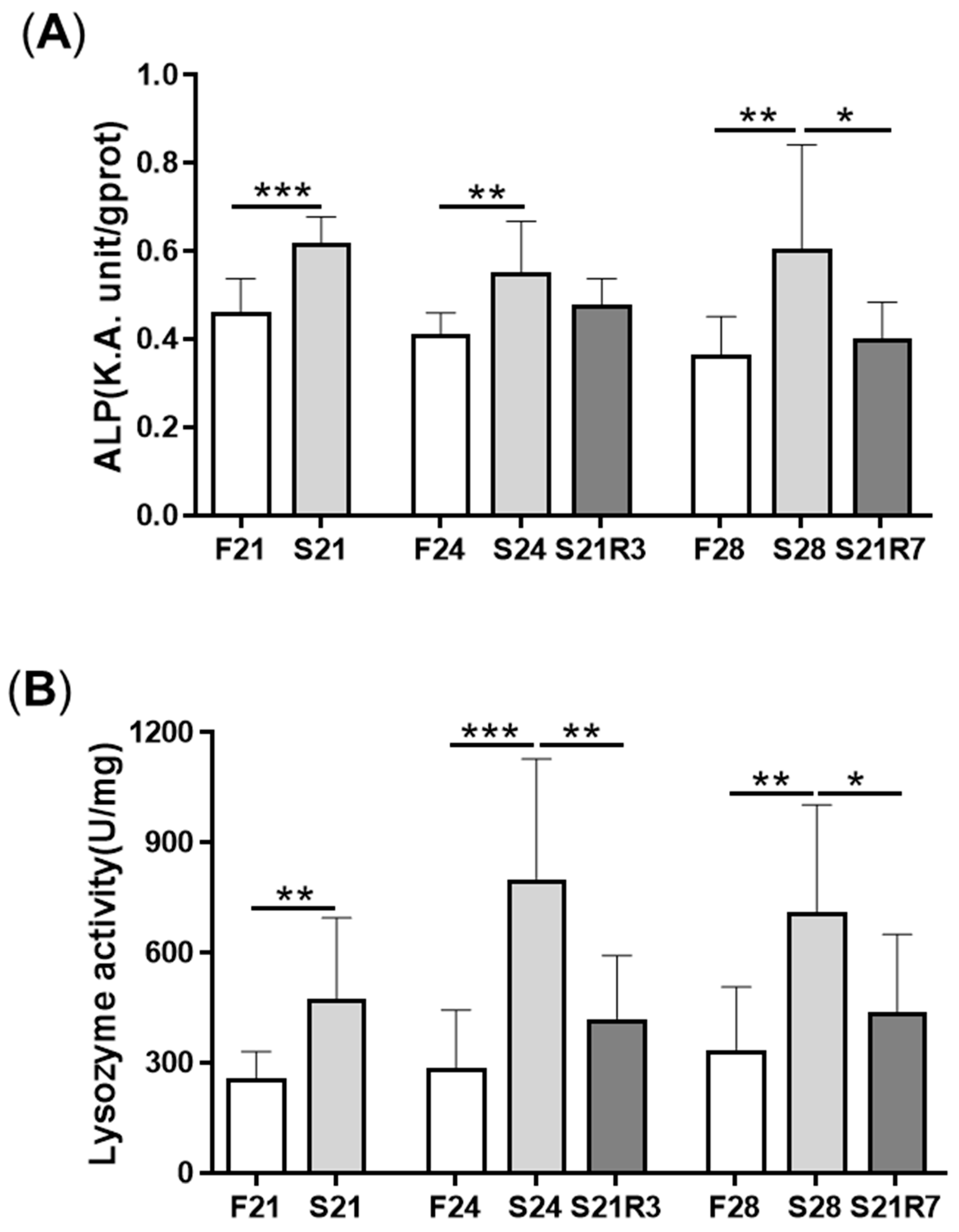
3.1. Changes in Body Weight and Antibacterial Properties of Zebrafish during Fasting and Refeeding
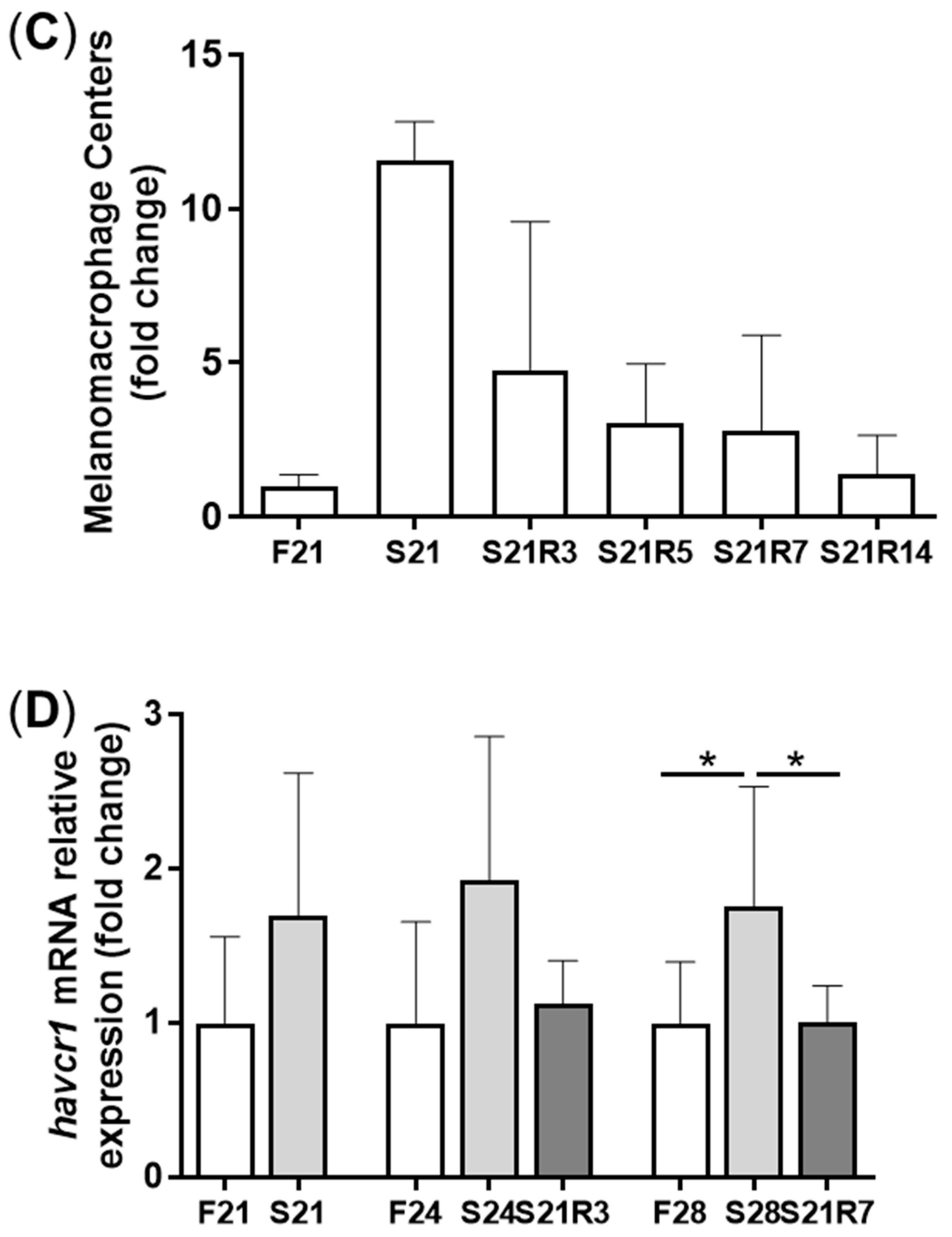
3.2. Morphologic Alterations of the Kidneys of Zebrafish during Fasting and Refeeding
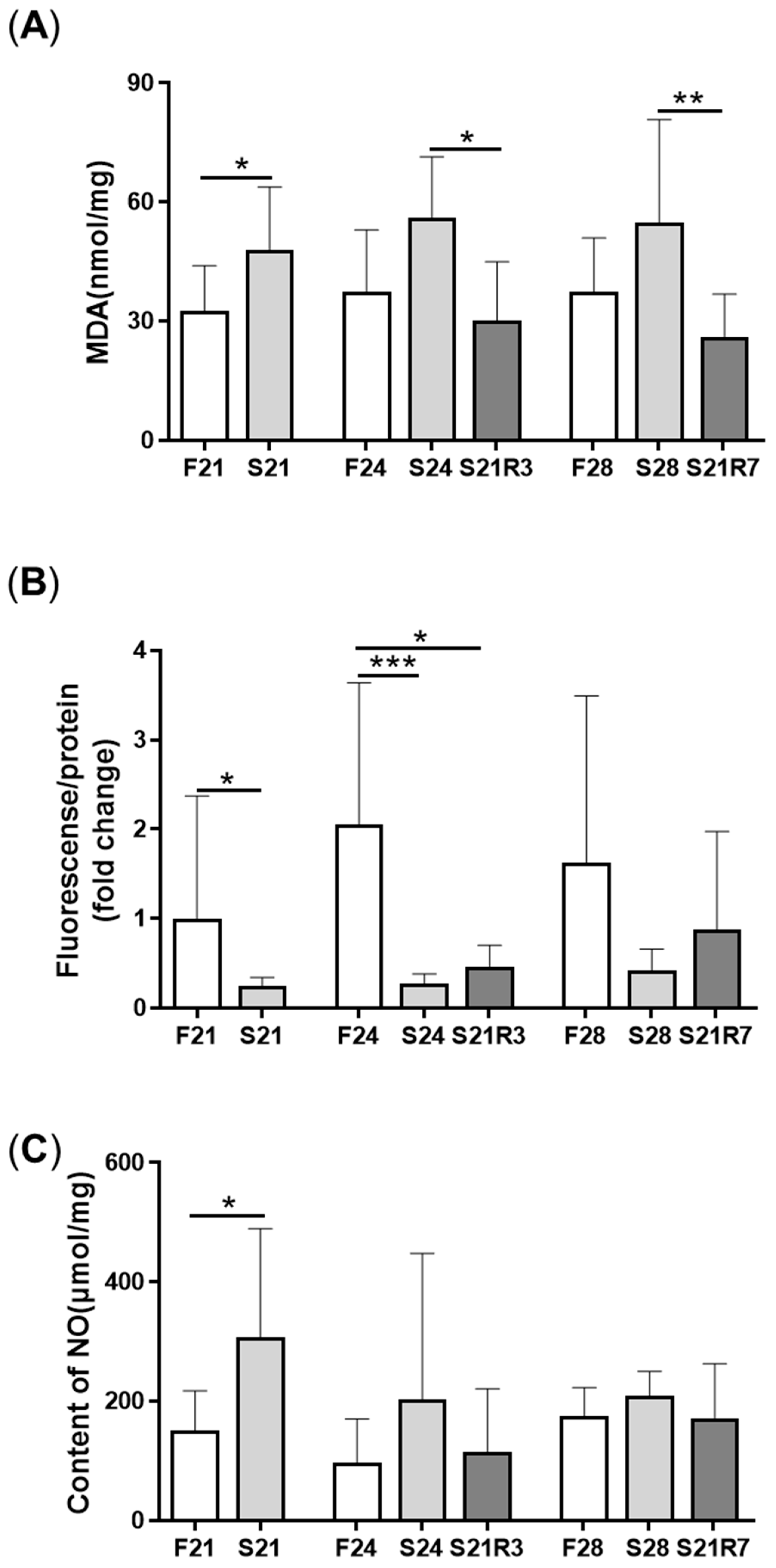
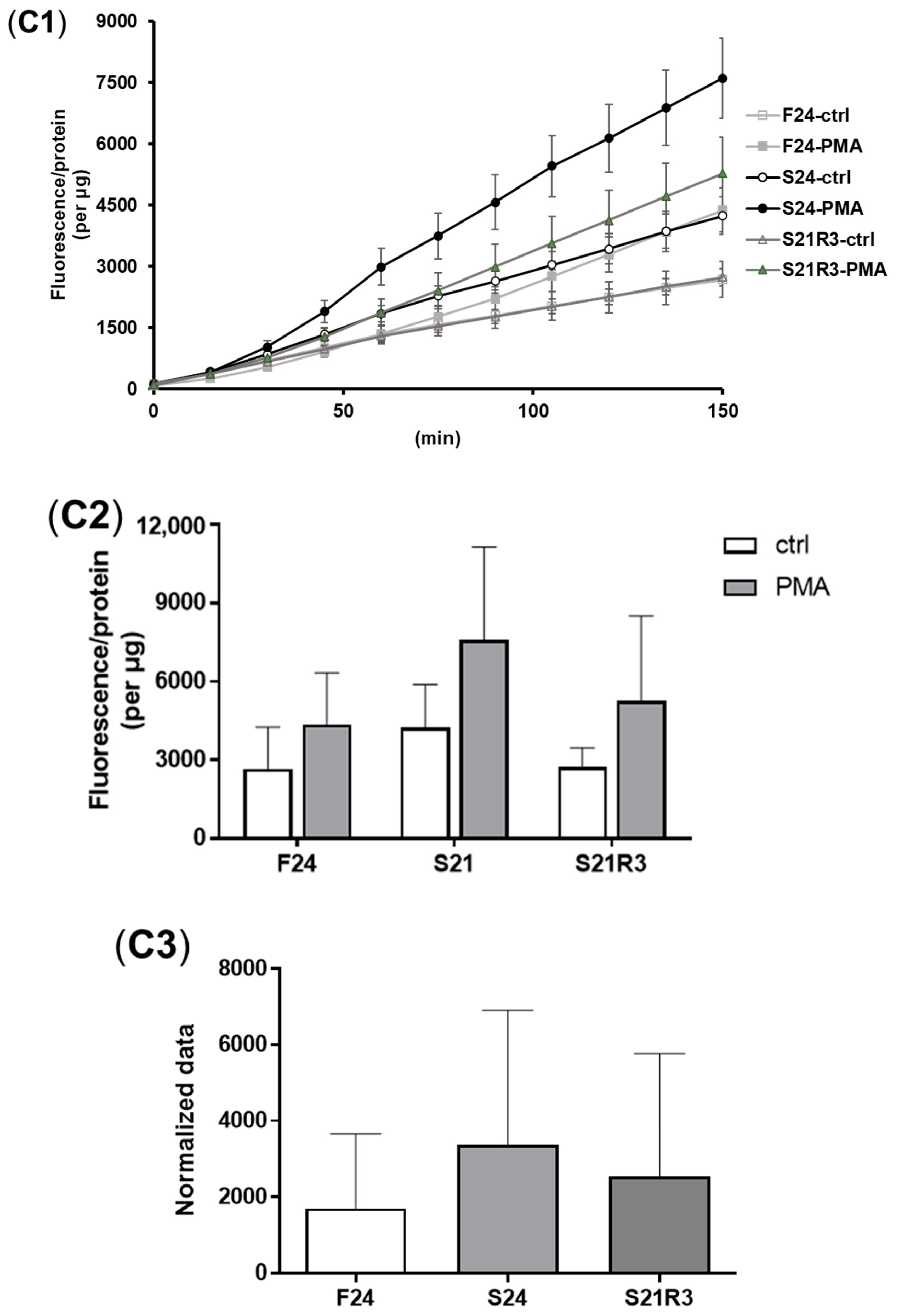
3.3. Oxidative Stress Was Triggered during Fasting and Refeeding
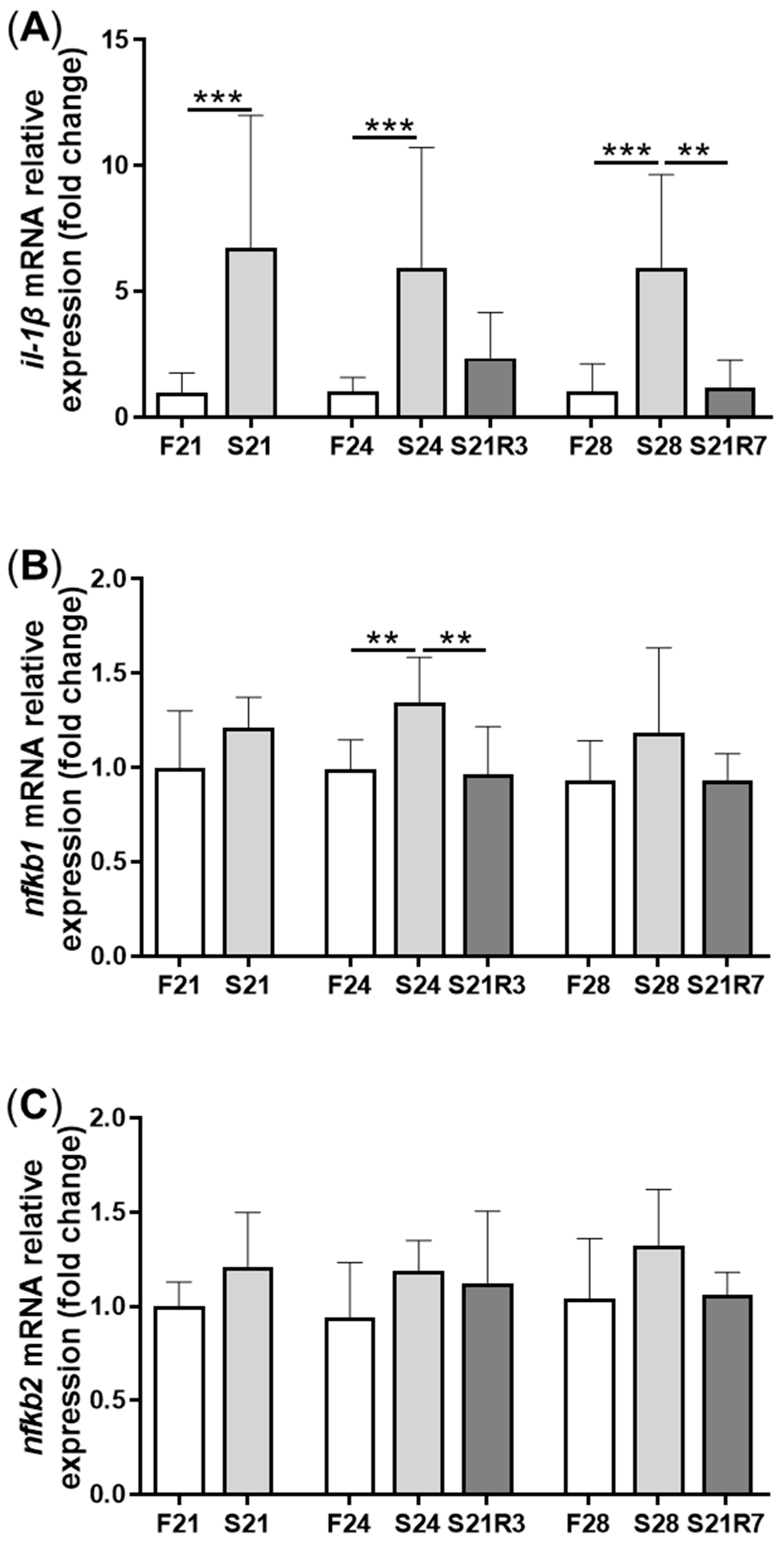
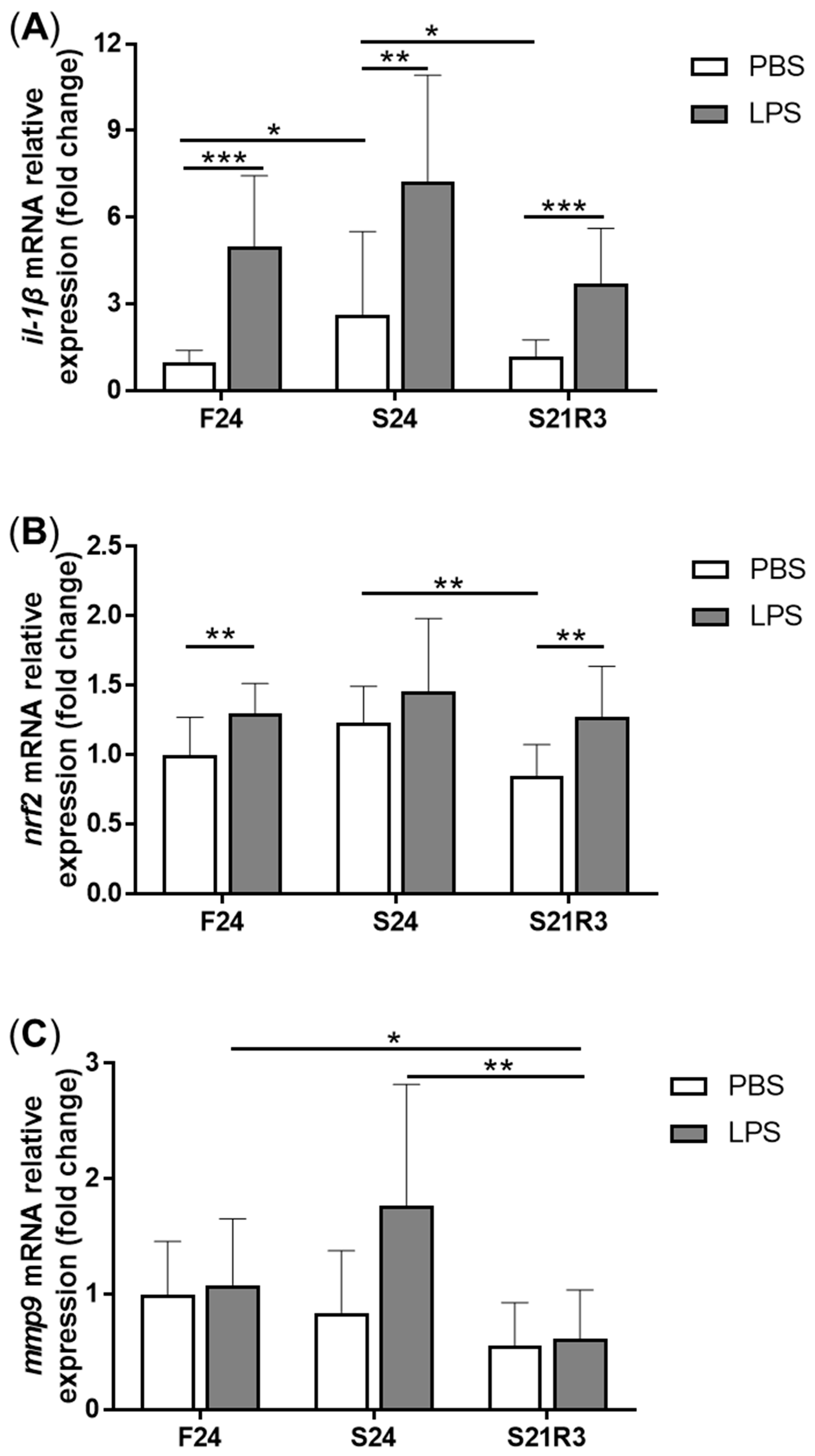
3.4. The Innate Immune System of the Zebrafish Kidney Was Affected by Fasting and Refeeding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alwarawrah, Y.; Kiernan, K.; Maciver, N.J. Changes in Nutritional Status Impact Immune Cell Metabolism and Function. Front. Immunol. 2018, 9, 1055. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Isik, M.; Moroz, N.; Steinbaugh, M.J.; Zhang, P.; Blackwell, T.K. Dietary Restriction Extends Lifespan through Metabolic Regulation of Innate Immunity. Cell Metab. 2019, 29, 1192–1205.e8. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.N.; Osbourn, D.F. Compensatory growth after undernutrition in mammals and birds. Biol. Rev. 1960, 35, 324–363. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Nicieza, A.; Wootton, R.J. Compensatory growth in fishes: A response to growth depression. Fish. Fish. 2003, 4, 147–190. [Google Scholar] [CrossRef]
- Bilton, H.T.; Robins, G.L. The Effects of Starvation and Subsequent Feeding on Survival and Growth of Fulton Channel Sockeye Salmon Fry (Oncorhynchus nerka). J. Fish. Res. Board Can. 1973, 30, 1–5. [Google Scholar] [CrossRef]
