Regulatory Action of Plasma from Patients with Obesity and Diabetes towards Muscle Cells Differentiation and Bioenergetics Revealed by the C2C12 Cell Model and MicroRNA Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
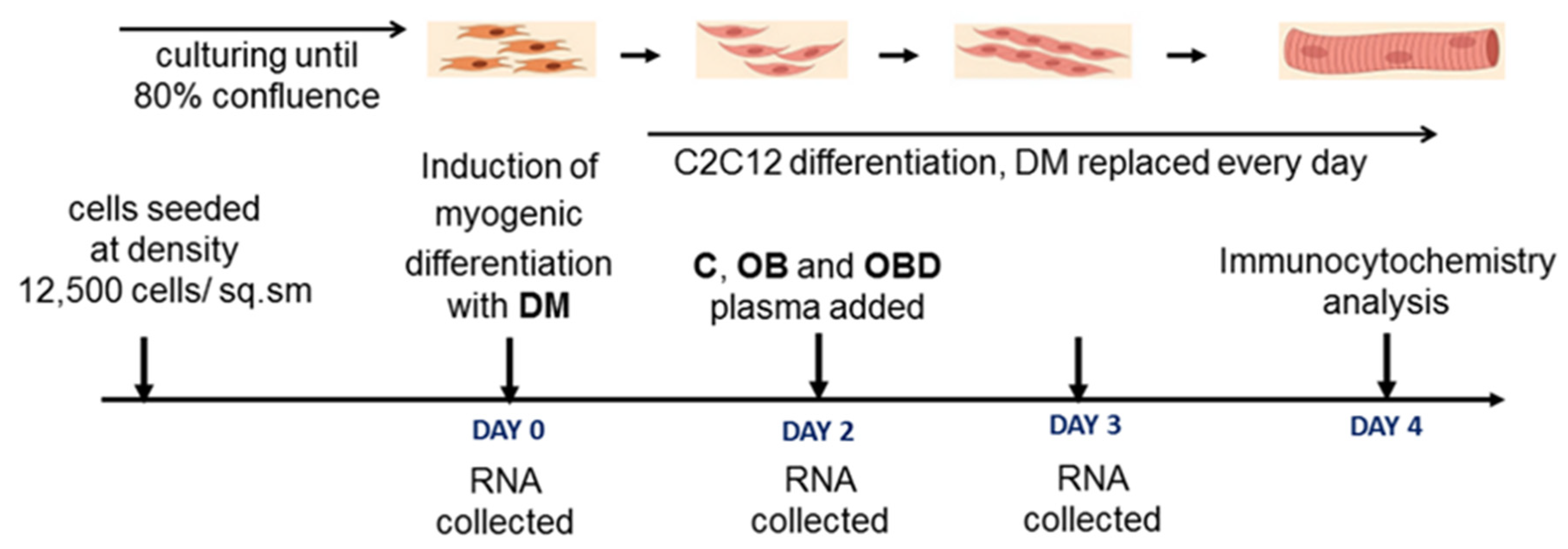
2.2. Cell Culture and Myogenic Differentiation
2.3. Plasma Samples Preparation
2.4. C2C12 Treatment with Plasma
2.5. Immunocytochemistry
2.6. RNA Isolation and Quantification of mRNA of Protein-Coding Genes
2.7. MicroRNA Quantification
2.8. Bioinformatic Analysis and Statististics
3. Results
3.1. Description of Patients, Baseline Characteristics
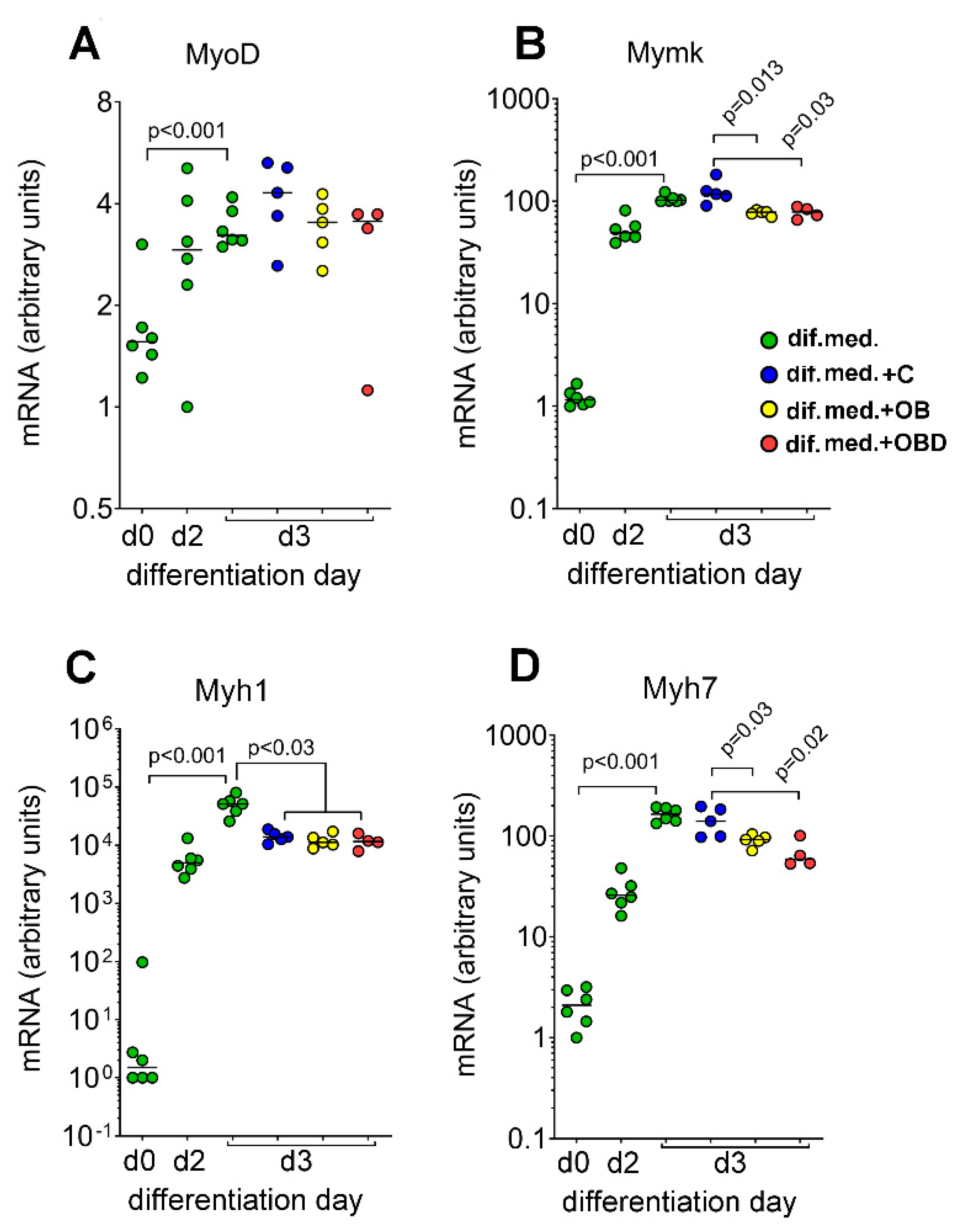
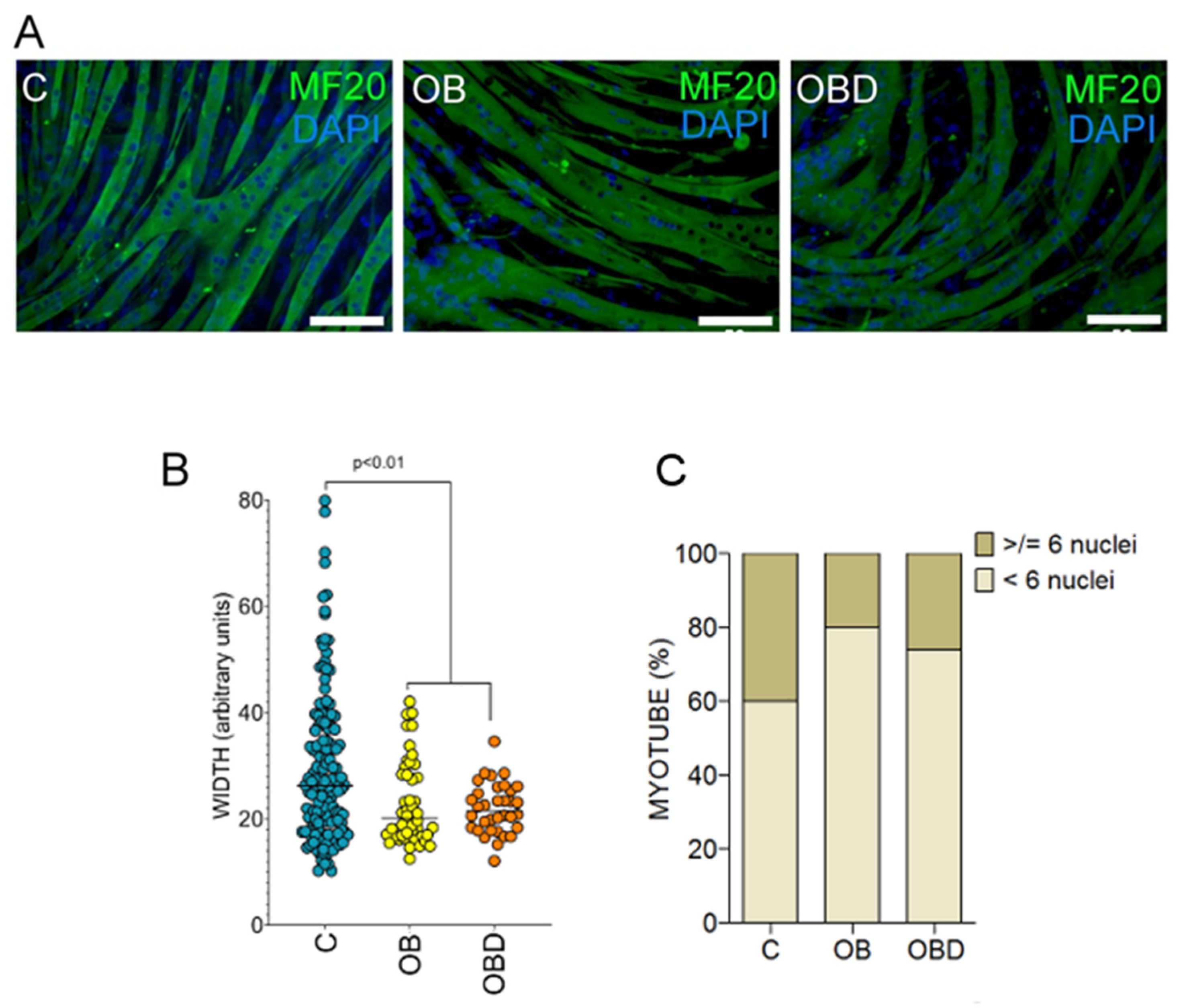
3.2. Plasma from Obese Patients (OB) and Patients with Type 2 Diabetes Comorbid with Obesity (OBD) Affects the Expression of Mymk and the Composition of Myofiber Types during Skeletal Muscle Differentiation
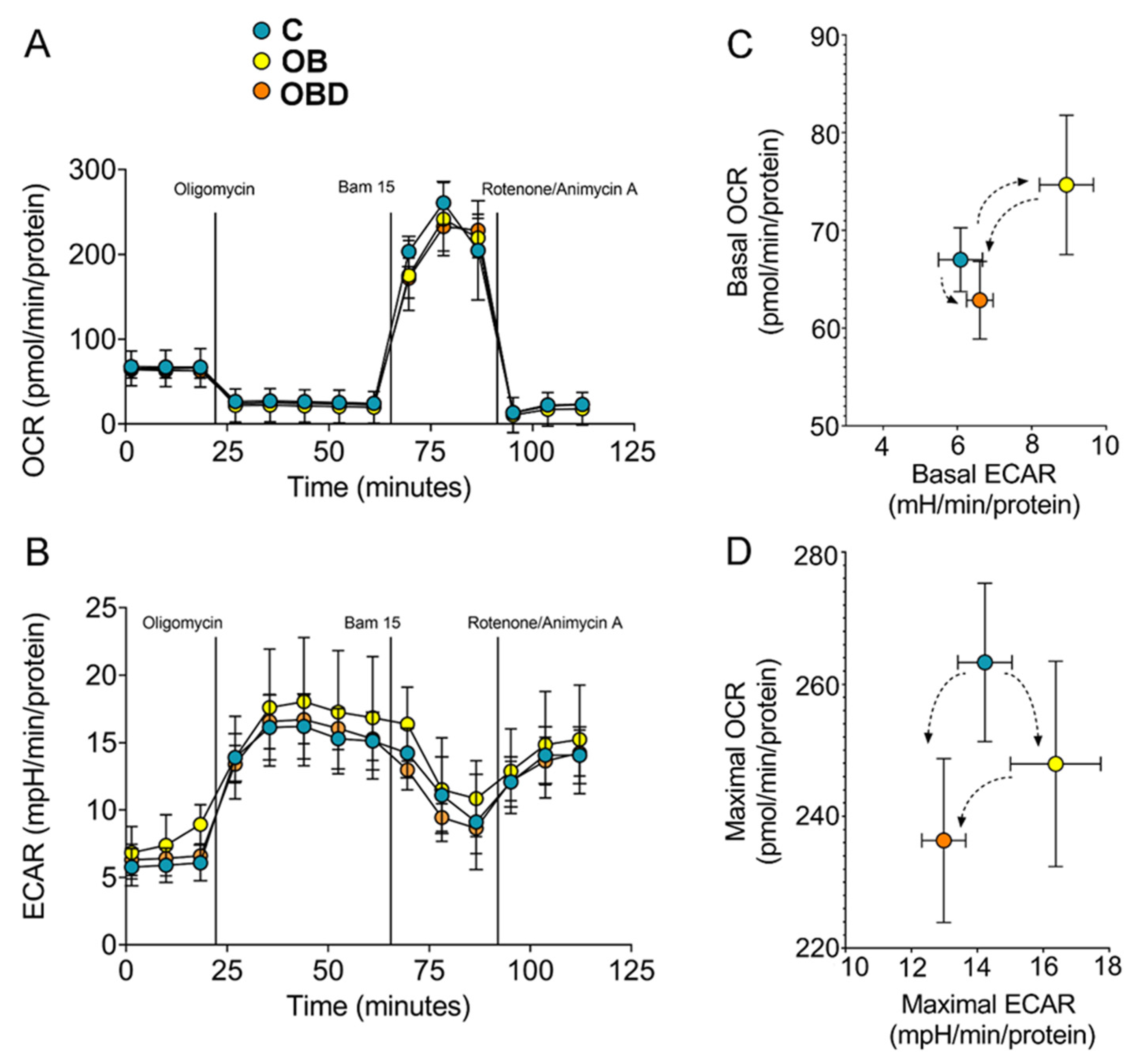
3.3. Plasma from Obese Patients (OB) and from OB Patients Comorbid with Type 2 Diabetes (OBD) Affects C2C12 Myotubes Bioenergetics
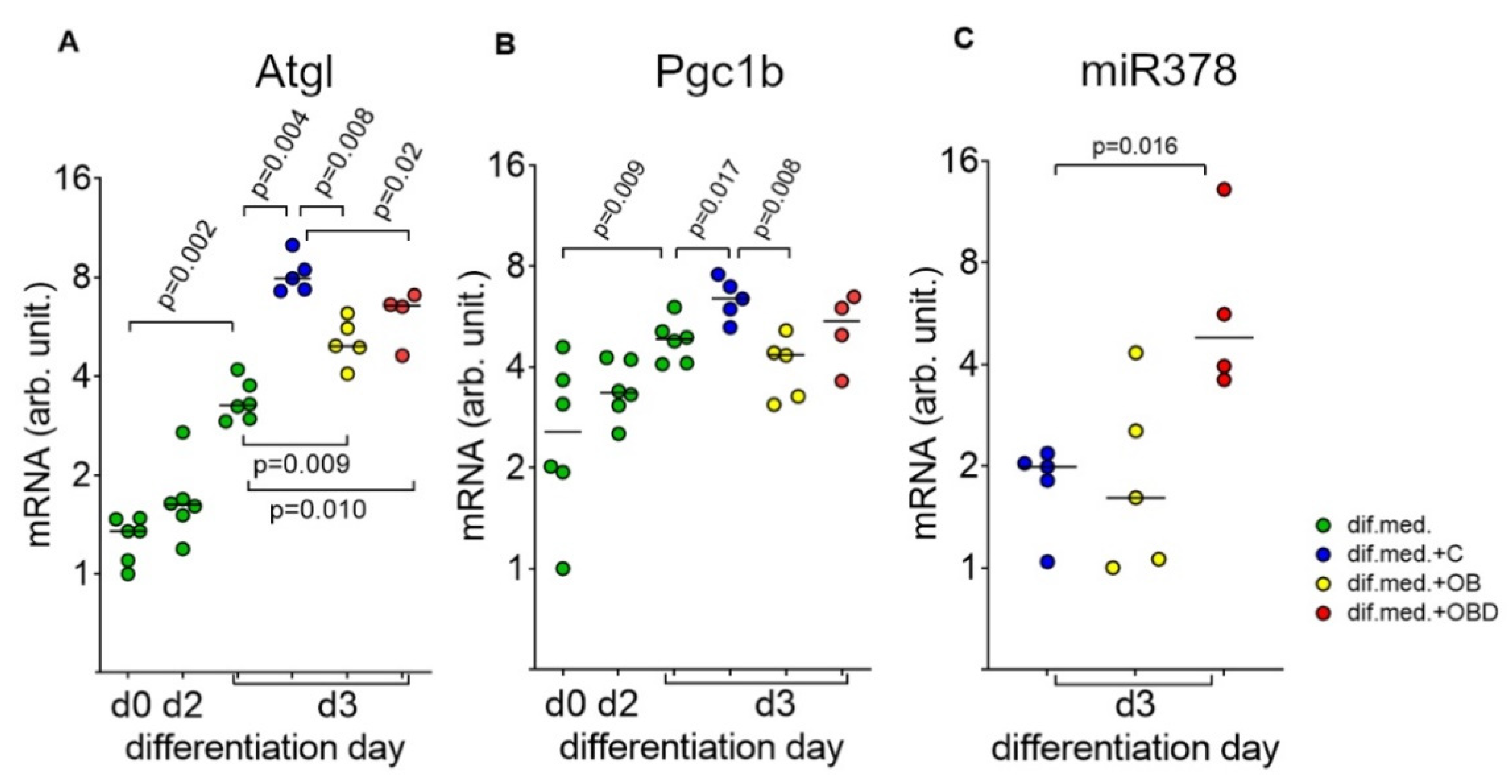
3.4. Plasma from Obese Patients (OB) and Patients with Type 2 Diabetes Comorbid with Obesity (OBD) Affects the Expression of the Mitochondria Regulators Atgl/Pgc1b/miR378-a-3p
3.5. Analysis of Existing Databases of mir-378a-3p Potential Targets to Predict Downstream Cellular Processes
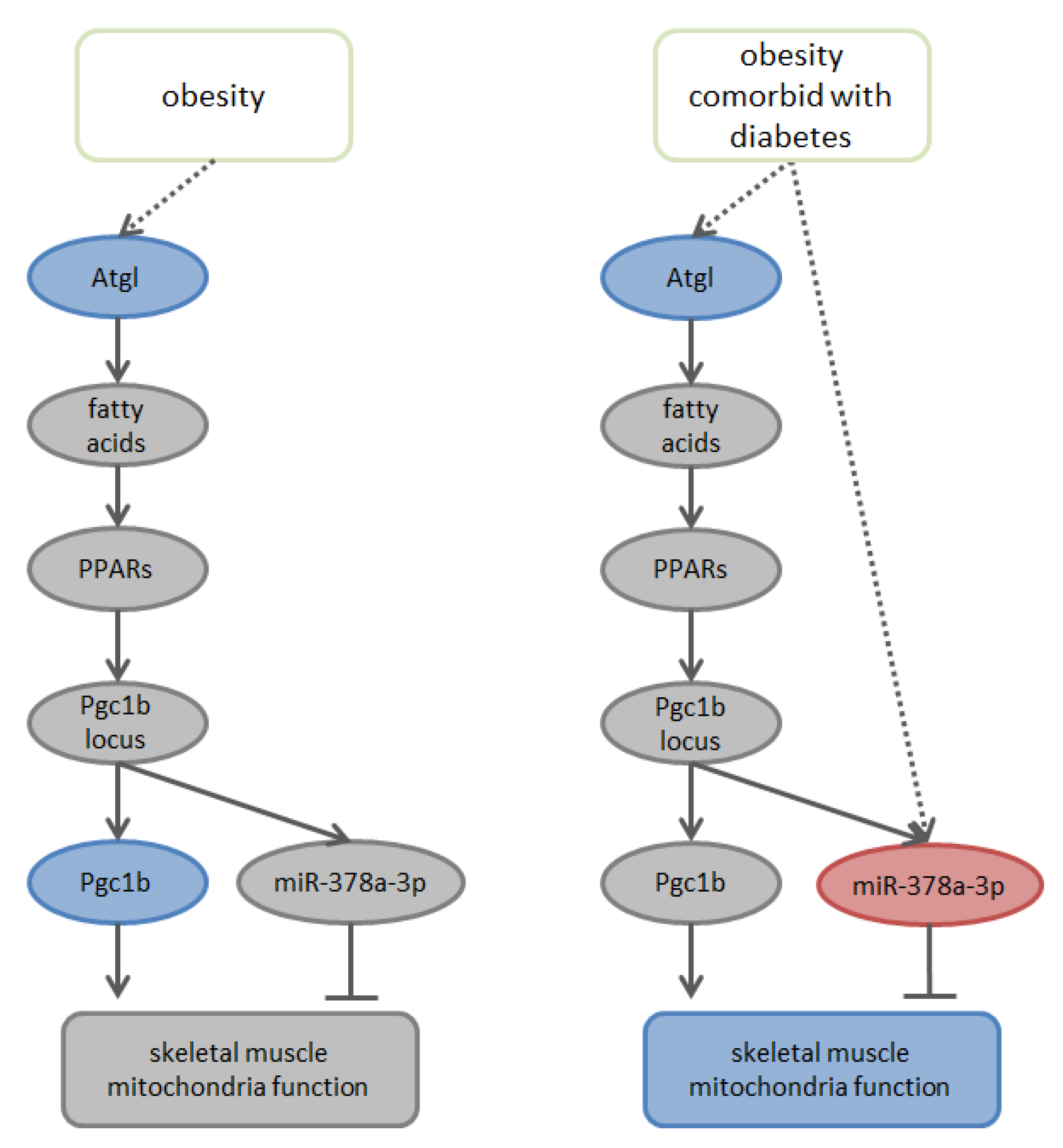
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arroyo-Johnson, C.; Mincey, K.D. Obesity Epidemiology Worldwide. Gastroenterol. Clin. N. Am. 2016, 45, 571–579. [Google Scholar] [CrossRef]
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Lehr, S.; Hartwig, S.; Sell, H. Adipokines: A treasure trove for the discovery of biomarkers for metabolic disorders. Proteom. Clin. Appl. 2012, 6, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Fasshauer, M.; Blüher, M. Adipokines in health and disease. Trends Pharmacol. Sci. 2015, 36, 461–470. [Google Scholar] [CrossRef]
- Jallut, D.; Golay, A.; Munger, R.; Frascarolo, P.; Schutz, Y.; Jéquier, E.; Felber, J.P. Impaired glucose tolerance and diabetes in obesity: A 6-year follow-up study of glucose metabolism. Metabolism 1990, 39, 1068–1075. [Google Scholar] [CrossRef]
- Teasdale, N.; Martin, S.; Philippe, C.; Grant, H.; Angelo, T.; Olivier, H. Obesity alters balance and movement control. Curr. Obes. Rep. 2013, 2, 235–240. [Google Scholar] [CrossRef]
- Prado, C.; Lieffers, J.; McCargar, L.; Reiman, T.; Sawyer, M.; Martin, L.; Baracos, V. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: A population-based study. Lancet 2008, 9, 629–635. [Google Scholar] [CrossRef]
