Zebrafish mafbb Mutants Display Osteoclast Over-Activation and Bone Deformity Resembling Osteolysis in MCTO Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance and Embryo Handling
2.2. Generation and Analysis of Tg(mafbb:GFP) and Tg(ctsk:mGFP)
2.3. Generation of mafbb Knockout Mutants
2.4. Synthesis of Antisense Probes and Whole-Mount In Situ Hybridization (WISH)
2.5. Neutral Red Staining and Benzidine Staining
2.6. Tartrate-Resistant Acid Phosphatase (TRAcP) Staining
2.7. MicroCT Scans
2.8. Alcian Blue Staining and Alizarin Red Staining
2.9. Whole Kidney Marrow Cell Collection and Cytology
2.10. Flow Cytometry and Cell Sorting and Counting
2.11. Gene Expression by Real-Time qPCR
2.12. Alendronate Treatment
2.13. mRNA Injection
2.14. Statistical Analysis
3. Results
3.1. Generation of Zebrafish mafbb Mutants Using the CRISPR/Cas9 System
3.2. Osteoclast Cell Development Is Enhanced in mafbb Deficient Embryos
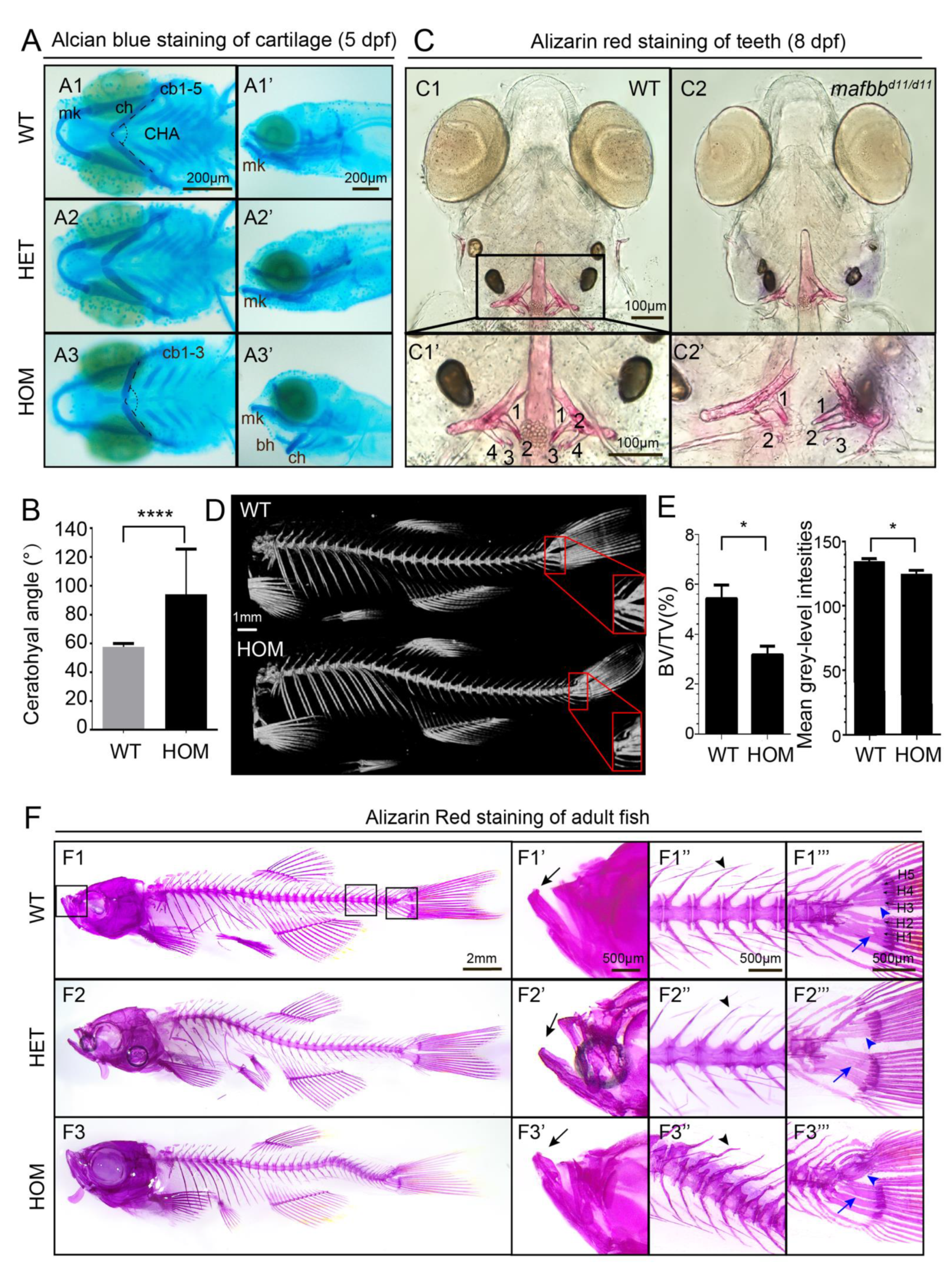
3.3. mafbb Deficiency Results in Abnormal Cartilage and Bone Development
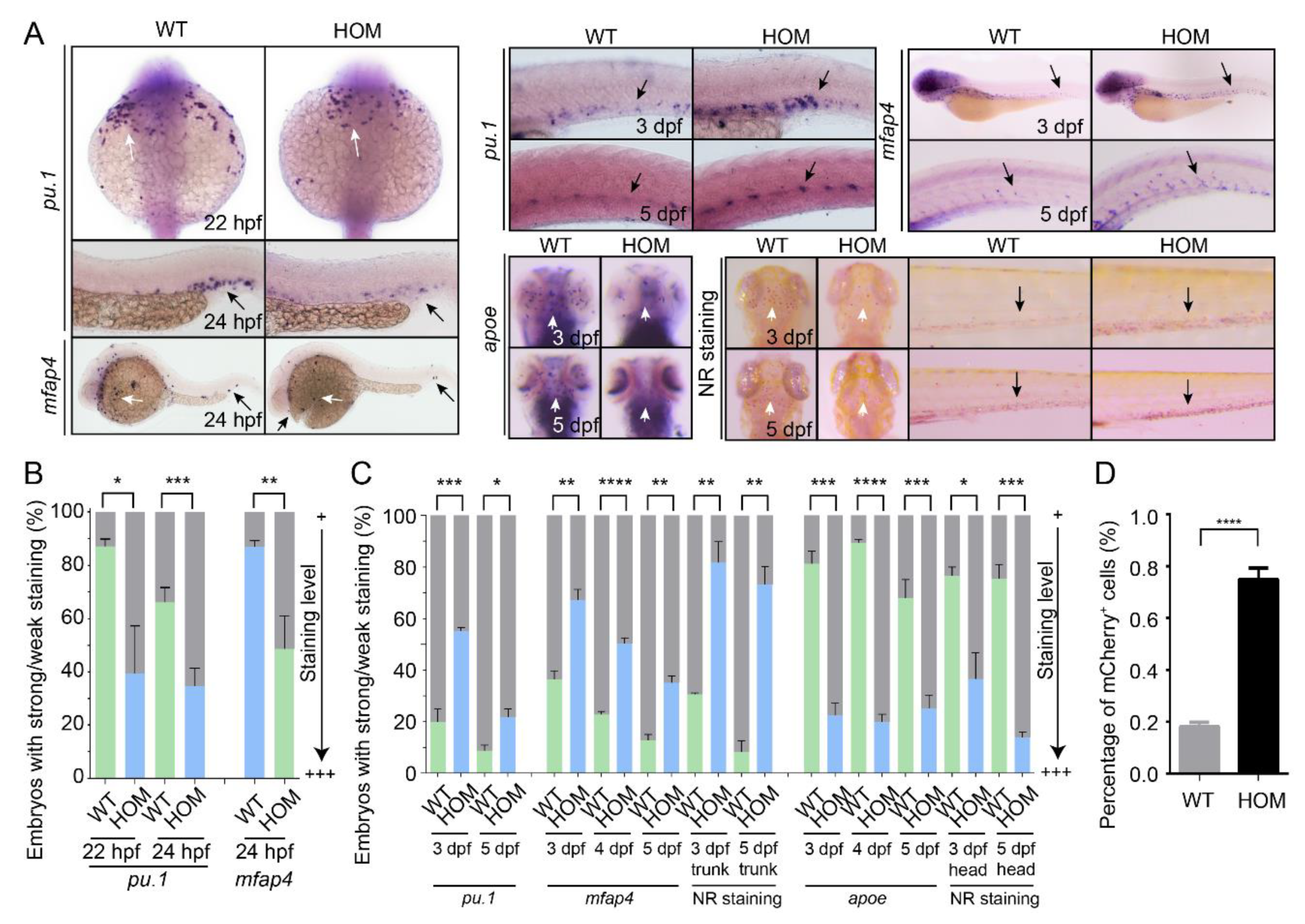
3.4. Macrophage Differentiation Is Altered in mafbb Mutants
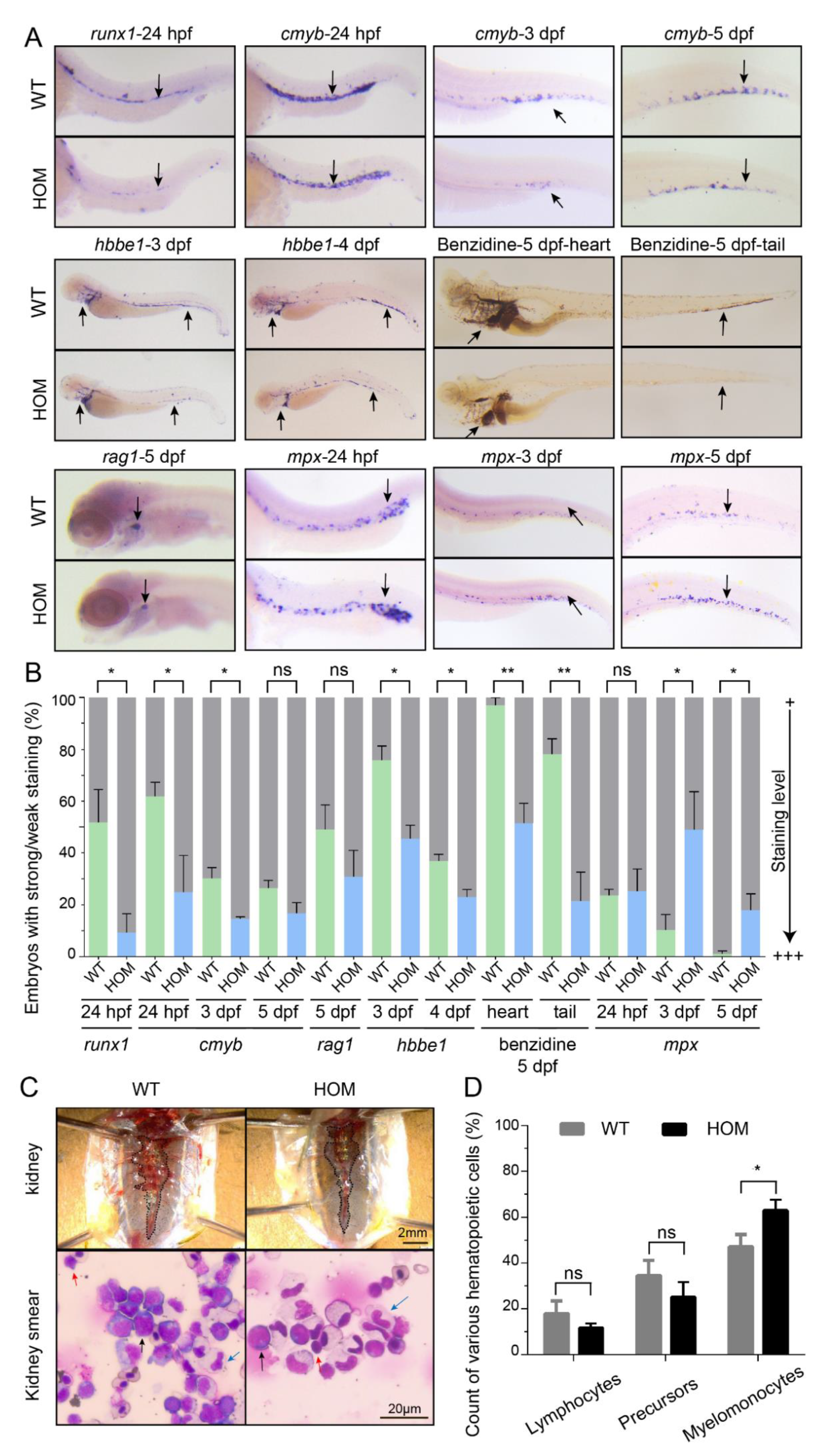
3.5. Expansion of Definitive Myelopoiesis in mafbb Mutants
3.6. MCTO Mutant MAFB Does Not Rescue Osteolysis in mafbb−/− Embryos
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer | Nucleotide Sequence 5′-3′ |
|---|---|
| DET F | GCGACGACAAACAGGCTAAT |
| DET R | GGGTGTGCATGCATGAGATT |
| Primer | Nucleotide Sequence 5′-3′ |
|---|---|
| rank F | AAAA TCTAGA TGGGACTTTGCTGCAGTAGA |
| rank R | AAAA GAATTC GCCGTGATGCTGAGATTGAG |
| ctsk F | AAAA TCTAGA CTGGCTCACTCTCTGGACAA |
| ctsk R | AAAA GAATTC AGCTCTCACATGACGGGAAA |
| Primer | Nucleotide Sequence 5′-3′ |
|---|---|
| ctsk F | ACCCAAACTGCAACAAGG |
| ctsk R | TAGCCCTTCTTTCCCCAC |
| fosab F | GGAGCAAAGACCTCCAACAA |
| fosab R | TCTTGTTTCGTTCACGACGTA |
| nfatc1 F | CCGAGAGCAACATGAGAGC |
| nfatc1 R | AGCTCGATGTCTGAGTTACGC |
| opg F | GTGAGTGTGAGGAGGGCTTC |
| opg R | TGTCACTGTACGGCGTTCC |
| acp5a F | CCATGTAGGAAACGTCAAAGC |
| acp5a R | GAATGCGGAAGTTCATCTCAT |