- Won, E.T.; Borski, R.J. Endocrine regulation of compensatory growth in fish. Front. Endocrinol. 2013, 4, 74. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M. Are compensatory growth and catch-up growth two sides of the same coin? Aquac. Int. 2009, 18, 501–510. [Google Scholar] [CrossRef]
- Whyte, S.K. The innate immune response of finfish—A review of current knowledge. Fish. Shellfish. Immunol. 2007, 23, 1127–1151. [Google Scholar] [CrossRef]
- Novoa, B.; Figueras, A. Zebrafish: Model for the Study of Inflammation and the Innate Immune Response to Infectious Diseases. Curr. Top. Innate Immun. 2012, 946, 253–275. [Google Scholar]
- Mathis, D.; Shoelson, S.E. Foreword Immunometabolism: An emerging frontier. Nat. Rev. Immunol. 2011, 11, 81–83. [Google Scholar] [CrossRef]
- Walrand, S.; Moreau, K.; Caldefie, F.; Tridon, A.; Chassagne, J.; Portefaix, G.; Cynober, L.; Beaufrère, B.; Vasson, M.-P.; Boirie, Y. Specific and nonspecific immune responses to fasting and refeeding differ in healthy young adult and elderly persons. Am. J. Clin. Nutr. 2001, 74, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Oarada, M.; Kamei, K.; Gonoi, T.; Tsuzuki, T.; Toyotome, T.; Hirasaka, K.; Nikawa, T.; Sato, A.; Kurita, N. Beneficial effects of a low-protein diet on host resistance to Paracoccidioides brasiliensis in mice. Nutrition 2009, 25, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.J.; Carrillo, O.V.; Llauradó, G.; Alonso, M.E.; Bermúdez, R.C.; Lebeque, Y.; Fontaine, R.; Soria, N.E.; Venet, G. Effect of starvation and refeeding on biochemical and immunological status of Balb/c mice: An experimental model of malnutrition. Immunopharmacol. Immunotoxicol. 