Antimicrobial Photodynamic Inactivation Affects the Antibiotic Susceptibility of Enterococcus spp. Clinical Isolates in Biofilm and Planktonic Cultures
Abstract
1. Introduction
2. Results
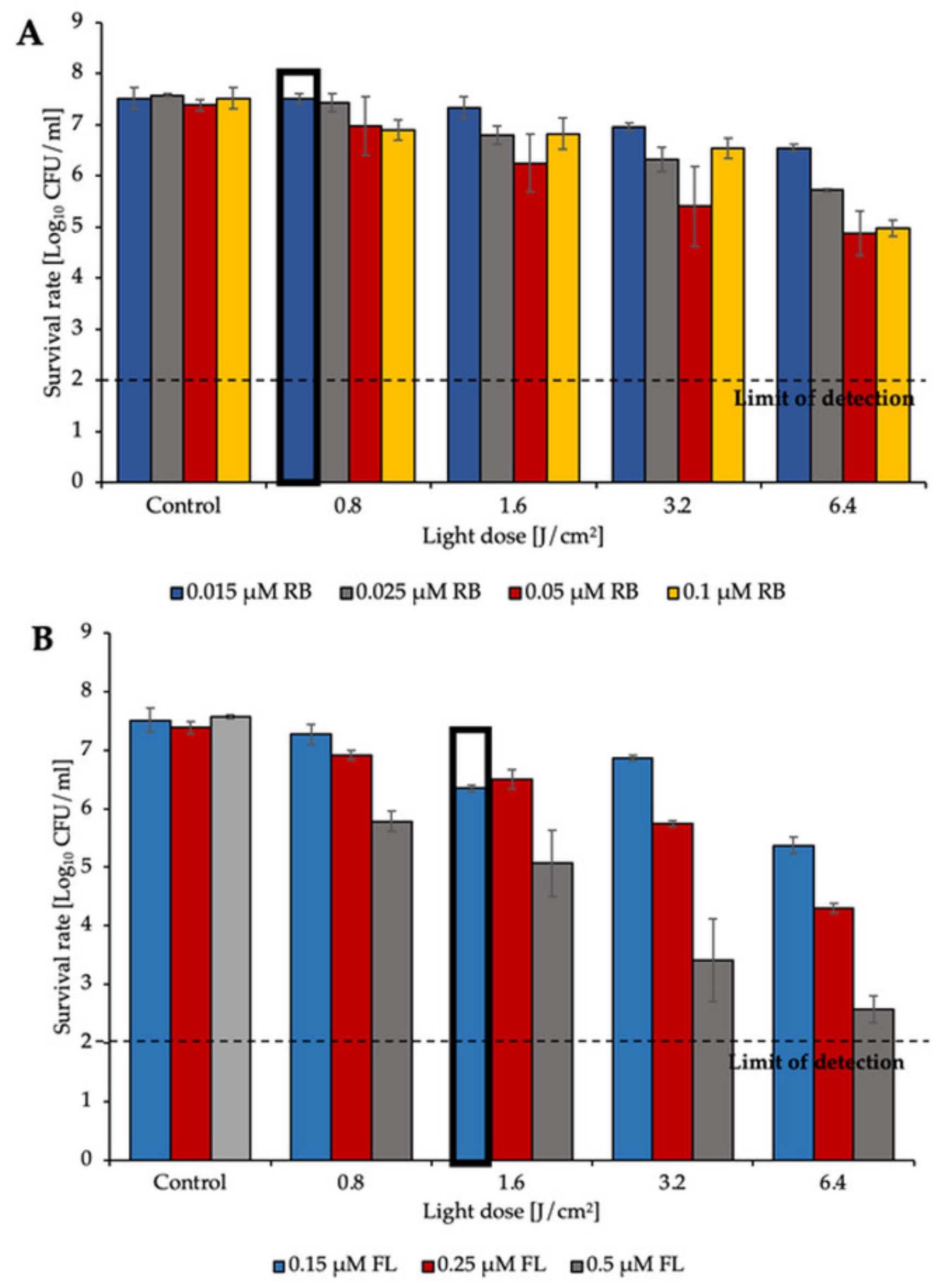
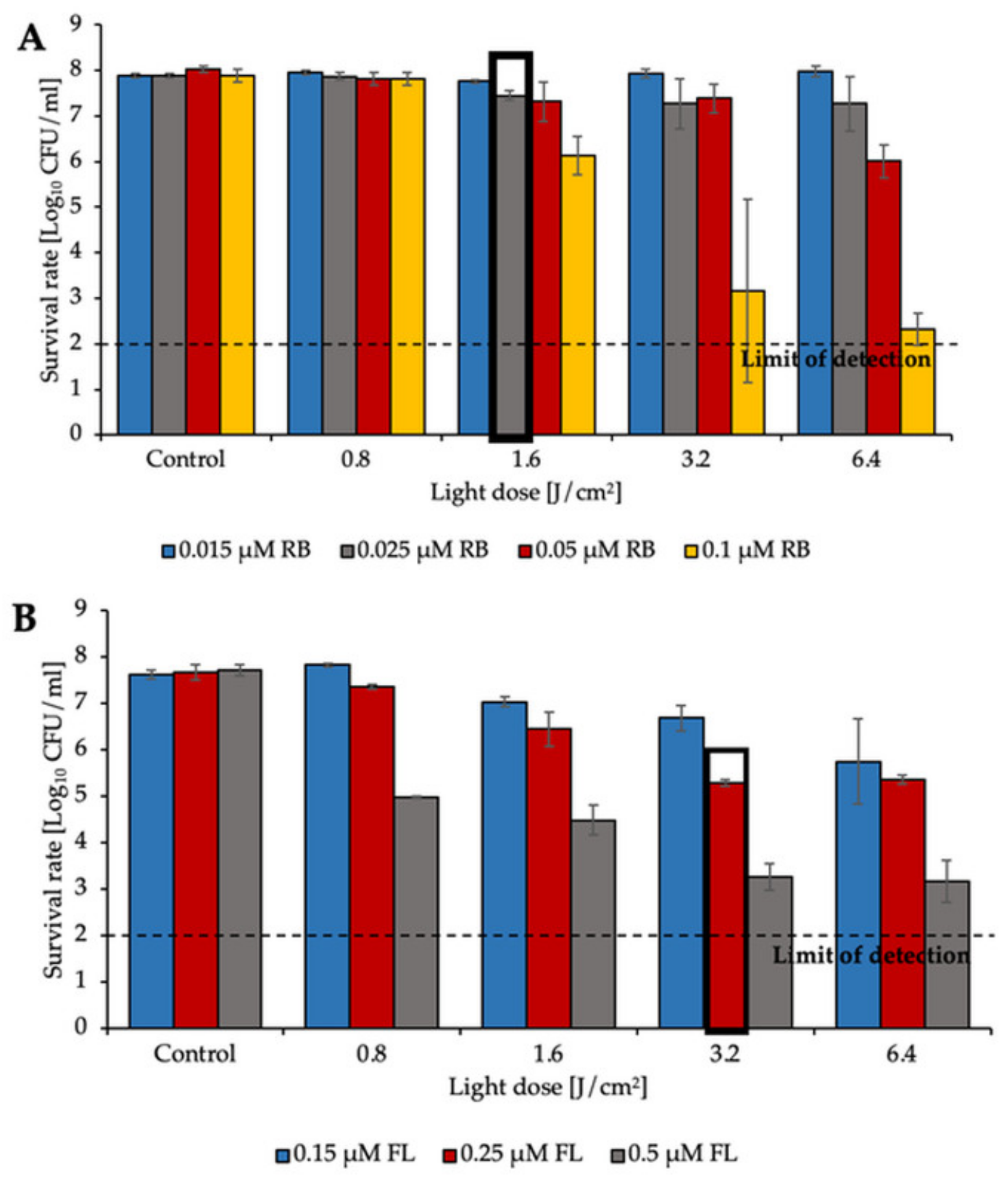
2.1. aPDI Significantly Influences the Survival Rate of Planktonic Cultures of Enterococcus Species
2.2. Identification of MIC of Treatments
2.3. Diffusion-Based Assays Confirm aPDI/Antimicrobial Synergy
2.4. Serial Dilution Methods Demonstrate aPDI/Antimicrobial Synergy
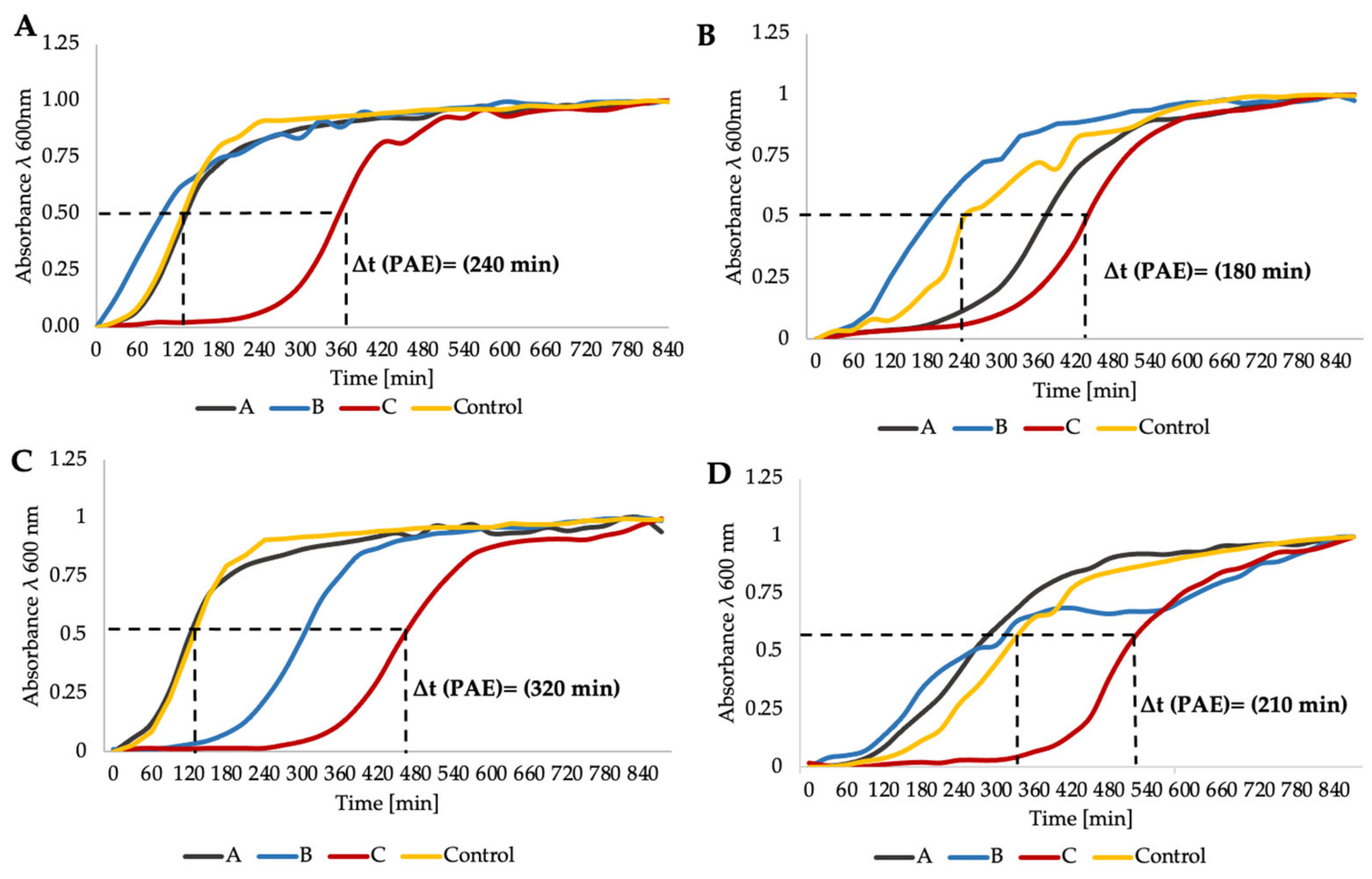
2.5. Time–Kill Curve Assay Confirms aPDI/Antimicrobial Synergy
2.6. aPDI/Antimicrobials Exerts Numerous Synergies
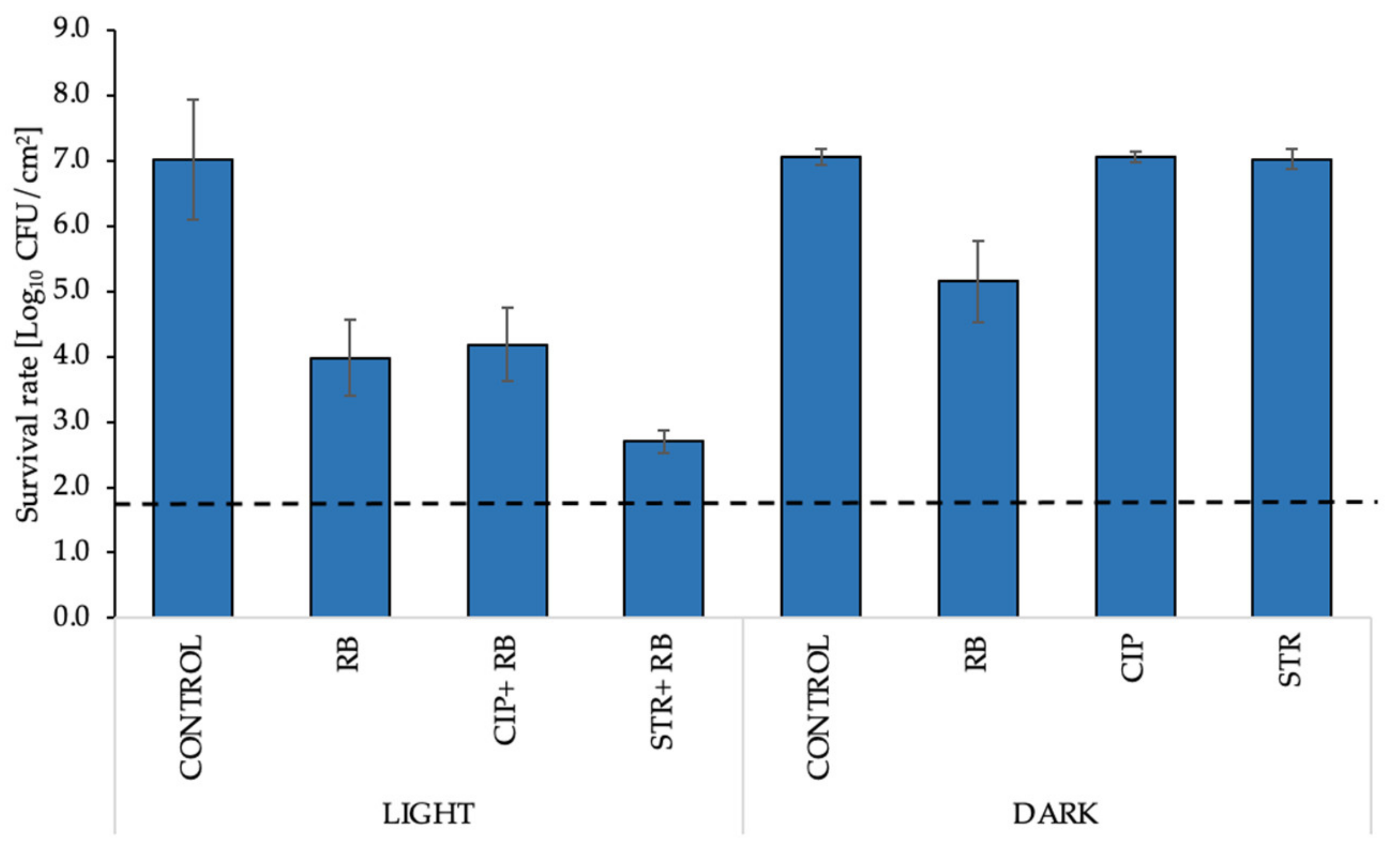
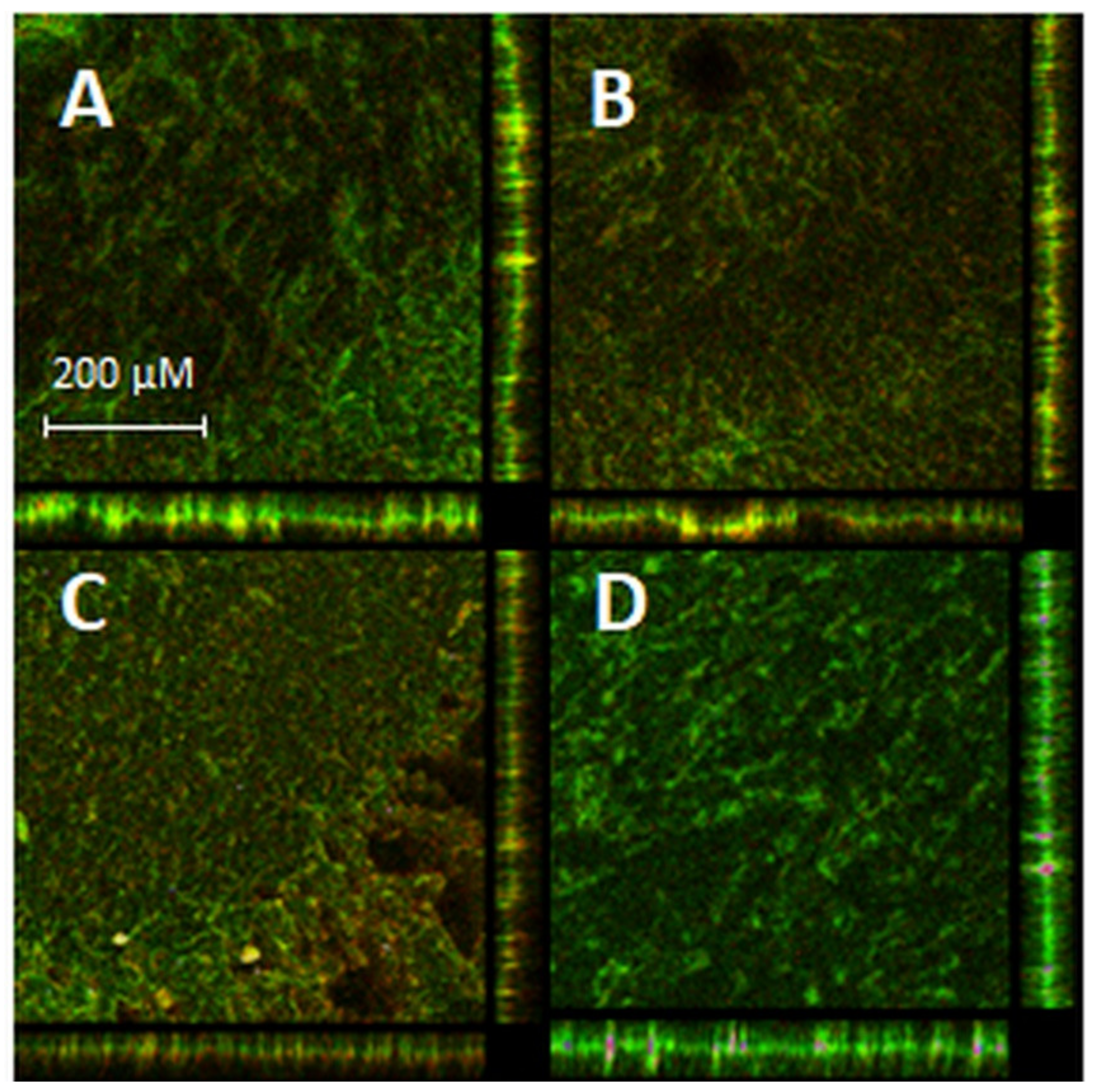
2.7. aPDI/Antimicrobial Synergy Can Be Reached in the Mature Biofilm Model
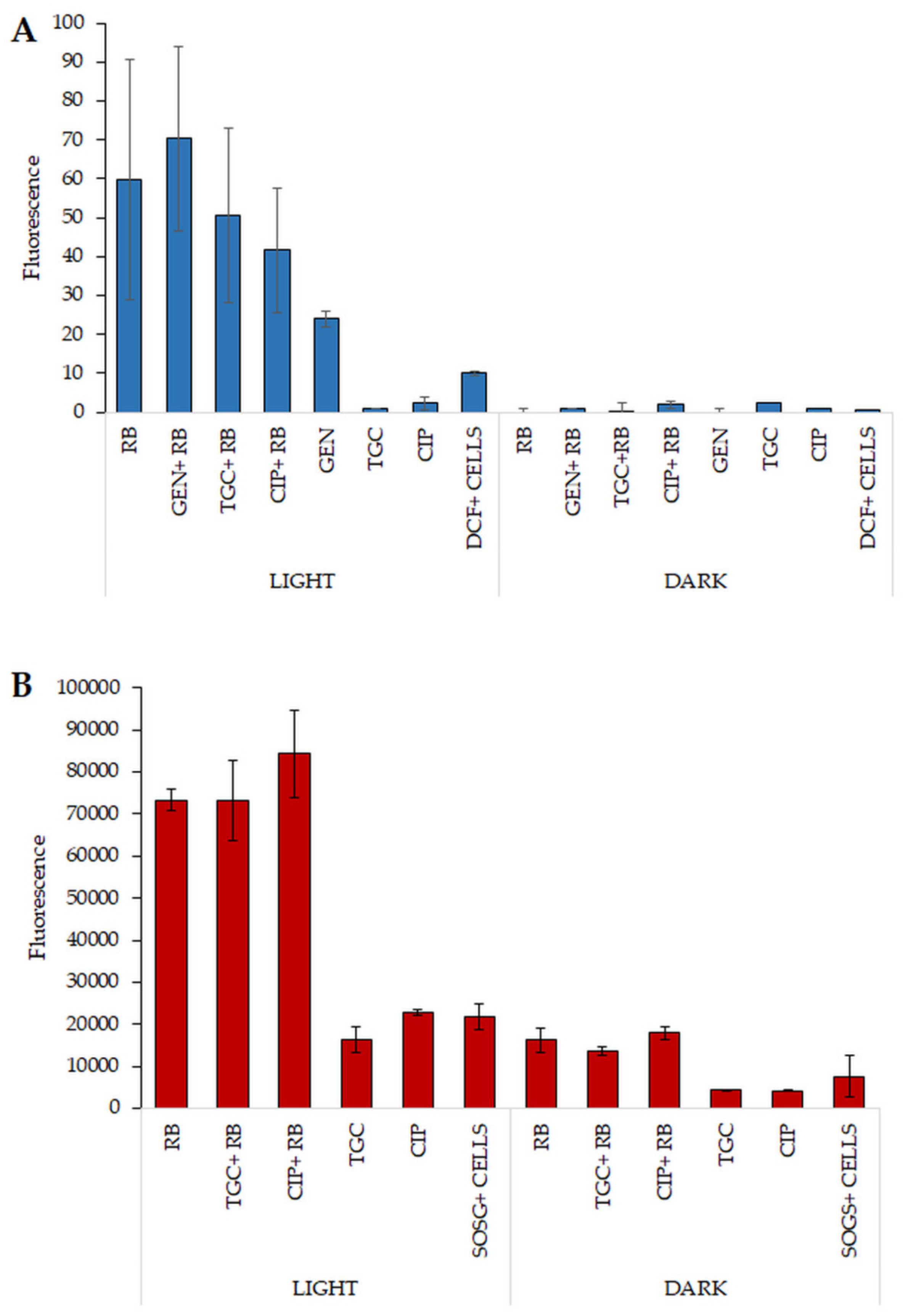
2.8. Increased ROS Generation Can Explain the Mechanism Underlying the Observed Synergies
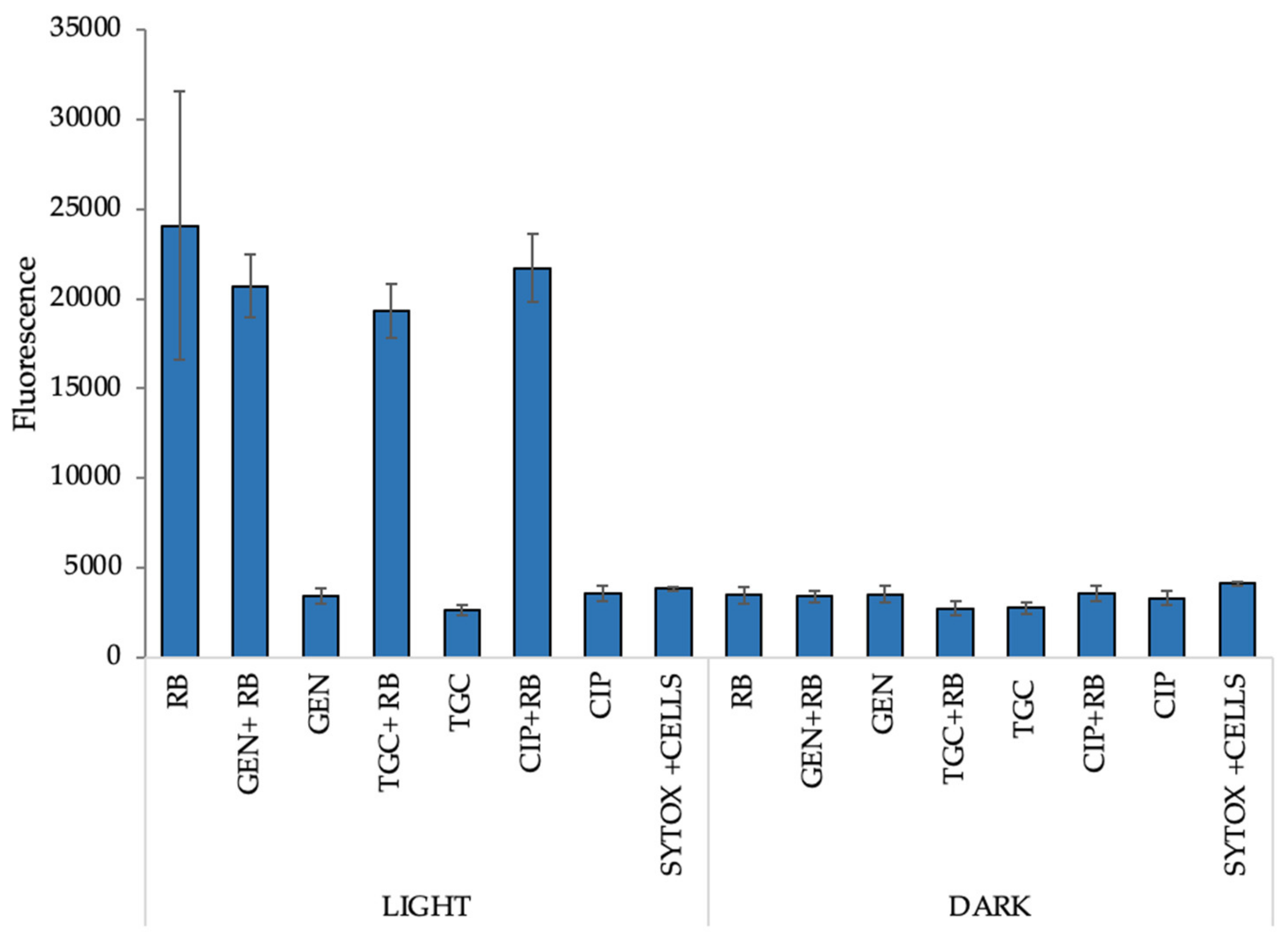
2.9. Increased Permeabilization Could Explain the Mechanism of the Observed Synergies
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Photosensitizers
4.3. Antibiotics
4.4. Light Sources
4.5. Photodynamic Inactivation of Planktonic Cultures
4.6. Determination of Sublethal and Lethal Doses of aPDI for Planktonic Cultures
4.7. Determination of MIC Doses of aPDI
4.8. MIC Determination of Tested Antibiotics
4.9. Synergy Testing
4.9.1. Antimicrobial Susceptibility Testing (Disk Diffusion Method and E-Tests)
4.9.2. Checkerboard Assay
4.9.3. Postantibiotic Effect
4.10. Determination of Singlet Oxygen Production
4.11. Determination of Production of ROS/Radicals
4.12. Cell Membrane Integrity Assay
4.13. Materials and Methods Referring to Biofilm Culture
4.13.1. Biofilm Culture Conditions
4.13.2. Biofilm Treatment
4.13.3. Biofilm Visualization
