Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Horticultural and Flowering Characteristics Assessmenet from Different Sources
2.3. RNA-Seq Library Preparation and Sequencing
2.4. Analysis of DEGs
2.5. GO Biological Process Enrichment for Flowering
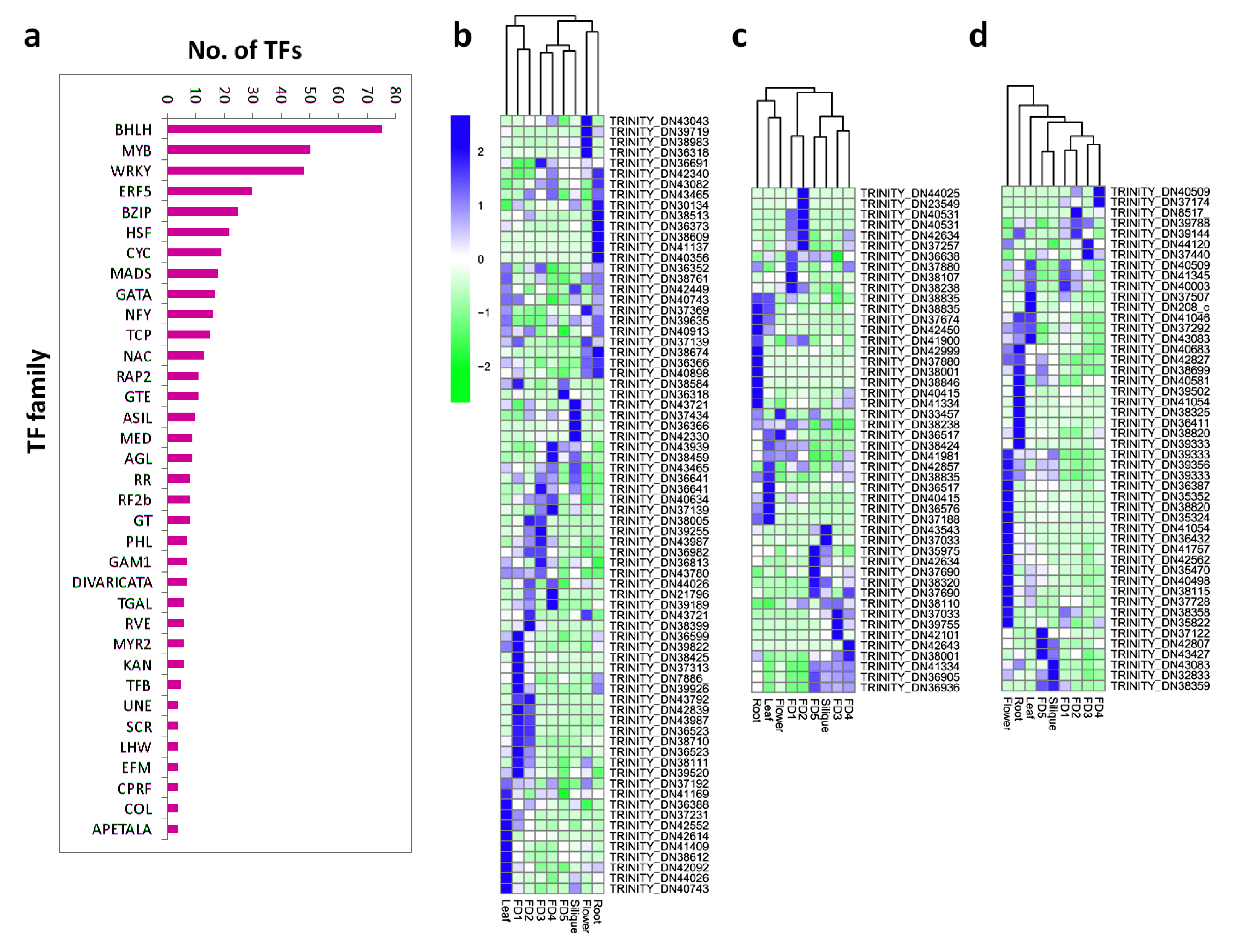
2.6. Filtering of TF Families
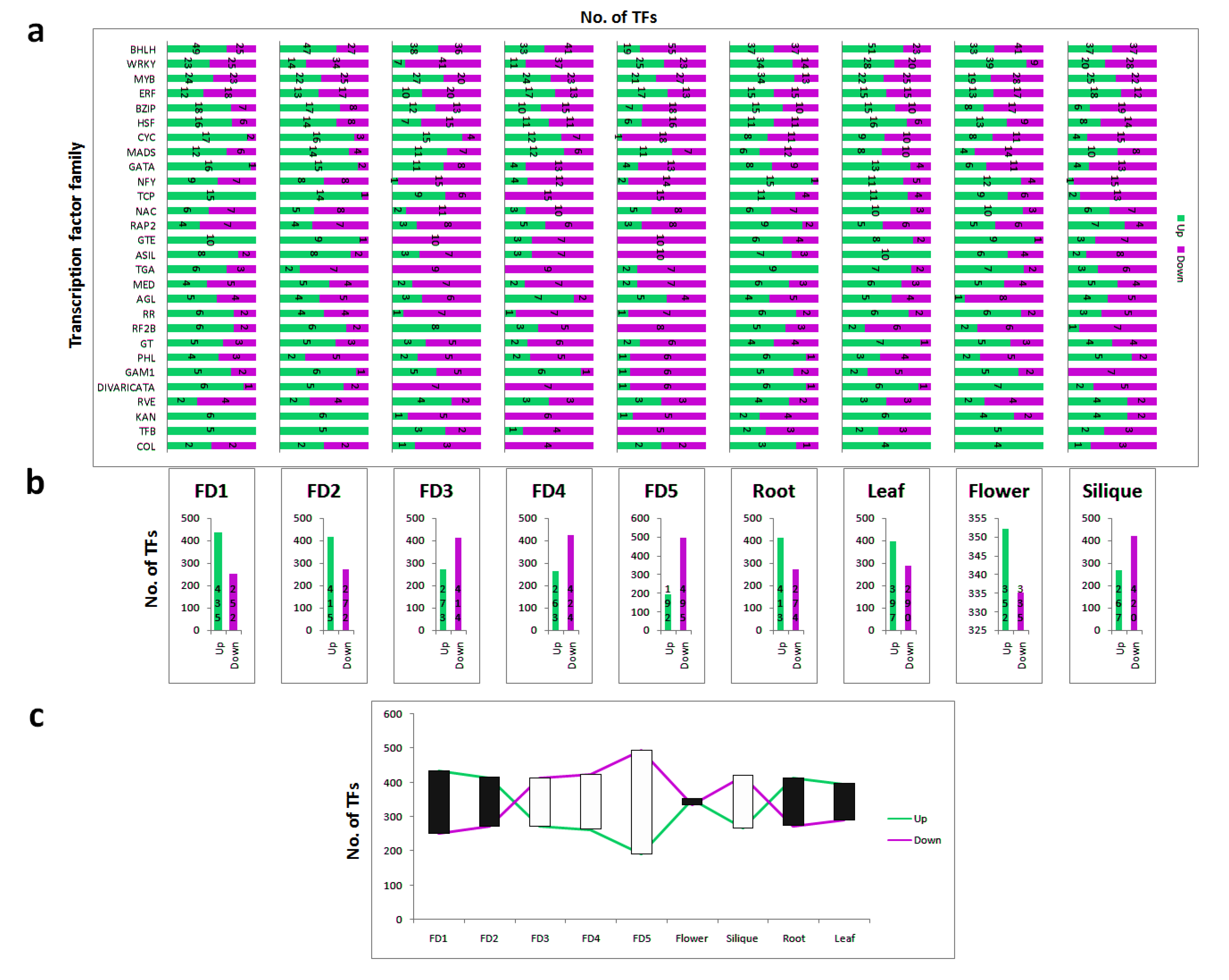
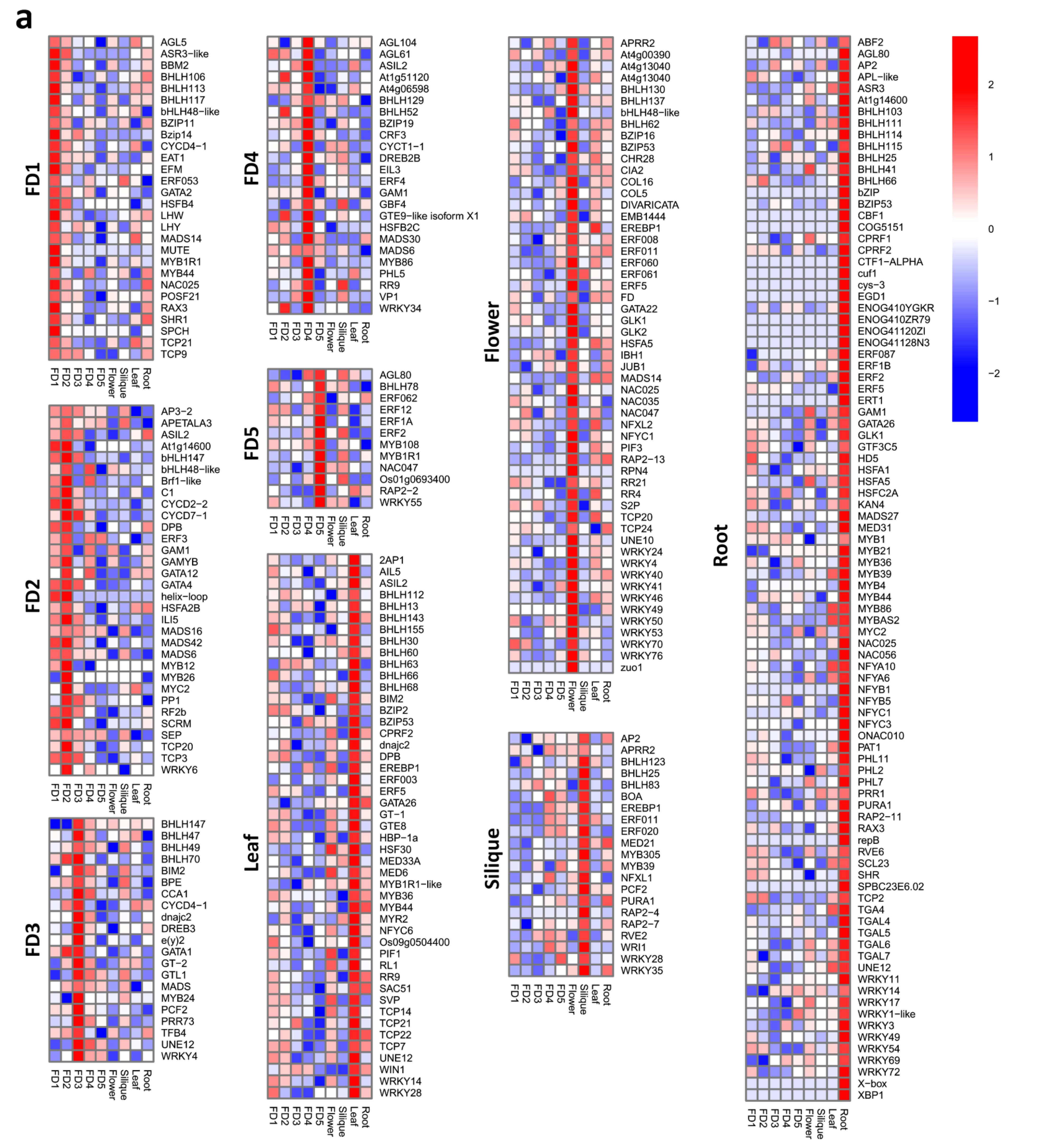
2.7. Identification of Significantly Differential and Stage Specific Tfs
2.8. Quantitative Real-Time PCR Analysis
2.9. Statistical Analysis
3. Results
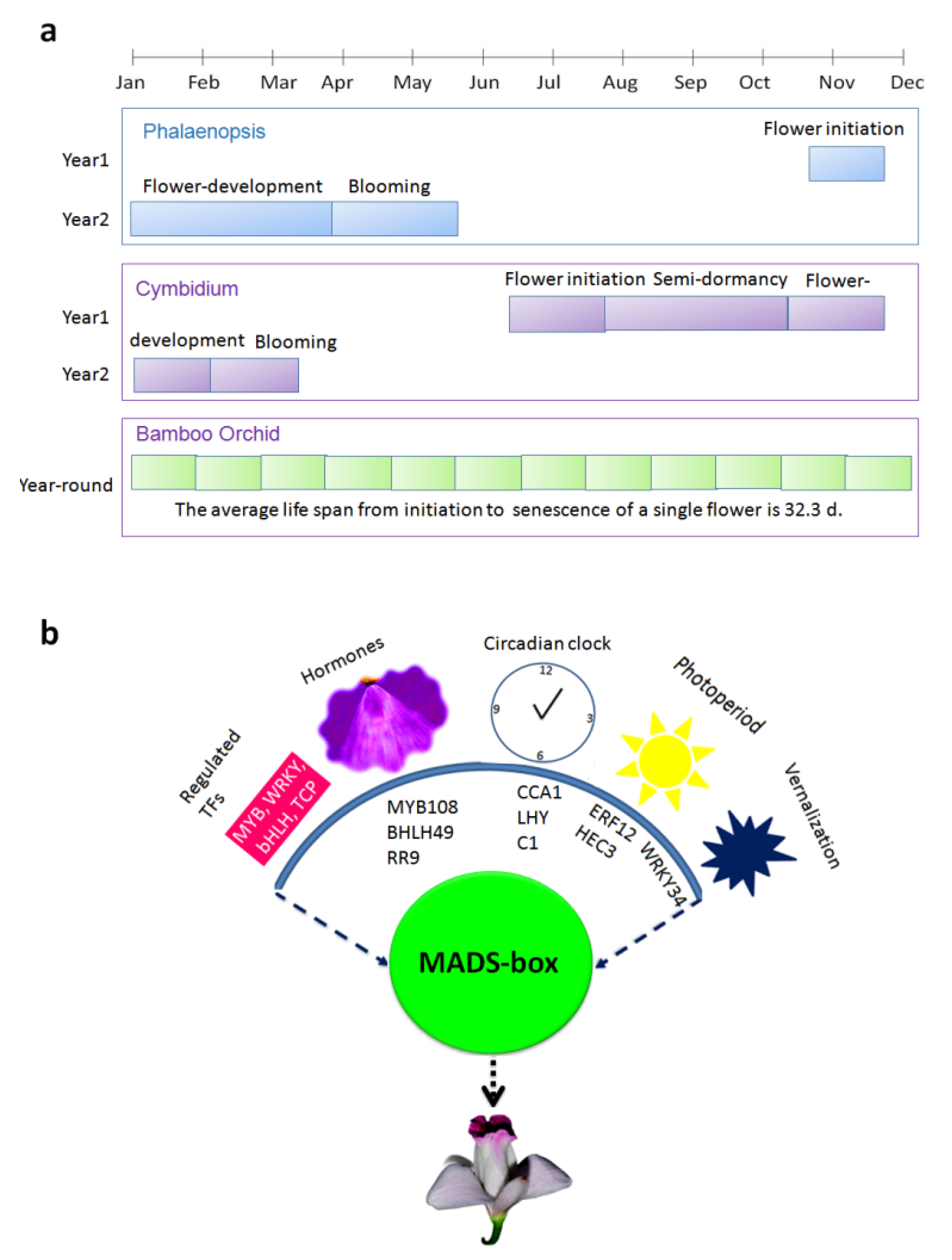
3.1. Flowering Habit of A. graminifolia
3.2. RNA-Seq and Functional Annotation
3.3. Biological Process Enrichment of Hormonal and Flowering Related DEGs
3.4. Filtering of Transcription Factor (TF) Families
3.5. Tissue-Specific Up and Down Regulation of TFs
3.6. Top 20 Highly Differential TFs
3.7. Stage-Specific Highly Upregulated TFs
3.8. The qRT-PCR Validation of Some Selected TFs in Flower Regulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bäurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef]
- Zhou, A.; Sun, H.; Dai, S.; Feng, S.; Zhang, J.; Gong, S.; Wang, J. Identification of Transcription Factors Involved in the Regulation of Flowering in Adonis Amurensis Through Combined RNA-seq Transcriptomics and iTRAQ Proteomics. Genes 2019, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550–550.e2. [Google Scholar] [CrossRef]
- Ausin, I.; Alonso-Blanco, C.; Martinez-Zapater, J.-M. Environmental regulation of flowering. Int. J. Dev. Biol. 2004, 49, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Mutasa-Göttgens, E.; Hedden, P. Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 2009, 60, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 2004, 7, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, K.; Zeng, S.; Teixeira da Silva, J.A.; Zhao, X.; Tian, C.E.; Xia, H.; Duan, J. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genom. 2013, 14, 279. [Google Scholar] [CrossRef]
- Komeda, Y. Genetic regulation of time to flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 521–535. [Google Scholar] [CrossRef]
- Imaizumi, T.; Kay, S.A. Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 2006, 11, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Ito, S.; Nakamichi, N.; Mizoguchi, T.; Niinuma, K.; Yamashino, T.; Mizuno, T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.; Dean, C. Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J. 2015, 83, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.C.; Robertson, M.; Tanner, G.; Peacock, W.J.; Dennis, E.S.; Helliwell, C.A. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. USA 2006, 103, 14631–14636. [Google Scholar] [CrossRef]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef]
- Wang, J.-W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef]
- Wang, M.; Hoekstra, S.; van Bergen, S.; Lamers, G.E.; Oppedijk, B.J.; van der Heijden, M.W.; de Priester, W.; Schilperoort, R.A. Apoptosis in developing anthers and the role of ABA in this process during androgenesis in Hordeum vulgare L. Plant Mol. Biol. 1999, 39, 489–501. [Google Scholar] [CrossRef]
- Yu, H.; Ito, T.; Zhao, Y.; Peng, J.; Kumar, P.; Meyerowitz, E.M. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 2004, 101, 7827–7832. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of auxin in regulating Arabidopsis flower development. Planta 2006, 223, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.L.; Sawhney, V.K.; Bonham-Smith, P.C. Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. 2006, 170, 1111–1117. [Google Scholar] [CrossRef]