- Tallis, J.; James, R.; Seebacher, F. The effects of obesity on skeletal muscle contractile function. J. Exp. Biol. 2018, 221, jeb163840. [Google Scholar] [CrossRef]
- Tanner, C.J.; Barakat, H.; Dohm, G.; Pories, W.; MacDonald, K.; Cunningham, P.; Swanson, M.; Houmard, J. Muscle fiber type is associated with obesity and weight loss. Am. J. Physiol. Endocrinol. Metab. 2002, 282, E1191–E1196. [Google Scholar] [CrossRef]
- De Wilde, J.; Mohren, R.; van den Berg, S.; Boekschoten, M.; Willems-Van Dijk, K.; de Groot, P.; Müller, M.; Mariman, E.; Smit, E. Short-term high fat-feeding results in morphological and metabolic adaptations in the skeletal muscle of C57BL/6J mice. Physiol. Genom. 2008, 32, 360–369. [Google Scholar] [CrossRef]
- Helge, J.; Fraser, A.; Kriketos, A.; Jenkins, A.; Calvert, G.; Ayere, K.; Storlien, L. Interrelationships between muscle fibre type, substrate oxidation and body fat. Int. J. Obes. 1999, 23, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Stuart, C.; McCurry, M.; Marino, A.; South, M.; Howell, M.; Layne, A.; Ramsey, M.; Stone, M. Slow-twitch fiber proportion in skeletal muscle correlates with insulin responsiveness. J. Clin. Endocrinol. Metab. 2013, 98, 2027–2036. [Google Scholar] [CrossRef] [PubMed]
- Akhmedov, D.; Berdeaux, R. The effects of obesity on skeletal muscle regeneration. Front. Physiol. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.; Trajcevski, K.; Al-Sajee, D.; Wang, D.; Thomas, M. Diet-induced obesity impairs muscle satellite cell activation and muscle repair through alterations in hepatocyte growth factor signaling. Physiol. Rep. 2015, 3, e12506. [Google Scholar] [CrossRef] [PubMed]
- Sexton, W.L.; Poole, D.C.; Mathieu-Costello, O. Microcirculatory structure-function relationships in skeletal muscle of diabetic rats. Am. J. Physiol. 1994, 266, H1502–H1511. [Google Scholar] [PubMed]
- Andersen, H.; Gadeberg, P.C.; Brock, B.; Jakobsen, J. Muscular atrophy in diabetic neuropathy: A stereological magnetic resonance imaging study. Diabetologia 1997, 40, 1062–1069. [Google Scholar] [CrossRef]
- Kamei, Y.; Miura, S.; Suzuki, M.; Kai, Y.; Mizukami, J.; Taniguchi, T.; Mochida, K.; Hata, T.; Matsuda, J.; Aburatani, H.; et al. Skeletal muscle foxo1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004, 279, 41114–41123. [Google Scholar] [CrossRef]
- Hickey, M.S.; Carey, J.O.; Azevedo, J.L.; Houmard, J.A.; Pories, W.J.; Israel, R.G.; Dohm, G.L. Skeletal muscle fiber composition is related to adiposity and in vitro glucose transport rate in humans. Am. J. Phys. 1995, 268, E453–E457. [Google Scholar] [CrossRef]