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Růžičková, M.; Vítězová, M.; Kushkevych, I. The characterization of Enterococcus genus: Resistance mechanisms and inflammatory bowel disease. Open Med. 2020, 15, 211–224. [Google Scholar] [CrossRef]
- Massier, S.; Bouffartigues, E.; Rincé, A.; Maillot, O.; Feuilloley, M.G.J.; Orange, N.; Chevalier, S. Effects of a pulsed light-induced stress on Enterococcus faecalis. J. Appl. Microbiol. 2013, 114, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Nakonieczna, J.; Wozniak, A.; Pieranski, M.; Rapacka-Zdonczyk, A.; Ogonowska, P.; Grinholc, M. Photoinactivation of ESKAPE pathogens: Overview of novel therapeutic strategy. Future Med. Chem. 2019, 11, 443–461. [Google Scholar] [CrossRef]
- Cheng, X.; Tian, Y.; Zhao, C.; Qu, T.; Ma, C.; Liu, X.; Yu, Q. Bactericidal effect of strong acid electrolyzed water against flow enterococcus faecalis biofilms. J. Endod. 2016, 42, 1120–1125. [Google Scholar] [CrossRef]
- Cieplik, F.; Deng, D.; Crielaard, W.; Buchalla, W.; Hellwig, E.; Al-Ahmad, A.; Maisch, T. Antimicrobial photodynamic therapy—What we know and what we don’t. Crit. Rev. Microbiol. 2018, 7828, 1–19. [Google Scholar] [CrossRef]
- Maisch, T.; Hackbarth, S.; Regensburger, J.; Felgenträger, A.; Bäumler, W.; Landthaler, M.; Röder, B. Photodynamische inaktivierung von multiresistenten bakterien (PIB)—Ein neuer ansatz zur behandlung oberflächlicher infektionen im 21. jahrhundert. JDDG J. Ger. Soc. Dermatology 2011, 9, 360–367. [Google Scholar] [CrossRef]
- Pidot, S.J.; Gao, W.; Buultjens, A.H.; Monk, I.R.; Guerillot, R.; Carter, G.P.; Lee, J.Y.H.; Lam, M.M.C.; Grayson, M.L.; Ballard, S.A.; et al. Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci. Transl. Med. 2018, 10, eaar6115. [Google Scholar] [CrossRef] [PubMed]