- McConn, M. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Rieu, I.; Wolters-Arts, M.; Derksen, J.; Mariani, C.; Weterings, K. Ethylene regulates the timing of anther dehiscence in tobacco. Planta 2003, 217, 131–137. [Google Scholar] [CrossRef]
- Auberon, F.; Olatunji, O.J.; Krisa, S.; Antheaume, C.; Herbette, G.; Bonté, F.; Mérillon, J.-M.; Lobstein, A. Two new stilbenoids from the aerial parts of Arundina graminifolia (Orchidaceae). Molecules 2016, 21, 1430. [Google Scholar] [CrossRef]
- Hooker, J. Gnetaceae. Flora Br. India 1890, 5, 640–643. [Google Scholar]
- Seidenfaden, G.; Wood, J.J.; Holttum, R.E. The Orchids of Peninsular Malaysia and Singapore; Olsen & Olsen: Fredensborg, Denamrk, 1992. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Ahmad, S.; Yuan, C.; Yang, Q.; Yang, Y.; Cheng, T.; Wang, J.; Pan, H.; Zhang, Q. Morpho-physiological integrators, transcriptome and coexpression network analyses signify the novel molecular signatures associated with axillary bud in chrysanthemum. BMC Plant Biol. 2020, 20, 145. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, X.; Wang, X.; Zeng, J.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zhou, Y. RNA-Seq and iTRAQ reveal the dwarfing mechanism of dwarf polish wheat (Triticum polonicum L.). Int. J. Biol. Sci. 2016, 12, 653. [Google Scholar] [CrossRef][Green Version]