| ocstamp F | TCAGGTGGTCCTTGGATTTC |
| ocstamp R | AATGGGTACTTTTGTTCCAACCT |
References
- Hamada, M.; Tsunakawa, Y.; Jeon, H.; Yadav, M.K.; Takahashi, S. Role of MafB in macrophages. Exp. Anim. 2020, 69, 1–10. [Google Scholar] [CrossRef]
- Miyai, M.; Hamada, M.; Moriguchi, T.; Hiruma, J.; Kamitani-Kawamoto, A.; Watanabe, H.; Hara-Chikuma, M.; Takahashi, K.; Takahashi, S.; Kataoka, K. Transcription Factor MafB Coordinates Epidermal Keratinocyte Differentiation. J. Investig. Dermatol. 2016, 136, 1848–1857. [Google Scholar] [CrossRef]
- Michito, H. The mouse mafB 5′-upstream fragment directs gene expression in myelomonocytic cells, differentiated macrophages and the ventral spinal cord in transgenic mice. J. Biochem. 2003, 134, 203–210. [Google Scholar] [CrossRef]
- Kelly, L.M.; Englmeier, U.; Lafon, I.; Sieweke, M.H.; Graf, T. MafB is an inducer of monocytic differentiation. EMBO J. 2000, 19, 1987–1997. [Google Scholar] [CrossRef]
- Tran, M.T.N.; Hamada, M.; Jeon, H.; Shiraishi, R.; Asano, K.; Hattori, M.; Nakamura, M.; Imamura, Y.; Tsunakawa, Y.; Fujii, R.; et al. MafB is a critical regulator of complement component C1q. Nat. Commun. 2017, 8, 1700. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T. MafB is essential for renal development and F4/80 expression in macrophages. Mol. Cell Biol. 2006, 26, 5715–5727. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.H.; Lee, J.; Jin, H.M.; Kook, H.; Kim, K.K.; Lee, S.Y.; Kim, N. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 2007, 109, 3253–3259. [Google Scholar] [CrossRef]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef]
- Kylmaoja, E.; Nakamura, M.; Tuukkanen, J. Osteoclasts and Remodeling Based Bone Formation. Curr. Stem Cell Res. Ther. 2016, 11, 626–633. [Google Scholar] [CrossRef]
- Soysa, N.S.; Alles, N. Osteoclast function and bone-resorbing activity: An overview. Biochem. Biophys. Res. Commun. 2016, 476, 115–120. [Google Scholar] [CrossRef]
- Xu, F.; Teitelbaum, S.L. Osteoclasts: New Insights. Bone Res. 2013, 1, 11–26. [Google Scholar] [CrossRef]