- Sassoubre, L.M.; Nelson, K.L.; Boehm, A.B. Mechanisms for photoinactivation of Enterococcus faecalis in seawater. Appl. Environ. Microbiol. 2012, 78, 7776–7785. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vatansever, F.; de Melo, W.C.M.A.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial strategies centered around reactive oxygen species—Bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.R.; Viveiros, J.; Yang, C.; Ahmadi, A.; Ganz, R.A.; Tolkoff, M.J. Helicobacter pylori accumulates photoactive porphyrins and is killed by visible light. Antimicrob. Agents Chemother. 2005, 49, 2822–2827. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.; Rapacka-Zdonczyk, A.; Mutters, N.T.; Grinholc, M. Antimicrobials Are a Photodynamic Inactivation Adjuvant for the Eradication of Extensively Drug-Resistant Acinetobacter baumannii. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Imlay, J.A. Cell death from antibiotics without the involvement of reactive oxygen species. Science 2013, 339, 1210–1213. [Google Scholar] [CrossRef]
- Grinholc, M.; Nakonieczna, J.; Fila, G.; Taraszkiewicz, A.; Kawiak, A.; Szewczyk, G.; Sarna, T.; Lilge, L.; Bielawski, K.P. Antimicrobial photodynamic therapy with fulleropyrrolidine: Photoinactivation mechanism of Staphylococcus aureus, in vitro and in vivo studies. Appl. Microbiol. Biotechnol. 2015, 99, 4031–4043. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, F.L.; Willems, R.J.L.W.; Jansen, P.; Hendrickx, A.; Zhang, X.; Bonten, M.J.M.B.; Leavis, H.L. Enterococcus faecium biofilm formation: Identification of major autolysin AtlAefm, associated acm surface localization, and AtlAefm-independent extracellular DNA release. MBio 2013, 4. [Google Scholar] [CrossRef]
- Zhang, C.; Du, J.; Peng, Z. Correlation between Enterococcus faecalis and Persistent Intraradicular Infection Compared with Primary Intraradicular Infection: A Systematic Review. J. Endod. 2015, 41, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Tavares, A.; Carvalho, C.; Faustino, M.A.; Neves, M.G.; Tomé, J.P.; Tomé, A.C.; Cavaleiro, J.A.; Cunha, Â.; Gomes, N.; Alves, E.; et al. Antimicrobial photodynamic therapy: Study of bacterial recovery viability and potential development of resistance after treatment. Mar. Drugs 2010, 8, 91–105. [Google Scholar] [CrossRef]