- Ye, X.; Wang, H.; Chen, P.; Fu, B.; Zhang, M.; Li, J.; Zheng, X.; Tan, B.; Feng, J. Combination of iTRAQ proteomics and RNA-seq transcriptomics reveals multiple levels of regulation in phytoplasma-infected Ziziphus jujuba Mill. Hortic. Res. 2017, 4, 17080. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Abbas, H.M.K.; Huang, H.-X.; Wang, A.-J.; Wu, T.-Q.; Xue, S.-D.; Ahmad, A.; Xie, D.-S.; Li, J.-X.; Zhong, Y.-J. Metabolic and transcriptomic analysis of two Cucurbita moschata germplasms throughout fruit development. BMC Genomics 2020, 21, 365. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xin, R.; Bu, Q.; Shen, H.; Dang, J.; Huq, E. A negative feedback loop between PHYTOCHROME INTERACTING FACTORs and HECATE proteins fine-tunes photomorphogenesis in Arabidopsis. Plant Cell 2016, 28, 855–874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.-O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.-p. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef]
- Sarnowska, E.A.; Rolicka, A.T.; Bucior, E.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kamiya, Y.; Franzen, R.; Schmelzer, E. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 2013, 163, 305–317. [Google Scholar] [CrossRef]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP 2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Carbonero, P.; Iglesias-Fernández, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J. Exp. Bot. 2017, 68, 871–880. [Google Scholar] [CrossRef]
- Suzuki, M.; Kao, C.Y.; Cocciolone, S.; McCarty, D.R. Maize VP1 complements Arabidopsisabi3 and confers a novel ABA/auxin interaction in roots. Plant J. 2001, 28, 409–418. [Google Scholar] [CrossRef]
- Yan, Y.; Shen, L.; Chen, Y.; Bao, S.; Thong, Z.; Yu, H. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 2014, 30, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Z.; Wang, L.; Kim, S.G.