mTOR Signaling in Metabolic Stress Adaptation
Abstract
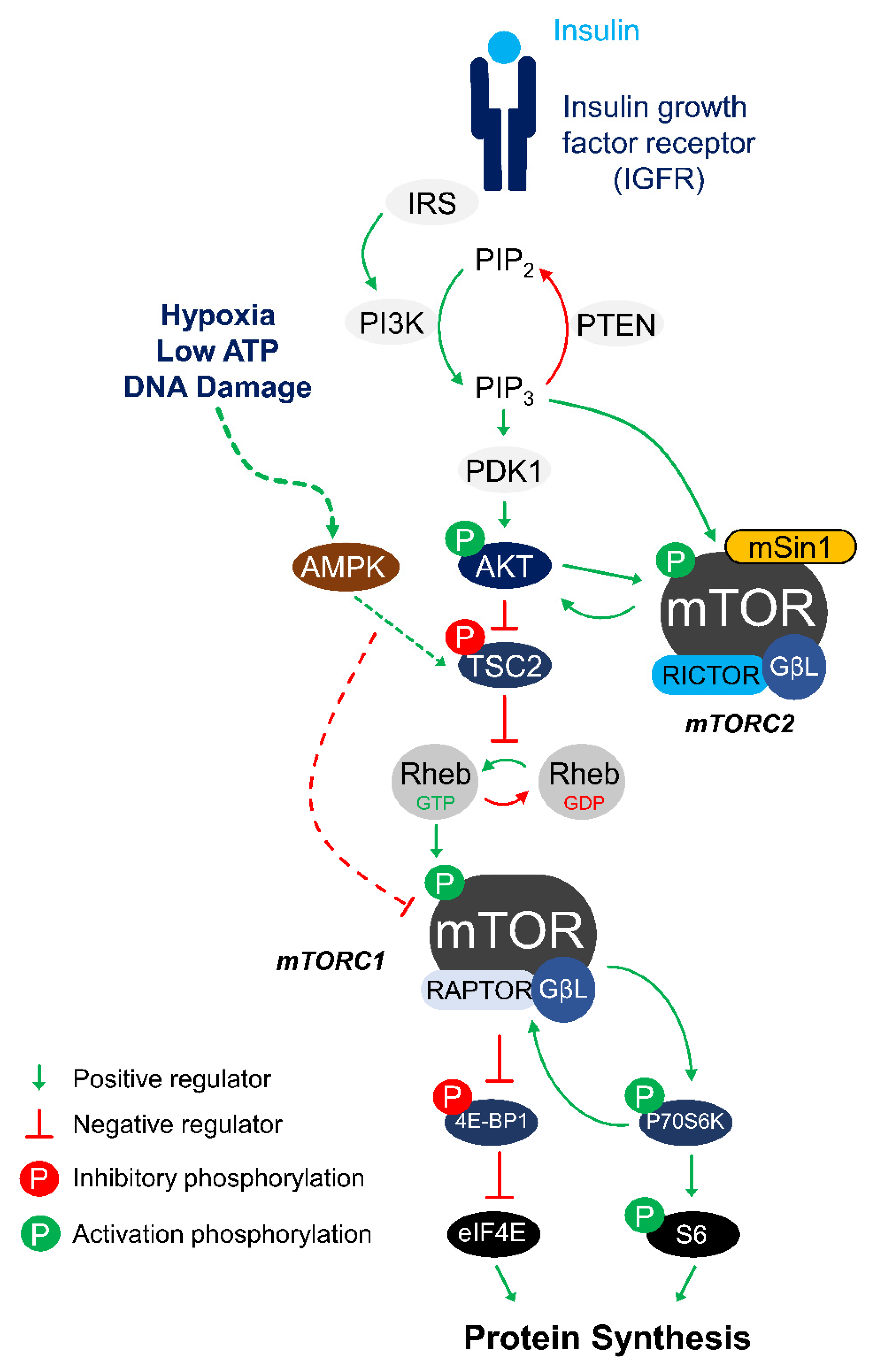
1. Historical Background
2. mTORC1 Control of Cell Growth under Metabolic Stress
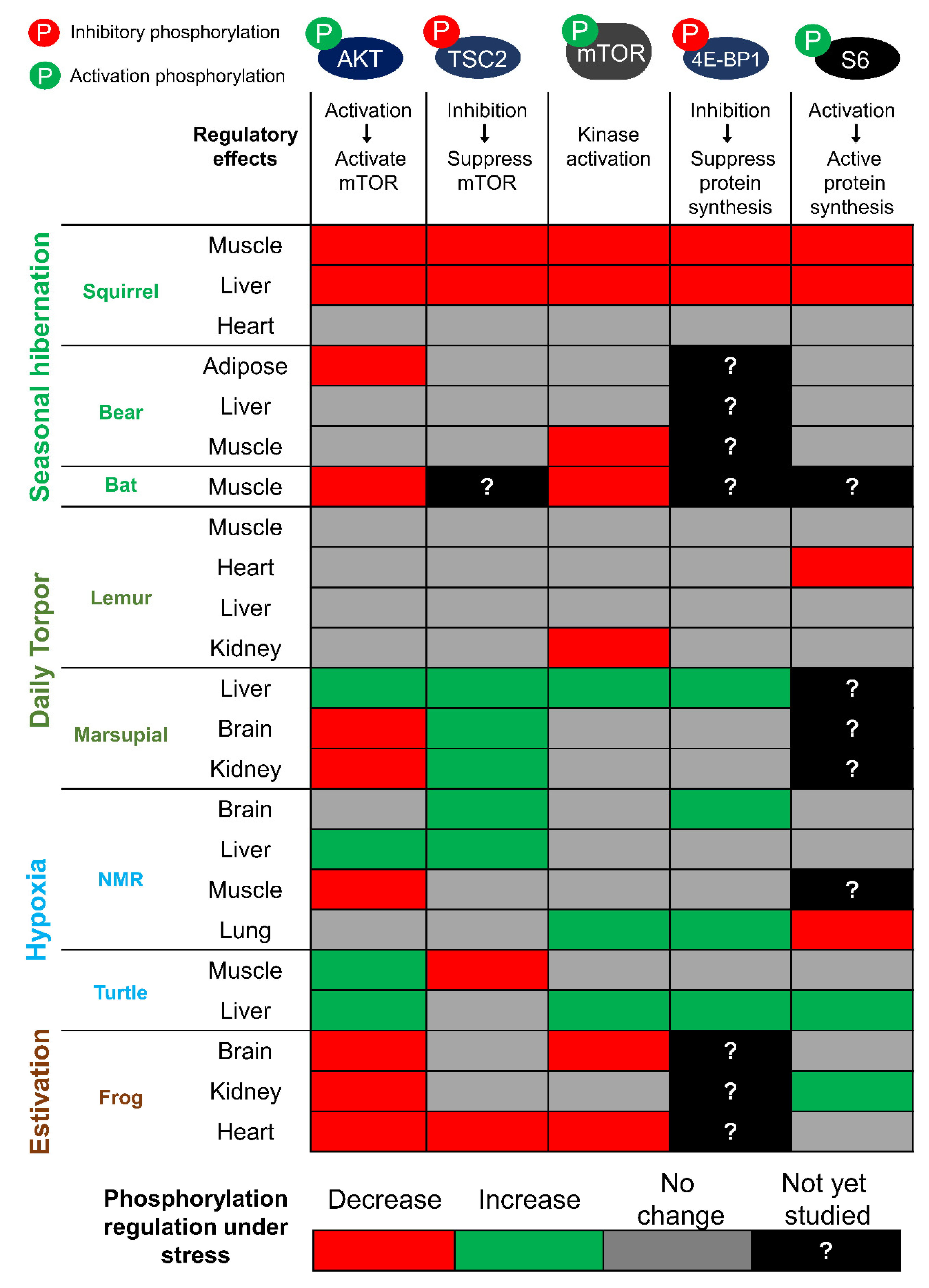
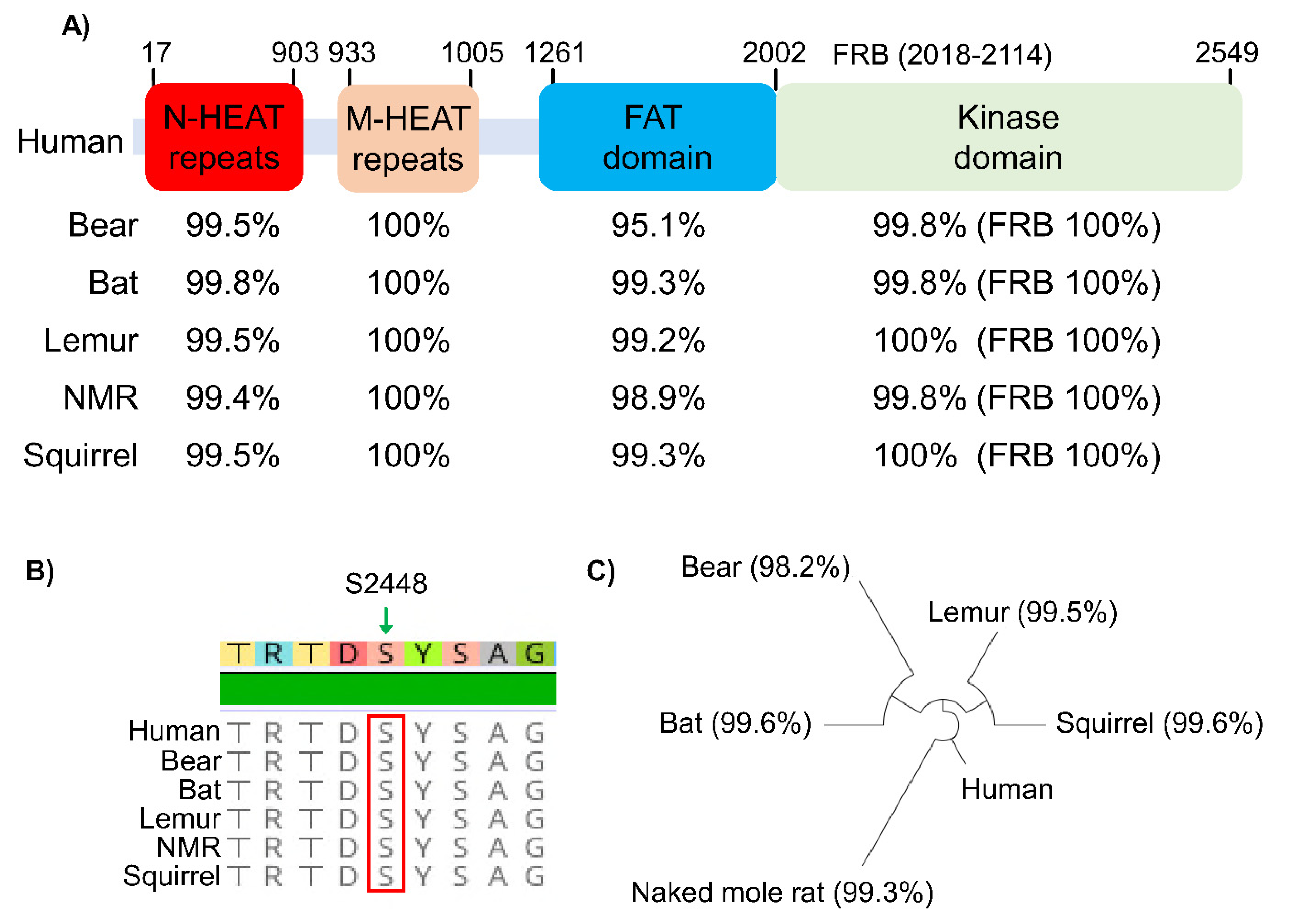
2.1. mTORC1 Regulation in Hibernation
2.2. mTORC1 Regulation in Hypoxia/Anoxia Tolerant Models
2.3. mTORC1 Regulation in Animal Models of Estivation
2.4. mTORC1 in Genetic Models of Metabolic Arrest
3. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sehgal, S.N.; Baker, H.; Vézina, C. Rapamycin (Ay-22,989), a New Antifungal Antibiotic. II.; Fermentation, Isolation and Characterization. J. Antibiot. (Tokyo) 1975, 28, 727–732. [Google Scholar] [CrossRef]