- Radunović, M.; Petrini, M.; Vlajic, T.; Iezzi, G.; Di Lodovico, S.; Piattelli, A.; D’Ercole, S. Effects of a novel gel containing 5-aminolevulinic acid and red LED against bacteria involved in peri-implantitis and other oral infections. J. Photochem. Photobiol. B Biol. 2020, 205. [Google Scholar] [CrossRef] [PubMed]
- Chibebe, J., Jr.; Fuchs, B.B.; Sabino, C.P.; Junqueira, J.C.; Jorge, A.O.C.; Ribeiro, M.S.; Gilmore, M.S.; Rice, L.B.; Tegos, G.P.; Hamblin, M.R.; et al. Photodynamic and Antibiotic Therapy Impair the Pathogenesis of Enterococcus faecium in a Whole Animal Insect Model. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Kang, S.M.; Jung, H.I.; Kim, B.I. Susceptibility of oral bacteria to antibacterial photodynamic therapy. J. Oral Microbiol. 2019, 11. [Google Scholar] [CrossRef]
- López-Jiménez, L.; Fusté, E.; Martínez-Garriga, B.; Arnabat-Domínguez, J.; Vinuesa, T.; Viñas, M. Effects of photodynamic therapy on Enterococcus faecalis biofilms. Lasers Med. Sci. 2015, 30, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Garcez, A.S.; Núñez, S.C.; Azambuja, N.; Fregnani, E.R.; Rodriguez, H.M.H.; Hamblin, M.R.; Suzuki, H.; Ribeiro, M.S. Effects of photodynamic therapy on gram-positive and gram-negative bacterial biofilms by bioluminescence imaging and scanning electron microscopic analysis. Photomed. Laser Surg. 2013, 31, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.B.; Tessare, P.O.; Pallota, R.C.; Filho, H.F.; Denardin, O.V.P.; Rapoport, A.; Dedivitis, R.A.; Veronezi, J.F.; Genovese, W.J.; Ricardo, A.L.F. Photodynamic therapy for root canals infected with Enterococcus faecalis. Photomed. Laser Surg. 2008, 26, 209–213. [Google Scholar] [CrossRef]