- Oberbach, A.; Bossenz, Y.; Lehmann, S.; Niebauer, J.; Adams, V.; Paschke, R.; Schon, M.R.; Bluher, M.; Punkt, K. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care 2006, 29, 895–900. [Google Scholar] [CrossRef]
- Greco, A.V.; Tataranni, P.A.; Mingrone, G.; de Gaetano, A.; Manto, A.; Cotroneo, P.; Ghirlanda, G. Daily energy metabolism in patients with type 1 diabetes mellitus. J. Am. Coll. Nutr. 1995, 14, 286–291. [Google Scholar] [CrossRef]
- Brandenburg, S.L.; Reusch, J.E.; Bauer, T.A.; Jeffers, B.W.; Hiatt, W.R.; Regensteiner, J.G. Effects of exercise training on oxygen uptake kinetic responses in women with type 2 diabetes. Diabetes Care 1999, 22, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Fujimaki, S.; Kuwabara, T. Diabetes-Induced Dysfunction of Mitochondria and Stem Cells in Skeletal Muscle and the Nervous System. Int. J. Mol. Sci. 2017, 18, 2147. [Google Scholar] [CrossRef] [PubMed]
- Regensteiner, J.G.; Sippel, J.; McFarling, E.T.; Wolfel, E.E.; Hiatt, W.R. Effects of non-insulin-dependent diabetes on oxygen consumption during treadmill exercise. Med. Sci. Sport. Exerc. 1995, 27, 875–881. [Google Scholar] [CrossRef]
- Yokota, T.; Kinugawa, S.; Hirabayashi, K.; Matsushima, S.; Inoue, N.; Ohta, Y.; Hamaguchi, S.; Sobirin, M.A.; Ono, T.; Suga, T.; et al. Oxidative stress in skeletal muscle impairs mitochondrial respiration and limits exercise capacity in type 2 diabetic mice. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, 1069–1077. [Google Scholar] [CrossRef]
- Pinti, M.V.; Fink, G.K.; Hathaway, Q.A.; Durr, A.J.; Kunovac, A.; Hollander, J.M. Mitochondrial dysfunction in type 2 diabetes mellitus: An organ-based analysis. Am. J. Physiol. Metab. 2019, 316, E268–E285. [Google Scholar] [CrossRef]
- Haemmerle, G.; Moustafa, T.; Woelkart, G. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat. Med. 2011, 17, 1076–1085. [Google Scholar] [CrossRef]
- Zechner, R.; Kienesberger, P.C.; Haemmerle, G.; Zimmermann, R.; Lass, A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 2009, 50, 3–21. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Xu, H.E.; Lambert, M.H.; Willson, T.M. Peroxisome proliferator-activated receptors: From genes to physiology. Recent Prog. Horm. Res. 2001, 56, 238–263. [Google Scholar] [CrossRef]
- Finck, B.N.; Kelly, D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Investig. 2006, 116, 615–622. [Google Scholar] [CrossRef]
- Carrer, M.; Liu, N.; Grueter, C.E.; Williams, A.H.; Frisard, M.I.; Hulver, M.W.; Bassel-Duby, R.; Olson, E.N. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378. Proc. Natl. Acad. Sci. USA 2012, 109, 15330–15335. [Google Scholar] [CrossRef]
