Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis
Abstract
1. Introduction
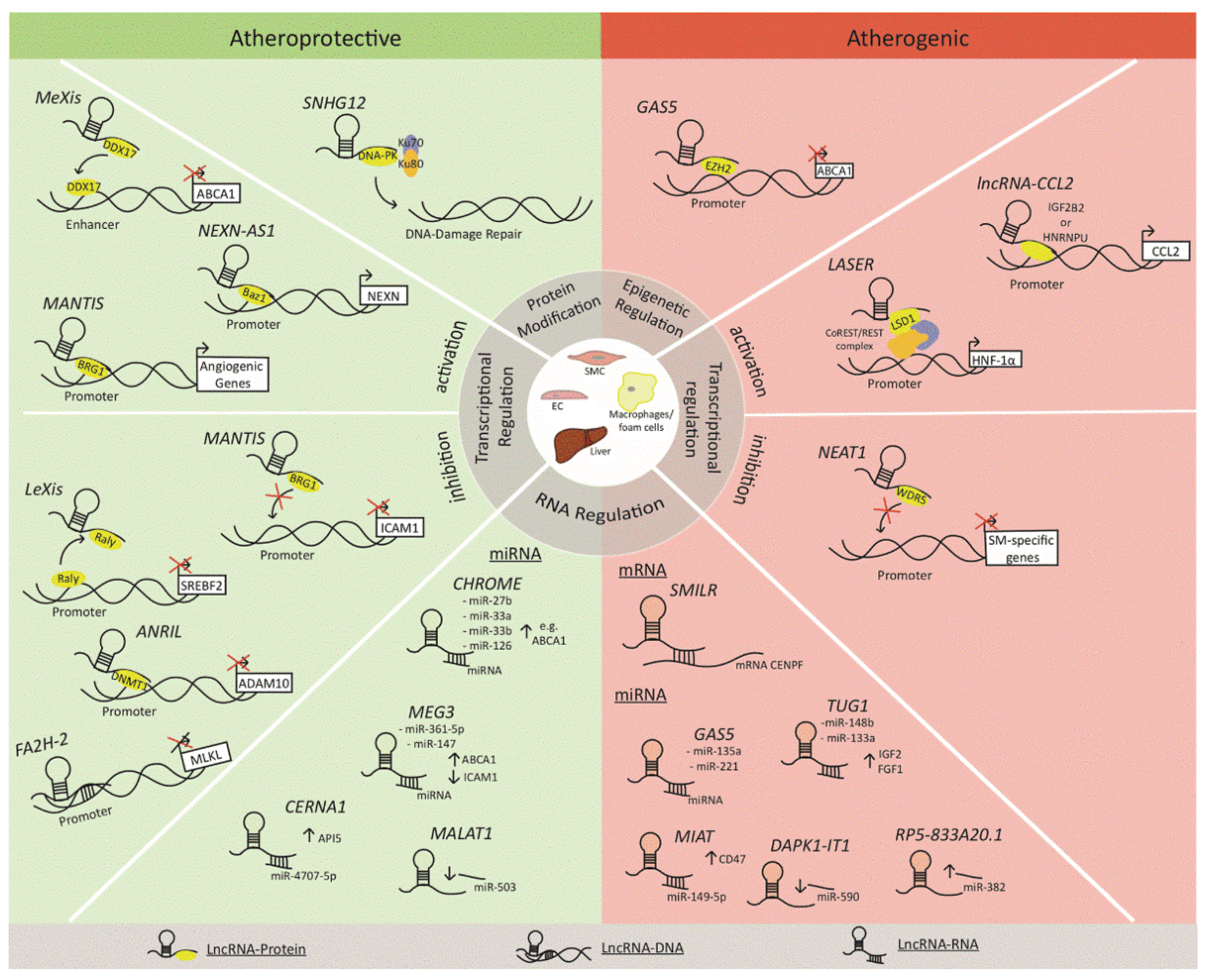
2. LncRNAs in Atherosclerosis: Mechanisms of Action
- Signaling lncRNAs act as molecular signals and regulate gene expression via interaction with chromatin-modifying complexes, transcriptional regulators, and DNA.