2010, 33, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Petersen, P.S.; Lei, X.; Seldin, M.M.; Rodriguez, S.; Byerly, M.S.; Wolfe, A.; Whitlock, S.; Wong, G.W. Dynamic and extensive metabolic state-dependent regulation of cytokine expression and circulating levels. Am. J. Physiol. Integr. Comp. Physiol. 2014, 307, R1458–R1470. [Google Scholar] [CrossRef]
- Li, T.; Qi, M.; Gatesoupe, F.-J.; Tian, D.; Jin, W.; Li, J.; Lin, Q.; Wu, S.; Li, H. Adaptation to Fasting in Crucian Carp (Carassius auratus): Gut Microbiota and Its Correlative Relationship with Immune Function. Microb. Ecol. 2018, 78, 6–19. [Google Scholar] [CrossRef]
- Martin, S.A.M.; Douglas, A.; Houlihan, D.F.; Secombes, C.J. Starvation alters the liver transcriptome of the innate immune response in Atlantic salmon (Salmo salar). BMC Genom. 2010, 11, 418. [Google Scholar] [CrossRef]
- Najafi, A.; Salati, A.P.; Yavari, V.; Asadi, F. Effects of short term fasting and refeeding on some hematological and immune parameters in Mesopotamichthys sharpeyi (Gunther, 1874) fingerlings. Iran J. Sci. Technol. A 2015, 39, 383–389. [Google Scholar]
- Gimbo, R.Y.; Favero, G.C.; Montoya, L.N.F.; Urbinati, E.C. Energy deficit does not affect immune responses of experimentally infected pacu (Piaractus mesopotamicus). Fish. Shellfish. Immunol. 2015, 43, 295–300. [Google Scholar] [CrossRef]
- Caruso, G.; Denaro, M.G.; Caruso, R.; Mancari, F.; Genovese, L.; Maricchiolo, G. Response to short term starvation of growth, haematological, biochemical and non-specific immune parameters in European sea bass (Dicentrarchus labrax) and blackspot sea bream (Pagellus bogaraveo). Mar. Environ. Res. 2011, 72, 46–52. [Google Scholar] [CrossRef]
- Biller, J.D.; Takahashi, L.S. Oxidative stress and fish immune system: Phagocytosis and leukocyte respiratory burst activity. Anais Acad. Brasileira Ciências 2018, 90, 3403–3414. [Google Scholar] [CrossRef]