- Kondratov, K.; Nikitin, Y.; Fedorov, A.; Kostareva, A.; Mikhailovskii, V.; Isakov, D.; Ivanov, A.; Golovkin, A. Heterogeneity of the nucleic acid repertoire of plasma extracellular vesicles demonstrated using high-sensitivity fluorescence-activated sorting. J. Extracell. Vesicles 2020, 9, 1743139. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013, 14, 128. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building Muscle: Molecular Regulation of Myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Sampath, S.C.; Sampath, S.C.; Millay, D.P. Myoblast fusion confusion: The resolution begins. Skelet. Muscle 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.L.; Foletta, V.C.; Wright, C.R.; Sepulveda, P.V.; Konstantopoulos, N.; Sanigorski, A.; Della Gatta, P.; Cameron-Smith, D.; Kralli, A.; Russell, A.P. PGC-1α and PGC-1β increase protein synthesis via ERRα in C2C12 myotubes. Front. Physiol. 2018, 9, 1336. [Google Scholar] [CrossRef] [PubMed]
- Krist, B.; Florczyk, U.; Pietraszek-Gremplewicz, K.; Józkowicz, A.; Dulak, J. The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int. J. Endocrinol. 2015, 2015, 281756. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. MicroRNA-378 Targets the Myogenic Repressor MyoR during Myoblast Differentiation. J. Biol. Chem. 2011, 286, 19431–19438. [Google Scholar] [CrossRef]
- Hou, X.; Tang, Z.; Liu, H.; Wang, N.; Ju, H.; Li, K. Discovery of MicroRNAs Associated with Myogenesis by Deep Sequencing of Serial Developmental Skeletal Muscles in Pigs. PLoS ONE 2012, 7, e52123. [Google Scholar] [CrossRef]
- Hur, K.Y.; Lee, M. New mechanisms of metformin action: Focusing on mitochondria and the gut. J. Diabetes Investig. 2015, 6, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Andrzejewski, S.; Gravel, S.-P.; Pollak, M.; St-Pierre, J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Wessels, B.; Ciapaite, J.; Van Den Broek, N.M.A.; Nicolay, K.; Prompers, J.J. Metformin impairs mitochondrial function in skeletal muscle of both lean and diabetic rats in a Dose-dependent manner. PLoS ONE 2014, 9, e100525. [Google Scholar] [CrossRef]
- Zhang, T.; Duan, J.; Zhang, L.; Li, Z.; Steer, C.J.; Yan, G.; Song, G. LXRα Promotes Hepatosteatosis in Part Through Activation of MicroRNA-378 Transcription and Inhibition of Ppargc1β Expression. Hepatology 2019, 69, 1488–1503. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, M.; Sahlin, K.; Fernström, M.; Glintborg, D.; Vind, B.F.; Beck-Nielsen, H.; Højlund, K. Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes 2007, 56, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Rabøl, R.; Larsen, S.; Højberg, P.M.V.; Almdal, T.; Boushel, R.; Haugaard, S.B.; Andersen, J.L.; Madsbad, S.; Dela, F. Regional anatomic differences in skeletal muscle mitochondrial respiration in type 2 diabetes and obesity. J. Clin. Endocrinol. Metab. 2010, 95, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Hesselink, M.K.C.; Schrauwen-Hinderling, V.; Schrauwen, P. Skeletal muscle mitochondria as a target to prevent or treat type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2016, 12, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Chambers, K.T.; Chen, Z.; Crawford, P.A.; Fu, X.; Burgess, S.C.; Lai, L.; Leone, T.C.; Kelly, D.P.; Finck, B.N. Liver-Specific PGC-1beta Deficiency Leads to Impaired Mitochondrial Function and Lipogenic Response to Fasting-Refeeding. PLoS ONE 2012, 7, e52645. [Google Scholar] [CrossRef]
- Zorzano, A.; Hernández-Alvarez, M.I.; Palacín, M.; Mingrone, G. Alterations in the mitochondrial regulatory pathways constituted by the nuclear co-factors PGC-1α or PGC-1β and mitofusin 2 in skeletal muscle in type 2 diabetes. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 1028–1033. [Google Scholar] [CrossRef]
- Bruce, C.R.; Anderson, M.J.; Carey, A.L.; Newman, D.G.; Bonen, A.; Kriketos, A.D.; Cooney, G.J.; Hawley, J.A. Muscle Oxidative Capacity Is a Better Predictor of Insulin Sensitivity than Lipid Status. J. Clin. Endocrinol. Metab. 2003, 88, 5444–5451. [Google Scholar] [CrossRef]
- Park, S.S.; Seo, Y.K. Excess accumulation of lipid impairs insulin sensitivity in skeletal muscle. Int. J. Mol. Sci. 2020, 21, 1949. [Google Scholar] [CrossRef]
- Huijsman, E.; van de Par, C.; Economou, C. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E505–E513. [Google Scholar] [CrossRef]
- Sitnick, M.T.; Basantani, M.K.; Cai, L.; Schoiswohl, G.; Yazbeck, C.F.; Distefano, G.; Ritov, V.; DeLany, J.P.; Schreiber, R.; Stolz, D.B.; et al. Skeletal muscle triacylglycerol hydrolysis does not influence metabolic complications of obesity. Diabetes 2013, 62, 3350–3361. [Google Scholar] [CrossRef]
- Meex, R.C.R.; Hoy, A.J.; Mason, R.M. ATGL-mediated triglyceride turnover and the regulation of mitochondrial capacity in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E960–E970. [Google Scholar] [CrossRef] [PubMed]
- Lettieri-Barbato, D.; Cannata, S.M.; Casagrande, V.; Ciriolo, M.R.; Aquilano, K. Time-controlled fasting prevents aging-like mitochondrial changes induced by persistent dietary fat overload in skeletal muscle. PLoS ONE 2018, 13, e0195912. [Google Scholar] [CrossRef]