- Decoy lncRNAs function as decoy molecules that bind to transcriptional regulators and inhibit their interaction with target genes.
- Guide lncRNAs enhance downstream effector functions by helping transcriptional regulators to localize to specific regions.
- Scaffold lncRNAs mediate protein–protein interactions, resulting in the organization of nuclear subdomains, acting as enhancers at certain areas of DNA, or repressing gene expression by creating RNA–DNA structures.
2.1. Cholesterol Homeostasis
2.2. Vascular Inflammation
2.3. Plaque Destabilization
3. Statins: Mechanism of Action and Rationale for Regulation of LncRNAs
4. Statins and LncRNAs: Current Evidence
4.1. Cholesterol Homeostasis
4.2. Vascular Inflammation
5. Regulation of LncRNA and Determinants of Statin Efficacy
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Forouzanfar, M.H.; Moran, A.E.; Barber, R.; Nguyen, G.; Feigin, V.L.; Naghavi, M.; Mensah, G.A.; Murray, C.J. Demographic and Epidemiologic Drivers of Global Cardiovascular Mortality. N. Engl. J. Med. 2015, 372, 1333–1341. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk: The Task Force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS). Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef] [PubMed]
- Nissen, S.E.; Nicholls, S.J.; Sipahi, I.; Libby, P.; Raichlen, J.S.; Ballantyne, C.M.; Davignon, J.; Erbel, R.; Fruchart, J.C.; Tardif, J.-C.; et al. Effect of Very High-Intensity Statin Therapy on Regression of Coronary Atherosclerosis. JAMA 2006, 295, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Lin, F.; Hulley, S.B.; Blanche, P.J.; Waters, D.; Shiboski, S.; Rotter, J.I.; Nickerson, D.A.; Yang, H.; Saad, M.; et al. Phenotypic Predictors of Response to Simvastatin Therapy Among African-Americans and Caucasians: The Cholesterol and Pharmacogenetics (CAP) Study. Am. J. Cardiol. 2006, 97, 843–850. [Google Scholar] [CrossRef]
- Kajinami, K.; Takekoshi, N.; Brousseau, M.E.; Schaefer, E.J. Pharmacogenetics of HMG-CoA reductase inhibitors: Exploring the potential for genotype-based individualization of coronary heart disease management. Atherosclerosis 2004, 177, 219–234. [Google Scholar] [CrossRef]
- Kajinami, K.; Akao, H.; Polisecki, E.; Schaefer, E.J. Pharmacogenomics of Statin Responsiveness. Am. J. Cardiol. 2005, 96, 65–70. [Google Scholar] [CrossRef]
- Guo, X.; Gao, L.; Wang, Y.; Chiu, D.K.Y.; Wang, T.; Deng, Y. Advances in long noncoding RNAs: Identification, structure prediction and function annotation. Briefings Funct. Genom. 2016, 15, 38–46. [Google Scholar] [CrossRef]
- Josefs, T.; Boon, R.A. The Long Non-coding Road to Atherosclerosis. Curr. Atheroscler. Rep. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Moore, J.B.; Uchida, S. Functional characterization of long noncoding RNAs. Curr. Opin. Cardiol. 2020, 35, 199–206. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Günther, S.; et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef]
- Man, H.-S.J.; Marsden, P. LncRNAs and epigenetic regulation of vascular endothelium: Genome positioning system and regulators of chromatin modifiers. Curr. Opin. Pharmacol. 2019, 45, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hu, Z.; Zhang, W.; Yu, J.; Yang, Y.; Xu, Z.; Luo, H.; Liu, X.; Liu, Y.; Chen, C.; et al. Regulation of Cholesterol Homeostasis by a Novel Long Non-coding RNA LASER. Sci. Rep. 2019, 9, 7693–7705. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.; Schlüter, K.-D.; Laufs, U. Molecular and cellular function of the proprotein convertase subtilisin/kexin type 9 (PCSK9). Basic Res. Cardiol. 2015, 110, 1–19. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.S.X.W.T.Q.D.A.; Hong, C.; et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nat. Cell Biol. 2016, 534, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Wu, X.; Jones, M.; Zhang, Z.; Salisbury, D.; Sallam, T. Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation 2017, 136, 776–778. [Google Scholar] [CrossRef]