- Anesi, A.; Generali, L.; Sandoni, L.; Pozzi, S.; Grande, A. From Osteoclast Differentiation to Osteonecrosis of the Jaw: Molecular and Clinical Insights. Int. J. Mol. Sci. 2019, 20, 4925. [Google Scholar] [CrossRef]
- Lacey, D.; Timms, E.; Tan, H.-L.; Kelley, M.; Dunstan, C.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin Ligand Is a Cytokine that Regulates Osteoclast Differentiation and Activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef]
- Mumm, S.; Huskey, M.; Duan, S.; Wenkert, D.; Madson, K.L.; Gottesman, G.S.; Nenninger, A.R.; Laxer, R.M.; McAlister, W.H.; Whyte, M.P. Multicentric carpotarsal osteolysis syndrome is caused by only a few domain-specific mutations inMAFB, a negative regulator of RANKL-induced osteoclastogenesis. Am. J. Med. Genet. Part A 2014, 164, 2287–2293. [Google Scholar] [CrossRef]
- Zankl, A.; Duncan, E.L.; Leo, P.J.; Clark, G.R.; Glazov, E.A.; Addor, M.-C.; Herlin, T.; Kim, C.A.; Leheup, B.P.; McGill, J.; et al. Multicentric Carpotarsal Osteolysis Is Caused by Mutations Clustering in the Amino-Terminal Transcriptional Activation Domain of MAFB. Am. J. Hum. Genet. 2012, 90, 494–501. [Google Scholar] [CrossRef]
- Park, P.G.; Kim, K.H.; Hyun, H.S.; Lee, C.H.; Park, J.-S.; Kie, J.H.; Choi, Y.H.; Moon, K.C.; Cheong, H.I. Three cases of multicentric carpotarsal osteolysis syndrome: A case series. BMC Med. Genet. 2018, 19, 164. [Google Scholar] [CrossRef]
- Sato, Y.; Tsukaguchi, H.; Morita, H.; Higasa, K.; Tran, M.T.N.; Hamada, M.; Usui, T.; Morito, N.; Horita, S.; Hayashi, T.; et al. A mutation in transcription factor MAFB causes Focal Segmental Glomerulosclerosis with Duane Retraction Syndrome. Kidney Int. 2018, 94, 396–407. [Google Scholar] [CrossRef]
- Choochuen, P.; Rojneuangnit, K.; Khetkham, T.; Khositseth, S. The First Report of Multicentric Carpotarsal Osteolysis Syndrome Caused by MAFB Mutation in Asian. Case Rep. Med. 2018, 2018, 6783957. [Google Scholar] [CrossRef]
- Tsunakawa, Y.; Hamada, M.; Matsunaga, Y.; Fuseya, S.; Jeon, H.; Wakimoto, Y.; Usui, T.; Kanai, M.; Mizuno, S.; Morito, N.; et al. Mice harboring an MCTO mutation exhibit renal failure resembling nephropathy in human patients. Exp. Anim. 2019, 68, 103–111. [Google Scholar] [CrossRef]