- Shrestha, A.; Hamblin, M.R.; Kishen, A. Characterization of a conjugate between rose bengal and chitosan for targeted antibiofilm and tissue stabilization effects as a potential treatment of infected dentin. Antimicrob. Agents Chemother. 2012, 56, 4876–4884. [Google Scholar] [CrossRef] [PubMed]
- Manoil, D.; Filieri, A.; Schrenzel, J.; Bouillaguet, S. Rose bengal uptake by E. faecalis and F. nucleatum and light-mediated antibacterial activity measured by flow cytometry. J. Photochem. Photobiol. B Biol. 2016, 162, 258–265. [Google Scholar] [CrossRef]
- Wozniak, A.; Grinholc, M. Combined antimicrobial activity of photodynamic inactivation and antimicrobials-state of the art. Front. Microbiol. 2018, 9, 930. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococc. In Expert Review of Anti-Infective Therapy; Expert Reviews Ltd. (United Kingdom): London, UK, 2014; Volume 12, pp. 1221–1236. [Google Scholar]
- Fiedler, S.; Bender, J.K.; Klare, I.; Halbedel, S.; Grohmann, E.; Szewzyk, U.; Werner, G. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 2016, 71, 871–881. [Google Scholar] [CrossRef]
- Gagetti, P.; Bonofiglio, L.; García Gabarrot, G.; Kaufman, S.; Mollerach, M.; Vigliarolo, L.; von Specht, M.; Toresani, I.; Lopardo, H.A. Resistance to β-lactams in enterococci. Rev. Argent. Microbiol. 2019, 51, 179–183. [Google Scholar] [CrossRef]
- Ono, S.; Muratani, T.; Matsumoto, T. Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis. Antimicrob. Agents Chemother. 2005, 49, 2954–2958. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Mukherjee, M. Reactive oxygen species and uspA overexpession: An alternative bacterial response toward selection and maintenance of multidrug resistance in clinical isolates of uropathogenic E. coli. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Memar, M.Y.; Ghotaslou, R.; Samiei, M.; Adibkia, K. Antimicrobial use of reactive oxygen therapy: Current insights. Infect. Drug Resist. 2018, 11, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Pieranski, M.; Sitkiewicz, I.; Grinholc, M. Increased photoinactivation stress tolerance of Streptococcus agalactiae upon consecutive sublethal phototreatments. Free Radic. Biol. Med. 2020, 160, 657–669. [Google Scholar] [CrossRef]