- Li, G.; Gu, H.-M.; Zhang, D.-W. ATP-binding cassette transporters and cholesterol translocation. IUBMB Life 2013, 65, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Sallam, T.; Jones, M.; Thomas, B.J.; Wu, X.; Gilliland, T.; Qian, K.; Eskin, A.; Casero, D.; Zhang, Z.; Sandhu, J.; et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018, 24, 304–312. [Google Scholar] [CrossRef]
- Meng, X.-D.; Yao, H.-H.; Wang, L.-M.; Yu, M.; Shi, S.; Yuan, Z.-X.; Liu, J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE−/− Mice. Mol. Ther. Nucleic Acids 2020, 19, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Guo, F.-X.; Xu, Y.-J.; Li, P.; Lu, Z.-F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.H.; Kang, C.-M.; et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J. Clin. Investig. 2019, 129, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-M.; Wu, S.-G.; Chen, F.; Wu, Q.; Wu, C.-M.; Kang, C.-M.; He, X.; Zhang, R.-Y.; Lu, Z.-F.; Li, X.-H.; et al. Atorvastatin inhibits pyroptosis through the lncRNA NEXN-AS1/NEXN pathway in human vascular endothelial cells. Atherosclerosis 2020, 293, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Leisegang, M.S.; Bibli, S.-I.; Günther, S.; Pflüger-Müller, B.; Oo, J.; Höper, C.; Seredinski, S.; Yekelchyk, M.; Schmitz-Rixen, T.; Schürmann, C.; et al. Pleiotropic effects of laminar flow and statins depend on the Krüppel-like factor-induced lncRNA MANTIS. Eur. Heart J. 2019, 40, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Sluimer, J.C.; Wang, Y.; Subramanian, M.; Brown, K.; Pattison, J.S.; Robbins, J.; Martinez, J.; Tabas, I. Macrophage Autophagy Plays a Protective Role in Advanced Atherosclerosis. Cell Metab. 2012, 15, 545–553. [Google Scholar] [CrossRef]
- Guo, F.-X.; Wu, Q.; Li, P.; Zheng, L.; Ye, S.; Dai, X.-Y.; Kang, C.-M.; Lu, J.-B.; Xu, B.-M.; Xu, Y.-J.; et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. 2019, 26, 1670–1687. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yang, C.; Shi, J.; Gao, T. Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur. J. Pharmacol. 2019, 858, 172338–172348. [Google Scholar] [CrossRef]
- Song, Y.; Yang, L.; Guo, R.; Lu, N.; Shi, Y.; Wang, X. Long noncoding RNA MALAT1 promotes high glucose-induced human endothelial cells pyroptosis by affecting NLRP3 expression through competitively binding miR-22. Biochem. Biophys. Res. Commun. 2019, 509, 359–366. [Google Scholar] [CrossRef]
- Cremer, S.; Michalik, K.M.; Fischer, A.; Pfisterer, L.; Jaé, N.; Winter, C.; Boon, R.A.; Muhly-Reinholz, M.; John, D.; Uchida, S.; et al. Hematopoietic Deficiency of the Long Noncoding RNA MALAT1 Promotes Atherosclerosis and Plaque Inflammation. Circ. 2019, 139, 1320–1334. [Google Scholar] [CrossRef]
- Khyzha, N.; Khor, M.; Distefano, P.V.; Wang, L.; Matic, L.; Hedin, U.; Wilson, M.D.; Maegdefessel, L.; Fish, J.E. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 16410–16419. [Google Scholar] [CrossRef]