- Lleras-Forero, L.; Winkler, C.; Schulte-Merker, S. Zebrafish and medaka as models for biomedical research of bone diseases. Dev. Biol. 2020, 457, 191–205. [Google Scholar] [CrossRef]
- Bergen, D.J.M.; Kague, E.; Hammond, C.L. Zebrafish as an Emerging Model for Osteoporosis: A Primary Testing Platform for Screening New Osteo-Active Compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef]
- Witten, P.E.; Hansen, A.; Hall, B.K. Features of mono- and multinucleated bone resorbing cells of the zebrafishDanio rerio and their contribution to skeletal development, remodeling, and growth. J. Morphol. 2001, 250, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Busse, B.; Galloway, J.L.; Gray, R.S.; Harris, M.P.; Kwon, R.Y. Zebrafish: An Emerging Model for Orthopedic Research. J. Orthop. Res. 2019, 38, 925–936. [Google Scholar] [CrossRef]
- Gomez, G.A.; Veldman, M.B.; Zhao, Y.; Burgess, S.; Lin, S. Discovery and Characterization of Novel Vascular and Hematopoietic Genes Downstream of Etsrp in Zebrafish. PLoS ONE 2009, 4, e4994. [Google Scholar] [CrossRef]
- Schvarzstein, M.; Kirn, A.; Haffter, P.; Cordes, S.P. Expression of Zkrml2, a homologue of the Krml1/val segmentation gene, during embryonic patterning of the zebrafish (Danio rerio). Mech. Dev. 1999, 80, 223–226. [Google Scholar] [CrossRef]
- Kajihara, M.; Kawauchi, S.; Kobayashi, M.; Ogino, H.; Takahashi, S.; Yasuda, K. Isolation, characterization, and expression analysis of zebrafish large Mafs. J. Biochem. 2001, 129, 139–146. [Google Scholar] [CrossRef]
- Vejnar, C.E.; Moreno-Mateos, M.A.; Cifuentes, D.; Bazzini, A.A.; Giraldez, A.J. Optimized CRISPR–Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb. Protoc. 2016, 10, 1101. [Google Scholar] [CrossRef]
- Sharif, F.; De Bakker, M.A.; Richardson, M.K. Osteoclast-like Cells in Early Zebrafish Embryos. Cell J. 2014, 16, 211–224. [Google Scholar]
- Wilkinson, D.G. In situ hybridization: A practical approach. Methods Cell Biol. 1992, 35, 259–262. [Google Scholar]