- Rapacka-Zdonczyk, A.; Wozniak, A.; Pieranski, M.; Woziwodzka, A.; Bielawski, K.P.; Grinholc, M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci. Rep. 2019, 9, 9423. [Google Scholar] [CrossRef] [PubMed]
- Di Poto, A.; Sbarra, M.S.; Provenza, G.; Visai, L.; Speziale, P. The effect of photodynamic treatment combined with antibiotic action or host defence mechanisms on Staphylococcus aureus biofilms. Biomaterials 2009, 30, 3158–3166. [Google Scholar] [CrossRef] [PubMed]

| Antibiotic Target | Antimicrobial Category | Antibiotic | E. faecalis | E. faecium |
|---|---|---|---|---|
| MIC (μg/mL) | MIC (μg/mL) | |||
| Protein synthesis (30S) | Aminoglycosides | Gentamycin | 64 | 32 |
| Tetracyclines | Doxycycline | 16/8 | 32 | |
| Streptomycin | Streptomycin | 256 (R) | 1024 (R) | |
| Glycylcyclines | Tigecycline | 1 (R) | 8/4 (R) | |
| 70S initiation complex | Oxazolidinones | Linezolid | 2 | 1 |
| DNA gyrase | Fluoroquinolones | Ciprofloxacin | 2 | 2 |
| Cell-wall synthesis | Carbapenems Glycopeptides Penicillins | Imipenem Vancomycin Ampicillin | 1/0.5 1 >64 (R) | 16/8 (R) 2 >1024 (R) |
| Cell membrane | Lipopeptides | Daptomycin | 128 (R) | 64 (R) |
| Phototherapy | aPDI (FL) | Green light + FL | 28.6 J/cm2 + 10 μM FL (3.6 J/cm2 + 0.625 μM FL) 1 | 28.6 J/cm2 + 10 μM FL (3.6 J/cm2 + 0.312 μM FL) |
| aPDI (RB) | Green light + RB | 15.9 J/cm2 + 1 μM RB (7.95 J/cm2 + 0.5 μM RB) | 15.9 J/cm2 + 1 μM RB (15.9 J/cm2 + 0.5 μM RB) |
| Antibiotic | E. faecalis | E. faecium | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | aPDI (RB) | aPDI (FL) | Control | aPDI (RB) | aPDI (FL) | |||||||
| DF 1 | E-Test | DF | E-Test | DF | E-Test | DF | E-test | DF | E-Test | DF | E-Test | |