- Ahmed, A.S.I.; Dong, K.; Liu, J.; Wen, T.; Yu, L.; Xu, F.; Kang, X.; Osman, I.; Hu, G.; Bunting, K.M.; et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA 2018, 115, 8660–8667. [Google Scholar] [CrossRef]
- Mahmoud, A.D.; Ballantyne, M.D.; Miscianinov, V.; Pinel, K.; Hung, J.; Scanlon, J.P.; Iyinikkel, J.; Kaczynski, J.; Tavares, A.S.; Bradshaw, A.C.; et al. The Human-Specific and Smooth Muscle Cell-Enriched LncRNA SMILR Promotes Proliferation by Regulating Mitotic CENPF mRNA and Drives Cell-Cycle Progression Which Can Be Targeted to Limit Vascular Remodeling. Circ. Res. 2019, 125, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Oesterle, A.; Laufs, U.; Liao, J.K. Pleiotropic Effects of Statins on the Cardiovascular System. Circ. Res. 2017, 120, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Hellberg, M.E.; Balch, D.P.; Roy, K. Structural Mechanism for Statin Inhibition of HMG-CoA Reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Kureishi, Y.; Luo, Z.; Shiojima, I.; Bialik, A.; Fulton, D.; Lefer, D.J.; Sessa, W.C.; Walsh, K. The HMG-CoA reductase inhibitor simvastatin activates the protein kinase Akt and promotes angiogenesis in normocholesterolemic animals. Nat. Med. 2000, 6, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Rikitake, Y.; Liao, J.K. Rho GTPases, Statins, and Nitric Oxide. Circ. Res. 2005, 97, 1232–1235. [Google Scholar] [CrossRef]
- Rezaie-Majd, A.; Maca, T.; Bucek, R.A.; Valent, P.; Müller, M.R.; Husslein, P.; Kashanipour, A.; Minar, E.; Baghestanian, M. Simvastatin Reduces Expression of Cytokines Interleukin-6, Interleukin-8, and Monocyte Chemoattractant Protein-1 in Circulating Monocytes From Hypercholesterolemic Patients. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1194–1199. [Google Scholar] [CrossRef]
- Niwa, S.; Totsuka, T.; Hayashi, S. Inhibitory effect of fluvastatin, an HMG-CoA reductase inhibitor, on the expression of adhesion molecules on human monocyte cell line. Int. J. Immunopharmacol. 1996, 18, 669–675. [Google Scholar] [CrossRef]
- Ichiki, T.; Takeda, K.; Tokunou, T.; Iino, N.; Egashira, K.; Shimokawa, H.; Hirano, K.; Kanaide, H.; Takeshita, A. Downregulation of Angiotensin II Type 1 Receptor by Hydrophobic 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Inhibitors in Vascular Smooth Muscle Cells. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1896–1901. [Google Scholar] [CrossRef]
- Hernández-Perera, O.; Perez-Sala, D.; Navarro-Antolin, J.; Sánchez-Pascuala, R.; Hernández, G.; Díaz, C.; Lamas, S. Effects of the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors, atorvastatin and simvastatin, on the expression of endothelin-1 and endothelial nitric oxide synthase in vascular endothelial cells. J. Clin. Investig. 1998, 101, 2711–2719. [Google Scholar] [CrossRef]
- Karlson, B.W.; Wiklund, O.; Palmer, M.K.; Nicholls, S.J.; Lundman, P.; Barter, P.J. Variability of low-density lipoprotein cholesterol response with different doses of atorvastatin, rosuvastatin, and simvastatin: Results from VOYAGER. Eur. Heart J. Cardiovasc. Pharmacother. 2016, 2, 212–217. [Google Scholar] [CrossRef]
- Postmus, I.; Verschuren, J.J.; De Craen, A.J.; Slagboom, P.; Westendorp, R.G.; Jukema, J.W.; Trompet, S. Pharmacogenetics of statins: Achievements, whole-genome analyses and future perspectives. Pharmacogenomics 2012, 13, 831–840. [Google Scholar] [CrossRef]