- Herbomel, P.; Thisse, B.; Thisse, C. Zebrafish Early Macrophages Colonize Cephalic Mesenchyme and Developing Brain, Retina, and Epidermis through a M-CSF Receptor-Dependent Invasive Process. Dev. Biol. 2001, 238, 274–288. [Google Scholar] [CrossRef]
- Kaplow, L.S. Simplified myeloperoxidase stain using benzidine dihydrochloride. Blood 1965, 26, 215–219. [Google Scholar] [CrossRef]
- Tazaki, Y.; Sugitani, K.; Ogai, K.; Kobayashi, I.; Kawasaki, H.; Aoyama, T.; Suzuki, N.; Tabuchi, Y.; Hattori, A.; Kitamura, K. RANKL, Ephrin-Eph and Wnt10b are key intercellular communication molecules regulating bone remodeling in au-tologous transplanted goldfish scales. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2018, 225, 46–58. [Google Scholar] [CrossRef]
- Monma, Y.; Shimada, Y.; Nakayama, H.; Zang, L.; Nishimura, N.; Tanaka, T. Aging-associated microstructural deterioration of vertebra in zebrafish. Bone Rep. 2019, 11, 100215. [Google Scholar] [CrossRef]
- Charles, J.F.; Sury, M.; Tsang, K.; Urso, K.; Henke, K.; Huang, Y.; Russell, R.; Duryea, J.; Harris, M.P. Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis. Bone 2017, 101, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, J.-R.; Lai, Y.-H.; Tsai, J.-J.; Hsiao, C.-D. Live Fluorescent Staining Platform for Drug-Screening and Mechanism-Analysis in Zebrafish for Bone Mineralization. Molecules 2017, 22, 2068. [Google Scholar] [CrossRef]
- Quillien, A.; Abdalla, M.; Yu, J.; Ou, J.; Zhu, L.J.; Lawson, N.D. Robust Identification of Developmentally Active Endothelial Enhancers in Zebrafish Using FANS-Assisted ATAC-Seq. Cell Rep. 2017, 20, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [PubMed]
- To, T.T.; Witten, P.E.; Renn, J.; Bhattacharya, D.; Huysseune, A.; Winkler, C. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development 2011, 139, 141–150. [Google Scholar] [CrossRef]
- Ishii, T.; Ruiz-Torruella, M.; Ikeda, A.; Shindo, S.; Movila, A.; Mawardi, H.; Albassam, A.; Kayal, R.A.; Al-Dharrab, A.A.; Egashira, K.; et al. OC-STAMP promotes osteoclast fusion for pathogenic bone resorption in periodontitis via up-regulation of permissive fusogen CD9. FASEB J. 2018, 32, 4016–4030. [Google Scholar] [CrossRef]