| GEN | 10 2 | 12 3 | 9.8 | 8 | 10.8 | 8 | 16.5 | 6 | 17.8 | 3 | 15.8 | 4 |
| STR | 11 | 256 | 13.9 | 128 4 | 15 | 128 | 10 | 1024 | 10.9 | ≥1024 | 8.6 | ≥1024 |
| TGC | 22.4 | 19 | 24.7 | 0.64 | 25.5 | ≥256 | 28.5 | 0.064 | 32.6 | 0.047 | 26 | 0.64 |
| DOX | 9.4 | 32 | 11.3 | 16 | 10.9 | 16 | 13 | 32 | 10.9 | 32 | 11.6 | 16 |
| LZD | 24.7 | 2 | 24 | 1.5 | 25.9 | 2 | 29.6 | 1 | 31 | 0.75 | 28.7 | 1 |
| CIP | 20.5 | 0.75 | 21.8 | 0.5 | 22.7 | 0.5 | 22.1 | 0.5 | 22.7 | 0.5 | 21.4 | 0.5 |
| IMP | 29.3 | 0.75 | 31.2 | 0.75 | 29.5 | 0.75 | 10.5 | 32 | 6 | 32 | 8.9 | ≥32 |
| VAN | 13.4 | 2 | 14 | 2 | 13.8 | 2 | 21 | 0.5 | 21.1 | 0.38 | 18 | 0.38 |
| AMP | 8.4 | 0.5 | 12.2 | 0.5 | 11 | 0.75 | 6 | 2 | 6 | 3 | 6 | - |
| DAP | - | 1 | - | 1 | - | 1 | - | 2 | - | 1.5 | - | 1.5 |
| Q-D | 11.5 | - | 12.1 | - | 12.2 | - | 17.7 | - | 16.1 | - | 17.7 | - |
| Antibotic | E. faecalis | E. faecium | ||
|---|---|---|---|---|
| aPDI (RB) | aPDI (FL) | aPDI (RB) | aPDI (FL) | |
| GEN | 0.38 1 | >0.5 | 0.5 | >0.5 |
| STR | >0.5 | >0.5 | >0.5 | >0.5 |
| TGC | >0.5 | >0.5 | >0.5 | >0.5 |
| DOX | >0.5 | >0.5 | >0.5 | >0.5 |
| LZD | >0.5 | >0.5 | >0.5 | 0.5 |
| CIP | 0.38 | >0.5 | 0.5 | >0.5 |
| IMP | >0.5 | 0.25 | >0.5 | >0.5 |
| VAN | >0.5 | >0.5 | 8.5 | >0.5 |
| AMP | >0.5 | >0.5 | >0.5 | >0.5 |
| DAP | 0.16 | >0.5 | 5.25 | >0.5 |
| Q-D | - | - | - | - |
| Antibiotic | aPDI (RB) | aPDI (FL) | ||||||
|---|---|---|---|---|---|---|---|---|
| DF 1 | E-Test | Checkerboard Assay | PAE | DF | E-Test | Checkerboard Assay | PAE | |
| GEN | - | + | + | + | - | - | - | + |
| STR | - | - | - | + | - | - | - | +/- |
| TGC | + | - | - | + | -- | -- | - | + |
| DOX | - | - | - | +/- | - | + | - | + |
| LZD | - | - | - | - | - | - | + | + |
| CIP | - | - | + | - | - | - | - | + |
| IMP | -- | - | - | - | - | - | - | + |
| VAN | - | - | -- | - | -- | - | - | + |
| AMP | - | - | - | - | - | - | - | +/- |
| DAP | - | -- | +/- | - | - | +/- | ||