- L’Honoré, A.; Commère, P.H.; Negroni, E.; Pallafacchina, G.; Friguet, B.; Drouin, J.; Buckingham, M.; Montarras, D. The role of Pitx2 and Pitx3 in muscle 1 stem cells gives new insights into P38α MAP kinase and redox regulation of muscle regeneration. eLife 2018, 7, e32991. [Google Scholar] [CrossRef]
- Corbel, S.Y.; Llewellyn, M.E.; Mourkioti, F.; Porpiglia, E.; Delp, S.L.; Gilbert, P.M.; Blau, H.M.; Cosgrove, B.D.; Lee, S.P. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 2014, 20, 255–264. [Google Scholar]

| Target | Sequence 5′–3′ |
|---|---|
| Myh1 forward | GCTGAGAGAAGCTACCACATT |
| Myh1 reverse | ACAAAGGCGTAGTCGTATGG |
| Myh7 forward | TGCCCGATGACAAAGAAGAG |
| Myh7 reverse | GTCACCGTCTTGCCATTCT |
| Mymk forward | CCTGTGATGGGCCTGGTTTGTC |
| Mymk reverse | GGTTCATCAAAGTCGGCCAGTGC |
| MyoD forward | TGCCTTCTACGCACCTGGA |
| MyoD reverse | CGCTGTAATCCATCATGCCATC |
| Atgl forward | CCACTTTAGCTCCAAGGATGAG |
| Atgl reverse | TTGGAGGGTAGGAGGAATGA |
| Pgc1b forward | CTTTATCTCTTCCTCTGACCCCAG |
| Pgc1b reverse | CCAGAGAGTTCCACACTTAAGGTT |
| Gapdh forward | GGATCTGACGTGCCGCCTG |
| Gapdh reverse | GAAGGTGGA AGAGTGGGAGTTGC |
| Group of Patients | Patiet ID | Age (Years) | BMI (kg/m2) | Obesity Grade | Diabetes Duration (Years) | HbA1c Levels (%) | Glucose Levels (mmol/L) | Pool_ID | Pool_BMI |
|---|---|---|---|---|---|---|---|---|---|
| Control (C) | c1 | 26 | 16.9 | NA | NA | 5.2 | 5.06 | C1 | 18.7 |
| c2 | 26 | 20.6 | NA | NA | 4.5 | 4.65 | C1 | 18.7 | |
| c8 | 43 | 20.7 | NA | NA | 5.5 | 5.24 | C2 | 21.0 | |
| c11 | 38 | 21.3 | NA | NA | 4.7 | 4.9 | C2 | 21.0 | |
| c9 | 45 | 22.2 | NA | NA | 5.1 | 5.0 | C3 | 22.2 | |
| c4 | 50 | 22.2 | NA | NA | 5.1 | 5.38 | C3 | 22.2 | |
| c10 | 32 | 22.4 | NA | NA | 5.0 | 5.38 | C4 | 22.9 | |
| c5 | 44 | 23.4 | NA | NA | 5.6 | 5.24 | C4 | 22.9 | |
| c7 | 33 | 24.1 | NA | NA | 4.5 | 4.39 | C5 | 24.2 | |
| c3 | 30 | 24.4 | NA | NA | 4.5 | 4.39 | C5 | 24.2 | |
| Obesity (OB) | B122 | 40 | 40.2 | III | NA | 5.75 | 6.29 | OB1 | 40.6 |
| B136 | 35 | 40.9 | III | NA | 5.14 | 6.18 | OB1 | 40.6 | |
| B130 | 57 | 41.2 | III | NA | 5.4 | 5.06 | OB2 | 41.7 | |
| B131 | 61 | 42.2 | III | NA | 5.7 | 5.9 | OB2 | 41.7 | |
| B141 | 40 | 43.9 | III | NA | 5.6 | 5.8 | OB3 | 44.4 | |
| B164 | 53 | 44.8 | III | NA | 5.5 | 5.63 | OB3 | 44.4 | |
| B160 | 34 | 46 | III | NA | 5.0 | 5.18 | OB4 | 48.2 | |
| B151 | 60 | 50.4 | III | NA | 5.3 | 5.99 | OB4 | 48.2 | |
| B147 | 56 | 56.4 | III | NA | 5.2 | 5.8 | OB5 | 61.9 | |
| B124 | 44 | 67.3 | III | NA | 6.4 | 6.84 | OB5 | 61.9 | |
| Obesity comorbid with diabetes (OBD) | B158 | 60 | 36.2 | II | 8 | 6.2 | 8.77 | OBD1 | 37.6 |
| B202 | 61 | 38.9 | II | 17 | 6.8 | 9.07 | OBD1 | 37.6 | |
| B174 | 46 | 47.2 | III | 13 | 6.8 | 6.63 | OBD2 | 48.1 | |