- Gill, R.; Tsung, A.; Billiar, T.R. Linking oxidative stress to inflammation: Toll-like receptors. Free. Radic. Biol. Med. 2010, 48, 1121–1132. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Gibson, S.B. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell. Signal. 2013, 25, 50–65. [Google Scholar] [CrossRef]
- Wu, C.-A.; Chao, Y.; Shiah, S.-G.; Lin, W.-W. Nutrient deprivation induces the Warburg effect through ROS/AMPK-dependent activation of pyruvate dehydrogenase kinase. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1833, 1147–1156. [Google Scholar] [CrossRef]
- Renshaw, S.A.; Trede, N.S. A model 450 million years in the making: Zebrafish and vertebrate immunity. Dis. Model. Mech. 2012, 5, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, V.S.; Ozer, J.S.; Dieterle, F.; Collings, F.B.; Ramirez, V.; Troth, S.; Muniappa, N.; Thudium, D.; Gerhold, D.; Holder, D.J.; et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat. Biotechnol. 2010, 28, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Naini, S.M.; Chen, G.; Hentschel, D.M.; Humphreys, B.D.; Bonventre, J.V. Mammalian Target of Rapamycin Mediates Kidney Injury Molecule 1-Dependent Tubule Injury in a Surrogate Model. J. Am. Soc. Nephrol. 2015, 27, 1943–1957. [Google Scholar] [CrossRef] [PubMed]
- Craig, P.M.; Moon, T.W. Fasted zebrafish mimic genetic and physiological responses in mammals: A model for obesity and diabetes? Zebrafish 2011, 8, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, Y.; Yuan, X.; Qin, J.; Yang, G.; Yu, X.; Wang, B.; Sun, C.; Li, W. Reactive oxygen species participate in liver function recovery during compensatory growth in zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 2018, 499, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.