- Postmus, I.; Welcome Trust Case Control Consortium; Trompet, S.; Deshmukh, H.A.; Barnes, M.R.; Li, X.; Warren, H.R.; Chasman, D.I.; Zhou, K.; Arsenault, B.J.; et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat. Commun. 2014, 5, 5068–5078. [Google Scholar] [CrossRef]
- Mitchel, K.; Theusch, E.; Cubitt, C.; Dosé, A.C.; Stevens, K.; Naidoo, D.; Medina, M.W. RP1-13D10.2 Is a Novel Modulator of Statin-Induced Changes in Cholesterol. Circ. Cardiovasc. Genet. 2016, 9, 223–230. [Google Scholar] [CrossRef]
- Paez, I.; Prado, Y.; Ubilla, C.G.; Zambrano, T.; Salazar, L.A. Atorvastatin Increases the Expression of Long Non-Coding RNAs ARSR and CHROME in Hypercholesterolemic Patients: A Pilot Study. Pharmaceuticals 2020, 13, 382. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Fang, M.; Tian, B.; Luo, J.; Jin, C.; Wang, X.; Ning, Z.; Li, X. Atorvastatin protects cardiac progenitor cells from hypoxia-induced cell growth inhibition via MEG3/miR-22/HMGB1 pathway. Acta Biochim. Biophys. Sin. 2018, 50, 1257–1265. [Google Scholar] [CrossRef]
- Huang, P.; Wang, L.; Li, Q.; Tian, X.; Xu, J.; Xu, J.; Xiong, Y.; Chen, G.; Qian, H.; Jin, C.; et al. Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc. Res. 2019, 116, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Josipovic, I.; Pflüger, B.; Fork, C.; Vasconez, A.E.; Oo, J.A.; Hitzel, J.; Seredinski, S.; Gamen, E.; zu Heringdorf, D.M.; Chen, W.; et al. Long noncoding RNA LISPR1 dis required for S1P signaling and endothelial cell function. J. Mol. Cell. Cardiol. 2018, 116, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Wang, B.Q. Clinical significance of lncRNA-AWPPH in coronary artery diseases. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11747–11751. [Google Scholar] [CrossRef]
- Lambert, G.; Sjouke, B.; Choque, B.; Kastelein, J.J.P.; Hovingh, G.K. The PCSK9 decade. J. Lipid Res. 2012, 53, 2515–2524. [Google Scholar] [CrossRef]
- Lawson, C.; Wolf, S. ICAM-1 signaling in endothelial cells. Pharmacol. Rep. 2009, 61, 22–32. [Google Scholar] [CrossRef]
- Bai, J.; Xu, J.; Zhao, J.; Zhang, R. Downregulation of lncRNA AWPPH inhibits colon cancer cell proliferation by downregulating GLUT-1. Oncol. Lett. 2019, 18, 2007–2012. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-Y.; Jiang, Y.-H.; Qi, Y.-Z.; Shao, L.-L.; Yang, C.-H. Integrated analysis of long noncoding RNA and mRNA profiling ox-LDL-induced endothelial dysfunction after atorvastatin administration. Medience 2018, 97, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Zhu, H.; Ning, X.; Li, L.; Yang, B.; Chen, S.; Wang, L.; Lu, X.; Gu, D. LncRNA ENST00000602558.1 regulates ABCG1 expression and cholesterol efflux from vascular smooth muscle cells through a p65-dependent pathway. Atherosclerosis 2019, 285, 31–39. [Google Scholar] [CrossRef]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Yin, R.; Shi, H.; Wang, X.; Shen, D.; Wang, X.; Pan, C. LncRNA ZFAS1 confers inflammatory responses and reduces cholesterol efflux in atherosclerosis through regulating miR-654-3p-ADAM10/RAB22A axis. Int. J. Cardiol. 2020, 315, 72–80. [Google Scholar] [CrossRef]
- Lan, X.; Yan, J.; Ren, J.; Zhong, B.; Li, J.; Li, Y.; Liu, L.; Yi, J.; Sun, Q.; Yang, X.; et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 2016, 64, 58–72. [Google Scholar] [CrossRef]