- Neuhauss, S.C.; Solnica-Krezel, L.; Schier, A.F.; Zwartkruis, F.; Stemple, D.L.; Malicki, J.; Abdelilah, S.; Stainier, D.Y.; Driever, W. Mutations affecting craniofacial development in zebrafish. Development 1996, 123, 357–367. [Google Scholar]
- Koch, B.E.; Spaink, H.P.; Meijer, A.H. A quantitative in vivo assay for craniofacial developmental toxicity of histone deacetylases. Toxicol. Lett. 2021, 342, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Nich, C.; Langlois, J.; Marchadier, A.; Vidal, C.; Cohen-Solal, M.; Petite, H.; Hamadouche, M. Oestrogen deficiency modulates particle-induced osteolysis. Arthritis Res. 2011, 13, R100. [Google Scholar] [CrossRef] [PubMed]
- Izawa, N.; Kurotaki, D.; Nomura, S.; Fujita, T.; Omata, Y.; Yasui, T.; Hirose, J.; Matsumoto, T.; Saito, T.; Kadono, Y.; et al. Cooperation of PU.1 With IRF8 and NFATc1 Defines Chromatin Landscapes During RANKL-Induced Osteoclasto-genesis. J. Bone Miner. Res. 2019, 34, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhu, L.; He, S.; Wu, Y.; Jin, W.; Yu, T.; Qu, J.Y.; Wen, Z. Temporal-Spatial Resolution Fate Mapping Reveals Distinct Origins for Embryonic and Adult Microglia in Zebrafish. Dev. Cell 2015, 34, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Sieweke, M.H.; Tekotte, H.; Frampton, J.; Graf, T. MafB Is an Interaction Partner and Repressor of Ets-1 That Inhibits Erythroid Differentiation. Cell 1996, 85, 49–60. [Google Scholar] [CrossRef]
- Gomes, A.; Weiser, P.; Descartes, M.; Upadia, J. A Familial Case of Multicentric Carpotarsal Osteolysis Syndrome and Treatment Outcome. J. Pediatr. Genet. 2018, 7, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.E.; Rogers, M.J.; Halasy, J.M.; Luckman, S.P.; Hughes, D.E.; Masarachia, P.J.; Wesolowski, G.; Russell, R.G.G.; Rodan, G.A.; Reszka, A.A. Alendronate mechanism of action: Geranylgeraniol, an intermediate in the mevalonate pathway, prevents inhibition of osteoclast formation, bone resorption, and kinase activation in