| Q-D | - | ND | ND | - | ND | ND | ||
| Antibiotic | aPDI (RB) | aPDI (FL) | ||||||
|---|---|---|---|---|---|---|---|---|
| DF 1 | E-Test | Checkerboard Assay | PAE | DF | E-Test | Checkerboard Assay | PAE | |
| GEN | - | - | + | - | - | - | - | + |
| STR | + | + | - | + | + | + | - | + |
| TGC | + | + | - | + | + | -- | - | + |
| DOX | +/- | + | - | + | - | + | - | + |
| LZD | - | - | - | - | - | - | - | + |
| CIP | - | - | + | - | + | - | - | + |
| IMP | +/- | - | - | - | - | - | + | + |
| VAN | - | - | - | - | - | -- | + | + |
| AMP | + | - | - | + | + | - | - | - |
| DAP | - | + | - | - | + | |||
| Q-D | - | ND | ND | - | - | ND | ND | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woźniak, A.; Kruszewska, B.; Pierański, M.K.; Rychłowski, M.; Grinholc, M. Antimicrobial Photodynamic Inactivation Affects the Antibiotic Susceptibility of Enterococcus spp. Clinical Isolates in Biofilm and Planktonic Cultures. Biomolecules 2021, 11, 693. https://doi.org/10.3390/biom11050693
Woźniak A, Kruszewska B, Pierański MK, Rychłowski M, Grinholc M. Antimicrobial Photodynamic Inactivation Affects the Antibiotic Susceptibility of Enterococcus spp. Clinical Isolates in Biofilm and Planktonic Cultures. Biomolecules. 2021; 11(5):693. https://doi.org/10.3390/biom11050693
Chicago/Turabian StyleWoźniak, Agata, Beata Kruszewska, Michał Karol Pierański, Michał Rychłowski, and Mariusz Grinholc. 2021. "Antimicrobial Photodynamic Inactivation Affects the Antibiotic Susceptibility of Enterococcus spp. Clinical Isolates in Biofilm and Planktonic Cultures" Biomolecules 11, no. 5: 693. https://doi.org/10.3390/biom11050693
APA StyleWoźniak, A., Kruszewska, B., Pierański, M. K., Rychłowski, M., & Grinholc, M. (2021). Antimicrobial Photodynamic Inactivation Affects the Antibiotic Susceptibility of Enterococcus spp. Clinical Isolates in Biofilm and Planktonic Cultures. Biomolecules, 11(5), 693. https://doi.org/10.3390/biom11050693