- Muret, K.; Désert, C.; Lagoutte, L.; Boutin, M.; Gondret, F.; Zerjal, T.; Lagarrigue, S. Long noncoding RNAs in lipid metabolism: Literature review and conservation analysis across species. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Chasman, D.I.; Giulianini, F.; MacFadyen, J.; Barratt, B.J.; Nyberg, F.; Ridker, P.M. Genetic Determinants of Statin-Induced Low-Density Lipoprotein Cholesterol Reduction. Circ. Cardiovasc. Genet. 2012, 5, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Liu, S.; Wang, C.; Han, X. A Functional Polymorphism rs145204276 in the Promoter of Long Noncoding RNA GAS5 Is Associated with an Increased Risk of Ischemic Stroke. J. Stroke Cerebrovasc. Dis. 2018, 27, 3535–3541. [Google Scholar] [CrossRef]
- Han, X.; Zheng, Z.; Wang, C.; Wang, L. Association between MEG3/miR-181b polymorphisms and risk of ischemic stroke. Lipids Health Dis. 2018, 17, 292–300. [Google Scholar] [CrossRef]
- Huang, Y.; Ye, H.; Hong, Q.; Xu, X.; Jiang, D.; Xu, L.; Dai, D.; Sun, J.; Gao, X.; Duan, S. Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis. Int. J. Mol. Sci. 2014, 15, 17478–17492. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Ali, I.S.; Riaz, M.; Younas, A.; Sadeque, A.; Niazi, A.K.; Niazi, S.H.; Ali, S.H.B.; Azam, M.; Qamar, R. Association of ANRIL polymorphism (rs1333049:C>G) with myocardial infarction and its pharmacogenomic role in hypercholesterolemia. Gene 2013, 515, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; Zhang, Y.; Xu, X.; Bi, L.; Zhang, M.; Yu, B.; Zhang, Y. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene 2020, 724, 143684–143690. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Definition | Humans (Plaques or Serum) | In Vivo (Experimental Models) | In Vitro |
|---|---|---|---|---|
| LASER | Lipid-associated single nucleotide polymorphism gene region | Positive correlation with Chol levels in PBMC patients | - | Deficiency: ↓ cholesterol in HepG2 cells |
| LeXis | Liver-expressed LXR-induced sequence | - | Overexpression: C57BL/6 mice, AV LeXis ↓total cholesterol and triglycerides, ↓ aortic root plaques on en face analysis | - |
| MeXis | Macrophage-expressed LXR-induced sequence | - | Deficiency: Ldlr−/− on WD +MeXis−/− bone marrow↓ Abca1 expression ↑ inflammatory gene expression ↑ lesion size ↑ CD68+ cell | Deficiency: Peritoneal macrophages (MeXis−/− mice fed WD) ↓ ABCA1 ↓ cholesterol efflux ↑ cholesterol accumulation |
| NEXN-AS1 | Nexilin F-actin binding protein antisense RNA 1 | ↓ atherosclerotic plaques ↓ NEXN in CAD patients (blood) | Deficiency: NEXN± /ApoE−/− on WD ↑ lesion area, macrophage abundance, expression of adhesion molecules ↑ inflammatory cytokines | Overexpression: HUVECs ↓ TLR4/NF-kB pathway ↓ inflammatory gene expression |
| MANTIS | - | ↓ atherosclerotic plaque | Deficiency: Retinal injection of siRNA MANTIS ↑ ICAM-1 | Deficiency: HUVECs ↓ angiogenic genes ↑ ICAM-1 ↑ monocyte adhesion ↑ apoptosis ↑ oxidative stress |
| CCL2 | C-C motif chemokine ligand 2 | ↑ unstable symptomatic atherosclerotic plaque | - | Deficiency: HUVEC (IL-1β) ↓ CCL2 |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 | - | Deficiency: NEAT1±, carotid artery ligation injury ↓ VSMC proliferation and migration ↓ Neointima formation | Overexpression: ↑ VSMC proliferation and migration Deficiency: ↓ VSMC proliferation and migration |
| SMILR | Smooth muscle-induced lncRNA enhances replication | ↑ unstable atherosclerotic plaque ↑in plasma from patients with high plasma C-reactive protein | - | Deficiency: ↑ Proliferation of arterial and venous SMCs |
| CHROME | Cholesterol homeostasis regulator of miRNA expression | ↑ CAD (plasma), ↑ symptomatic versus asymptomatic atherosclerotic plaques | - | Deficiency: HepG2 cells, primary human hepatocytes, THP-1 ↓ ABCA1 protein expression ↓ cholesterol efflux to exogenous apoA-1 |