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.; Park, C.M. WRKY 71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Li, X.; Liang, G.; Yu, D. Two DELLA-interacting proteins bHLH48 and bHLH60 regulate flowering under long-day conditions in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2757–2767. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.C.; Bracko, O.; Park, M.S.; Schwab, R.; Chun, H.J.; Park, K.M.; Seo, J.S.; Grbic, V.; Balasubramanian, S.; Schmid, M. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 2010, 62, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef]
- Kubota, A.; Ito, S.; Shim, J.S.; Johnson, R.S.; Song, Y.H.; Breton, G.; Goralogia, G.S.; Kwon, M.S.; Cintrón, D.L.; Koyama, T. TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 2017, 13, e1006856. [Google Scholar] [CrossRef]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef]
- Mandaokar, A.; Browse, J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009, 149, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Liu, H.; Tsuda, K.; Fukai, E.; Tanaka, K.; Sasaki, T.; Nonomura, K.-I. EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet. 2018, 14, e1007238. [Google Scholar] [CrossRef] [PubMed]
- Radoeva, T.; Lokerse, A.S.; Llavata-Peris, C.I.; Wendrich, J.R.; Xiang, D.; Liao, C.-Y.; Vlaar, L.; Boekschoten, M.; Hooiveld, G.; Datla, R. A robust auxin response network controls embryo and suspensor development through a basic helix loop helix transcriptional module. Plant Cell 2019, 31, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Worthen, J.M.; Yamburenko, M.V.; Lim, J.; Nimchuk, Z.L.; Kieber, J.J.; Schaller, G.E. Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 2019, 146, dev174870. [Google Scholar] [CrossRef]
- Hu, X.; Kong, X.; Wang, C.; Ma, L.; Zhao, J.; Wei, J.; Zhang, X.; Loake, G.J.; Zhang, T.; Huang, J. Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 2014, 26, 4763–4781. [Google Scholar] [CrossRef]