| B169 | 66 | 48.9 | III | 7 | 7.09 | 7.19 | OBD2 | 48.1 | |
| B168 | 56 | 49.8 | III | 3 | 7.87 | 8.35 | OBD3 | 51.8 | |
| B128 | 39 | 53.8 | III | 5 | 9.2 | 12.58 | OBD3 | 51.8 | |
| B112 | 53 | 63.2 | III | 7 | 6.56 | 5.25 | OBD4 | 64.2 | |
| B142 | 38 | 65.1 | III | 7 | 10.05 | 14.49 | OBD4 | 64.2 |
| GO Term | p-Value | Predicted Targets of miR-378a-3p |
|---|---|---|
| ERK1 and ERK2 cascade (GO:0070371) | 0.00005 | MAPK1; IGF1; ZFP36L2 |
| negative regulation of stem cell differentiation (GO:2000737) | 0.00052 | REST; ZFP36L2 |
| nervous system development (GO:0007399) | 0.00096 | NEUROD1; DSCAM; PAX8; DYRK1A; DCX; SHANK3; DACT1 |
| regulation of macromolecule metabolic process (GO:0060255) | 0.00103 | MAPK1; ALPK3; IGF1; TOB2 |
| positive regulation of muscle hypertrophy (GO:0014742) | 0.00104 | IGF1; IL6ST |
| cellular response to corticosteroid stimulus (GO:0071384) | 0.00120 | REST; ZFP36L2 |
| synapse assembly (GO:0007416) | 0.00135 | NPAS4; DSCAM; SHANK3 |
| positive regulation of cardiac muscle hypertrophy (GO:0010613) | 0.00136 | IGF1; IL6ST |
| myoblast differentiation (GO:0045445) | 0.00136 | REST; IGF1 |
| cellular response to glucocorticoid stimulus (GO:0071385) | 0.00154 | REST; ZFP36L2 |
| peptidyl-serine phosphorylation (GO:0018105) | 0.00159 | DYRK1A; MAPK1; CSNK1G2; CAMKK2 |
| peptidyl-threonine phosphorylation (GO:0018107) | 0.00168 | DYRK1A; MAPK1; CAMKK2 |
| positive regulation of transcription regulatory region DNA binding (GO:2000679) | 0.00193 | NEUROD1; IGF1 |
| phosphorylation (GO:0016310) | 0.00214 | TOLLIP; DYRK1A; MAPK1; ALPK3; CSNK1G2; CAMKK2 |
| regulation of cardiac muscle hypertrophy (GO:0010611) | 0.00236 | IGF1; IL6ST |
| MAPK cascade (GO:0000165) | 0.00272 | MAPK1; IGF1; SHANK3; ZFP36L2; CAMKK2 |
| peptidyl-serine modification (GO:0018209) | 0.00283 | DYRK1A; MAPK1; CSNK1G2; CAMKK2 |
| modulation of chemical synaptic transmission (GO:0050804) | 0.00286 | NPAS4; RIMS4; GRIK3 |
| response to glucocorticoid (GO:0051384) | 0.00308 | REST; ZFP36L2 |
| peptidyl-threonine modification (GO:0018210) | 0.00361 | DYRK1A; MAPK1; CAMKK2 |
| regulation of transcription regulatory region DNA binding (GO:2000677) | 0.00361 | NEUROD1; IGF1 |
| response to hexose (GO:0009746) | 0.00389 | NEUROD1; IGF1 |
| positive regulation of cell differentiation (GO:0045597) | 0.00454 | NEUROD1; PAX8; IGF1; IL6ST |
| positive regulation of gene expression (GO:0010628) | 0.00494 | NEUROD1; REST; PAX8; DYRK1A; MAPK1; IGF1; RBMS3; CAMKK2 |
| negative regulation of fat cell differentiation (GO:0045599) | 0.00544 | ZFPM2; ZFP36L2 |