| RP5-833A20.1 | - | - | Overexpression: ApoE−/− on HFD, LV-induced NFIA OE ↑ cholesterol efflux ↓ lesion size ↓ lipid accumulation | Overexpression: THP-1 (oxLDL) ↓ cholesterol efflux ↑ lipid accumulation ↑ miR-382-5p ↓ NFIA |
| GAS5 | Growth-arrest specific 5 | - | Overexpression: ApoE−/− on HFD, LV-induced OE ↓ HDL-C, ↑ LDL-C ↓ reduced cholesterol efflux ↑ lesion size ↑ inflammation | Overexpression: THP-1 (oxLDL) ↓ cholesterol efflux ↑ lipid accumulation ↓ ABCA1 ↑ inflammatory markers ↑ MMP-2, MMP-9 ↑ EZH ↓ miR-135 ↓ miR-221 |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 | ↓ atherosclerotic plaque, correlates with symptoms of plaque instability | Deficiency: ApoE−/− Malat1−/− bone marrow cells on HFD ↑ adhesion to endothelial cells ↑ proinflammatory mediators ↑ lesion size ↑ miR 503 | Deficiency: HUVECs (oxLDL) ↓ autophagy ↑ apoptosis ↑ miR-216a-5p EA.hy926 cells (high glucose) ↓ pryoptosis ↓ NLRP3 ↑ miR-22 |
| MEG3 | Maternally expressed 3 | - | Overexpression: Ldlr−/− on HFD ↓ CD68+, CD3+, ICAM-1 ↑ collagen content | Overexpression: HMEC-1 ↓ cell viability, migration, tube formation ↑ apoptosis via miR-147 suppression Deficiency: VSMCs ↑ proliferation ↓ apoptosis ↓ ABCA1 via ↓ miR-361-5p suppression |
| FA2H-2 | Fatty acid 2-hydroxylase 2 | ↓ atherosclerotic plaque | Deficiency: ApoE−/− + LV-si-lncRNA-FA2H-2 on WD ↑ autophagy flux ↑ inflammatory response ↑ increased lesion area | Deficiency: ECs and SMCs (oxLDL) ↑ autophagy flux ↑ increased inflammatory response |
| Author (year) | LncRNA | Definition | Regulation by Statin | Findings | Implications |
|---|---|---|---|---|---|
| Mitchel et al. (2016) [43] | RP1-13D10.2 | N/A | Simvastatin: Upregulation of RP1-13D10.2 in high responders to statin | -Statin induced expression of RP1-13D10.2 in lymphoblastoid cell lines was higher in the high vs. low responders -RP1-13D10.2 increased LDLR expression and stimulated LDL uptake | RP1-13D10.2 regulates LDLR and may contribute to LDLC response to statin treatment |
| Li et al. (2019) [13] | LASER | Lipid-associated single nucleotide polymorphism gene region | Atorvastatin: Upregulation of LASER in a dose-dependent manner | -Statin treatment increased LASER expression in HepG2 cells -LASER expression in HepG2 cells was positively correlated with plasma PCSK9 levels in statin-free patients -HNF-1α and PCSK9 were reduced after LASER knockdown in HepG2 cells | Targeting LASER might be an effective approach to enhance the effect of statins |
| Paez et al. (2020) [44] | ARSR CHROME LASER | ARSR: Activated in renal cell carcinoma (RCC) with sunitinib resistance CHROME: cholesterol homeostasis regulator of miRNA expression LASER: lipid-associated single nucleotide polymorphism gene region | Atorvastatin: Upregulation of ARSR and CHROME | -Statin increased the expression of lncRNAs ARSR and CHROME but not LASER in peripheral blood of hypercholesterolemic patients | Statins differentially regulate the expression of cholesterol-related lncRNAs |
| Su et al. (2018) [45] | MEG3 | Maternally expressed gene 3 | Atorvastatin: Downregulation of MEG3 | -Atorvastatin protected cardiac progenitor cells (CPCs) from hypoxia-induced injury through inhibiting MEG3 expression -Atorvastatin protected CPCs from hypoxia-induced injury through modulating the MEG3/miR-22/HMGB1 axis. | Molecular mechanism of atorvastatin under hypoxia may provide a target for developing effective drugs for MI patients |