- Chandler, J.; Werr, W. A phylogenetically conserved APETALA2/ethylene response factor, ERF12, regulates arabidopsis floral development. Plant Mol. Biol. 2020, 102, 39–54. [Google Scholar] [CrossRef]
- Yant, L.; Mathieu, J.; Dinh, T.T.; Ott, F.; Lanz, C.; Wollmann, H.; Chen, X.; Schmid, M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 2010, 22, 2156–2170. [Google Scholar] [CrossRef]
- Horvath, D.P.; Chao, W.S.; Suttle, J.C.; Thimmapuram, J.; Anderson, J.V. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genom. 2008, 9, 536. [Google Scholar] [CrossRef]
- Bai, S. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013, 54, 1132–1151. [Google Scholar] [CrossRef]
- Rinne, P.L. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 2011, 23, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Miskolczi, P.; Maurya, J.P.; Bhalerao, R.P. A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 2019, 29, 128–133.e122. [Google Scholar] [CrossRef]
- Yang, F.; Zhu, G.; Wei, Y.; Gao, J.; Liang, G.; Peng, L.; Lu, C.; Jin, J. Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genom. 2019, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Oda, A.; Yoshida, R.; Niinuma, K.; Miyata, K.; Tomozoe, Y.; Tajima, T.; Nakagawa, M.; Hayashi, K.; Coupland, G. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 2008, 20, 2960–2971. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Shen, L.; Liu, C.; Liu, L.; Yan, Y.; Yu, H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012, 70, 549–561. [Google Scholar] [CrossRef]
- Singh, R.K.; Maurya, J.P.; Azeez, A.; Miskolczi, P.; Tylewicz, S.; Stojkovič, K.; Delhomme, N.; Busov, V.; Bhalerao, R.P. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 2018, 9, 4173. [Google Scholar] [CrossRef]