-W.; Jiang, Y.-H.; Lin, L.-B.; Deng, X.-Y.; Zhang, Q. Genome-wide differential expression analysis explores antibacterial molecular mechanisms of zebrafish intestine upon pathogenic Streptococcus agalactiae challenge. Aquac. Rep. 2021, 19, 100639. [Google Scholar] [CrossRef]
- Li, W.; Su, Y.-L.; Mai, Y.-Z.; Li, Y.-W.; Mo, Z.-Q.; Li, A.-X. Comparative proteome analysis of two Streptococcus agalactiae strains from cultured tilapia with different virulence. Vet. Microbiol. 2014, 170, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Goody, M.F.; Peterman, E.; Sullivan, C.; Kim, C.H. Quantification of the respiratory burst response as an indicator of innate immune health in zebrafish. J. Vis. Exp. 2013, 2013, e50667. [Google Scholar] [CrossRef]
- Hermann, A.C.; Millard, P.J.; Blake, S.L.; Kim, C.H. Development of a respiratory burst assay using zebrafish kidneys and embryos. J. Immunol. Methods 2004, 292, 119–129. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bennett, C.M.; Kanki, J.P.; Rhodes, J.; Liu, T.X.; Paw, B.H.; Kieran, M.W.; Langenau, D.M.; Delahaye-Brown, A.; Zon, L.I.; Fleming, M.D.; et al. Myelopoiesis in the zebrafish, Danio rerio. Blood 2001, 98, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Traver, D.; Paw, B.H.; Poss, K.D.; Penberthy, W.T.; Lin, S.; Zon, L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003, 4, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 2017, 214, 2875–2887. [Google Scholar] [CrossRef] [PubMed]