vitro. Proc. Natl. Acad. Sci. USA 1999, 96, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, V.D.; Anta, L.; Samaniego, R.; Orta-Zavalza, E.; De La Rosa, J.V.; Baujat, G.; Domínguez-Soto, Á.; Sánchez-Mateos, P.; Escribese, M.M.; Castrillo, A.; et al. MAFB Determines Human Macrophage Anti-Inflammatory Polarization: Relevance for the Pathogenic Mechanisms Operating in Multicentric Carpotarsal Osteolysis. J. Immunol. 2017, 198, 2070–2081. [Google Scholar] [CrossRef]
- Aziz, A.; Soucie, E.; Sarrazin, S.; Sieweke, M.H. MafB/c-Maf Deficiency Enables Self-Renewal of Differentiated Functional Macrophages. Science 2009, 326, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Mossadegh-Keller, N.; Fukao, T.; Aziz, A.; Mourcin, F.; Vanhille, L.; Modis, L.K.; Kastner, P.; Chan, S.; Duprez, E.; et al. MafB Restricts M-CSF-Dependent Myeloid Commitment Divisions of Hematopoietic Stem Cells. Cell 2009, 138, 300–313. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Shao, W.; Zhong, D.; Ma, C.; Wei, X.; Ahmed, A.; Yu, T.; Jing, W.; Jing, L. Zebrafish mafbb Mutants Display Osteoclast Over-Activation and Bone Deformity Resembling Osteolysis in MCTO Patients. Biomolecules 2021, 11, 480. https://doi.org/10.3390/biom11030480
Han Y, Shao W, Zhong D, Ma C, Wei X, Ahmed A, Yu T, Jing W, Jing L. Zebrafish mafbb Mutants Display Osteoclast Over-Activation and Bone Deformity Resembling Osteolysis in MCTO Patients. Biomolecules. 2021; 11(3):480. https://doi.org/10.3390/biom11030480
Chicago/Turabian StyleHan, Yujie, Weihao Shao, Dan Zhong, Cui Ma, Xiaona Wei, Abrar Ahmed, Tingting Yu, Wei Jing, and Lili Jing. 2021. "Zebrafish mafbb Mutants Display Osteoclast Over-Activation and Bone Deformity Resembling Osteolysis in MCTO Patients" Biomolecules 11, no. 3: 480. https://doi.org/10.3390/biom11030480
APA StyleHan, Y., Shao, W., Zhong, D., Ma, C., Wei, X., Ahmed, A., Yu, T., Jing, W., & Jing, L. (2021). Zebrafish mafbb Mutants Display Osteoclast Over-Activation and Bone Deformity Resembling Osteolysis in MCTO Patients. Biomolecules, 11(3), 480. https://doi.org/10.3390/biom11030480