| Huang et al. (2020) [46] | H19 | N/A | Atorvastatin: Upregulation of H19 | -MSCATV-Exo resulted in improved recovery in cardiac function and reduced cardiomyocyte apoptosis compared to MSC-Exo. -MSCATV-Exo exhibited a significantly increased level of lncRNA H19 expression. -Silencing lncRNA H19 abolished the cardioprotective effects of MSCATV-Exo in a rat model of acute myocardial infarction | LncRNA H19 might mediate the cardioprotective effects of MSCATV-Exo on acutely infarcted hearts |
| Wu et al. (2020) [22] | NEXN-AS1 | Nexilin F-actin binding protein antisense RNA 1 | Atorvastatin: Upregulation of NEXN-AS1 in a dose- and time-dependent manner | -Atorvastatin upregulated lncRNA NEXN-AS1 and NEXN in HUVEC -Atorvastatin inhibited the canonical inflammasome pathway biomarkers of pyroptosis -Inhibition of pyroptosis was diminished by knockdown of lncRNA NEXN-AS1 in HUVEC | Regulation of pyroptosis through lncRNA NEXN might be a potential target against atherosclerosis |
| Leisegang et al. (2019) [23] | MANTIS | N/A | Cerivastatin, Fluvastatin, simvastatin, atorvastatin: Upregulation of MANTIS | -Statins upregulated lncRNA MANTIS in HUVEC and HAoEC -MANTIS limited the ICAM-1 expression in vivo in mice -expression of MANTIS in human carotid artery endarterectomy material was lower compared with healthy vessels -MANTIS was required to facilitate atorvastatin-induced changes in endothelial gene expression in HUVECs | Strategies to increase lncRNA MANTIS might improve vascular function in nonresponders to statin therapy |
| Josipovic et al. (2018) [47] | LISPR1 | Long intergenic noncoding RNA antisense to S1PR1 | Cerivastatin, Fluvastatin, simvastatin, atorvastatin: Upregulation of LISPR1 | -LISPR1 was downregulated in EC with vascular pathologies -LISPR1 was induced by statins in HUVECs | LISPR1 might be a potential target for statin-resistant patients |
| Tang et al. (2020) [48] | AWPPH | LncRNA associated with poor prognosis of hepatocellular carcinoma | Rosuvastatin, atorvastatin: Downregulation of AWPPH | -LncRNA AWPPH was highly expressed in CAD patients -Expression of LncRNA AWPPH was reduced with statin treatment, especially with rosuvastatin in CAD patients | LncRNA AWPPH can be a potential serum marker to predict prognosis of patients with CAD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsilimigras, D.I.; Bibli, S.-I.; Siasos, G.; Oikonomou, E.; Perrea, D.N.; Filis, K.; Tousoulis, D.; Sigala, F. Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis. Biomolecules 2021, 11, 623. https://doi.org/10.3390/biom11050623
Tsilimigras DI, Bibli S-I, Siasos G, Oikonomou E, Perrea DN, Filis K, Tousoulis D, Sigala F. Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis. Biomolecules. 2021; 11(5):623. https://doi.org/10.3390/biom11050623
Chicago/Turabian StyleTsilimigras, Diamantis I., Sofia-Iris Bibli, Gerasimos Siasos, Evangelos Oikonomou, Despina N. Perrea, Konstantinos Filis, Dimitrios Tousoulis, and Fragiska Sigala. 2021. "Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis" Biomolecules 11, no. 5: 623. https://doi.org/10.3390/biom11050623
APA StyleTsilimigras, D. I., Bibli, S.-I., Siasos, G., Oikonomou, E., Perrea, D. N., Filis, K., Tousoulis, D., & Sigala, F. (2021). Regulation of Long Non-Coding RNAs by Statins in Atherosclerosis. Biomolecules, 11(5), 623. https://doi.org/10.3390/biom11050623