- Agius, C.; Roberts, R.J. Effects of starvation on the melano-macrophage centres of fish. J. Fish. Biol. 1981, 19, 161–169. [Google Scholar] [CrossRef]
- Steinel, N.C.; Bolnick, D.I. Melanomacrophage Centers as a Histological Indicator of Immune Function in Fish and Other Poikilotherms. Front. Immunol. 2017, 8, 827. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, M.; Oshikata, T.; Katoku, K.; Tsuchitani, M.; Yamaguchi, R. Two types of deposits, hyaline droplets and eosinophilic bodies, associated with α2u-globulin accumulation in the rat kidney. J. Toxicol. Pathol. 2017, 30, 275–282. [Google Scholar] [CrossRef][Green Version]
- Decker, J.H.; Dochterman, L.W.; Niquette, A.L.; Brej, M. Association of Renal Tubular Hyaline Droplets with Lymphoma in CD-1 Mice. Toxicol. Pathol. 2012, 40, 651–655. [Google Scholar] [CrossRef]
- Cai, J.; Jiao, X.; Luo, W.; Chen, J.; Xu, X.; Fang, Y.; Ding, X.; Yu, X. Kidney injury molecule-1 expression predicts structural damage and outcome in histological acute tubular injury. Ren. Fail. 2019, 41, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Ushio-Fukai, M. Superoxide Dismutases: Role in Redox Signaling, Vascular Function, and Diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.E.; Pérez-Jiménez, A.; Hidalgo, M.C.; Abellán, E.; Cardenete, G. Oxidative stress and antioxidant defenses after prolonged starvation in Dentex dentex liver. Comp. Biochem. Physiol. Part. C 2004, 139, 153–161. [Google Scholar] [CrossRef]
- Hammock, B.G.; Ramírez-Duarte, W.F.; Garcia, P.A.T.; Schultz, A.A.; Avendano, L.I.; Hung, T.-C.; White, J.R.; Bong, Y.-T.; Teh, S.J. The health and condition responses of Delta Smelt to fasting: A time series experiment. PLoS ONE 2020, 15, e0239358. [Google Scholar] [CrossRef]