| Origin | Plant Height (cm) | Number of Blades (cm) | Leaf Length (cm) | Leaf Width (cm) | Stem Diameter (cm) | Flowering Period | Silique |
|---|---|---|---|---|---|---|---|
| Guangdong, China | 36.43 ± 1.53 | 18.74 ± 0.75 | 10.56 ± 0.48 | 0.81 ± 0.03 | 0.32 ± 0.02 | Throughout the year | Purple |
| Hainan, China | 31.09 ± 1.42 | 18.33 ± 0.84 | 11.59 ± 0.53 | 0.82 ± 0.03 | 0.31 ± 0.01 | Throughout the year | Green |
| Singapore | 80.34 ± 1.89 | 31.03 ± 0.97 | 12.30 ± 0.64 | 0.85 ± 0.04 | 0.42 ± 0.02 | Throughout the year | Green |
| Malaysia | 142.16 ± 3.32 | 34.17 ± 0.94 | 13.41 ± 0.49 | 0.79 ± 0.03 | 0.49 ± 0.01 | Throughout the year | Green |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, S.; Lu, C.; Wu, J.; Wei, Y.; Gao, J.; Jin, J.; Zheng, C.; Zhu, G.; Yang, F. Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia. Biomolecules 2021, 11, 771. https://doi.org/10.3390/biom11060771
Ahmad S, Lu C, Wu J, Wei Y, Gao J, Jin J, Zheng C, Zhu G, Yang F. Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia. Biomolecules. 2021; 11(6):771. https://doi.org/10.3390/biom11060771
Chicago/Turabian StyleAhmad, Sagheer, Chuqiao Lu, Jieqiu Wu, Yonglu Wei, Jie Gao, Jianpeng Jin, Chuanyuan Zheng, Genfa Zhu, and Fengxi Yang. 2021. "Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia" Biomolecules 11, no. 6: 771. https://doi.org/10.3390/biom11060771
APA StyleAhmad, S., Lu, C., Wu, J., Wei, Y., Gao, J., Jin, J., Zheng, C., Zhu, G., & Yang, F. (2021). Transcriptional Cascade in the Regulation of Flowering in the Bamboo Orchid Arundina graminifolia. Biomolecules, 11(6), 771. https://doi.org/10.3390/biom11060771