| positive regulation of signal transduction (GO:0009967) | 0.00561 | TSPAN17; IGF1; SHANK3; DACT1 |
| protein phosphorylation (GO:0006468) | 0.00561 | DYRK1A; MAPK1; ALPK3; CSNK1G2; CAMKK2; MAP2K6 |
| regulation of interferon-beta production (GO:0032648) | 0.00613 | YY1; TRAF3 |
| positive regulation of DNA binding (GO:0043388) | 0.00613 | NEUROD1; IGF1 |
| positive regulation of osteoblast differentiation (GO:0045669) | 0.00648 | IGF1; IL6ST |
| epithelium development (GO:0060429) | 0.00737 | PAX8; TOLLIP; DACT1 |
| sphingolipid metabolic process (GO:0006665) | 0.00754 | SERINC1; KDSR; CSNK1G2 |
| activation of MAPK activity (GO:0000187) | 0.00772 | MAPK1; IGF1; MAP2K6 |
| regulation of gene expression (GO:0010468) | 0.00905 | YY1; REST; ZNF514; MAPK1; ALPK3; IGF1; TOB2; SRSF10; RBMS3 |
| negative regulation of macromolecule metabolic process (GO:0010605) | 0.00905 | YY1; REST; RBMS3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khromova, N.V.; Fedorov, A.V.; Ma, Y.; Kondratov, K.A.; Prikhodko, S.S.; Ignatieva, E.V.; Artemyeva, M.S.; Anopova, A.D.; Neimark, A.E.; Kostareva, A.A.; et al. Regulatory Action of Plasma from Patients with Obesity and Diabetes towards Muscle Cells Differentiation and Bioenergetics Revealed by the C2C12 Cell Model and MicroRNA Analysis. Biomolecules 2021, 11, 769. https://doi.org/10.3390/biom11060769
Khromova NV, Fedorov AV, Ma Y, Kondratov KA, Prikhodko SS, Ignatieva EV, Artemyeva MS, Anopova AD, Neimark AE, Kostareva AA, et al. Regulatory Action of Plasma from Patients with Obesity and Diabetes towards Muscle Cells Differentiation and Bioenergetics Revealed by the C2C12 Cell Model and MicroRNA Analysis. Biomolecules. 2021; 11(6):769. https://doi.org/10.3390/biom11060769
Chicago/Turabian StyleKhromova, Natalya V., Anton V. Fedorov, Yi Ma, Kirill A. Kondratov, Stanislava S. Prikhodko, Elena V. Ignatieva, Marina S. Artemyeva, Anna D. Anopova, Aleksandr E. Neimark, Anna A. Kostareva, and et al. 2021. "Regulatory Action of Plasma from Patients with Obesity and Diabetes towards Muscle Cells Differentiation and Bioenergetics Revealed by the C2C12 Cell Model and MicroRNA Analysis" Biomolecules 11, no. 6: 769. https://doi.org/10.3390/biom11060769
APA StyleKhromova, N. V., Fedorov, A. V., Ma, Y., Kondratov, K. A., Prikhodko, S. S., Ignatieva, E. V., Artemyeva, M. S., Anopova, A. D., Neimark, A. E., Kostareva, A. A., Babenko, A. Y., & Dmitrieva, R. I. (2021). Regulatory Action of Plasma from Patients with Obesity and Diabetes towards Muscle Cells Differentiation and Bioenergetics Revealed by the C2C12 Cell Model and MicroRNA Analysis. Biomolecules, 11(6), 769. https://doi.org/10.3390/biom11060769