- Liu, X.; Shi, H.; He, Q.; Lin, F.; Wang, Q.; Xiao, S.; Dai, Y.; Zhang, Y.; Yang, H.; Zhao, H. Effect of starvation and refeeding on growth, gut microbiota and non-specific immunity in hybrid grouper (Epinephelus fuscoguttatus female x E. lanceolatus male). Fish Shellfish Immun. 2020, 97, 182–193. [Google Scholar] [CrossRef]
- Saurabh, S.; Sahoo, P.K. Lysozyme: An important defence molecule of fish innate immune system. Aquac. Res. 2008, 39, 223–239. [Google Scholar] [CrossRef]
- Rebl, A.; Goldammer, T. Under control: The innate immunity of fish from the inhibitors’ perspective. Fish. Shellfish. Immunol. 2018, 77, 328–349. [Google Scholar] [CrossRef]
- Yan, C.; Yang, Q.; Gong, Z. Transgenic expression of tgfb1a induces hepatic inflammation, fibrosis and metastasis in zebrafish. Biochem. Biophys. Res. Commun. 2019, 509, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Rajapakse, A.G.; Yepuri, G.; Xiong, Y.; Carvas, J.; Ruffieux, J.; Scerri, I.; Wu, Z.; Popp, K.; Li, J.; et al. Arginase II Promotes Macrophage Inflammatory Responses Through Mitochondrial Reactive Oxygen Species, Contributing to Insulin Resistance and Atherogenesis. J. Am. Heart Assoc. 2012, 1, e000992. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Eghan, K.; Lee, S.; Park, J.-S.; Yoon, S.; Pimtong, W.; Kim, W.-K. A Phenotypic and Genotypic Evaluation of Developmental Toxicity of Polyhexamethylene Guanidine Phosphate Using Zebrafish Embryo/Larvae. Toxics 2020, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Faggioni, R.; Moser, A.; Feingold, K.R.; Grunfeld, C. Reduced leptin levels in starvation increase susceptibility to endotoxic shock. Am. J. Pathol. 2000, 156, 1781–1787. [Google Scholar] [CrossRef]



| Experiment | Feeding Regime |
|---|---|
| body weight | group1: feeding for 35 days; group2: starving for 7 days and refeeding for 28 days; group3: starving for 14 days and refeeding for 21 days; group4: starving for 21 days and refeeding for 14 days; |
| histopathology | F21: feeding for 21 days; S21: starving for 21 days; S21R3/5/7/14: starving for days and refeeding for 3/5/7/14 days; |
| biochemical parameter assay | F21/24/28: feeding for 21/24/28 days; S21/24/28: starving for 21/24/28 days; S21R3/7: starving for days and refeeding for3/7 days; |
| ROS production | |
| immune-related gene expression | |
| Streptococcus agalactiae challenge | F24: feeding for24 days; S24: starving for 24 days; S21R3/7: starving for days and refeeding for3/7 days; |
| LPS injection | |
| respiratory burst assay |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, Z.; Lin, D.; Jia, J.; Cai, R.; Yu, Y.; Li, W. Innate Immune Response to Fasting and Refeeding in the Zebrafish Kidney. Biomolecules 2021, 11, 825. https://doi.org/10.3390/biom11060825
Liao Z, Lin D, Jia J, Cai R, Yu Y, Li W. Innate Immune Response to Fasting and Refeeding in the Zebrafish Kidney. Biomolecules. 2021; 11(6):825. https://doi.org/10.3390/biom11060825
Chicago/Turabian StyleLiao, Zongzhen, Dihang Lin, Jirong Jia, Ran Cai, Yang Yu, and Wensheng Li. 2021. "Innate Immune Response to Fasting and Refeeding in the Zebrafish Kidney" Biomolecules 11, no. 6: 825. https://doi.org/10.3390/biom11060825
APA StyleLiao, Z., Lin, D., Jia, J., Cai, R., Yu, Y., & Li, W. (2021). Innate Immune Response to Fasting and Refeeding in the Zebrafish Kidney. Biomolecules, 11(6), 825. https://doi.org/10.3390/biom11060825