- Martel, R.R.; Klicius, J.; Galet, S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can. J. Physiol. Pharmacol. 1977, 55, 48–51. [Google Scholar] [CrossRef]
- Eng, C.P.; Sehgal, S.N.; Vézina, C. Activity of rapamycin (ay-22,989) against transplanted tumors. J. Antibiot. (Tokyo) 1984, 37, 1231–1237. [Google Scholar] [CrossRef]
- Bierer, B.E.; Mattila, P.S.; Standaert, R.F.; Herzenberg, L.A.; Burakoff, S.J.; Crabtree, G.; Schreiber, S.L. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc. Natl. Acad. Sci. USA 1990, 87, 9231–9235. [Google Scholar] [CrossRef]
- Chung, J.; Kuo, C.J.; Crabtree, G.R.; Blenis, J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992, 69, 1227–1236. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science (80-) 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Katewa, S.D.; Kapahi, P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp. Gerontol. 2011, 46, 382–390. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Keith Blackwell, T.; Sewell, A.K.; Wu, Z.; Han, M. TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Genetics 2019, 213, 329–360. [Google Scholar] [CrossRef]
- Nojima, H.; Tokunaga, C.; Eguchi, S.; Oshiro, N.; Hidayat, S.; Yoshino, K.I.; Hara, K.; Tanaka, N.; Avruch, J.; Yonezawa, K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 2003, 278, 15461–15464. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic rate depression in animals: Transcriptional and translational controls. Biol. Rev. Camb. Philos. Soc. 2004, 79, 207–233. [Google Scholar] [CrossRef]
- Lee, K.; So, H.; Gwag, T.; Ju, H.; Lee, J.W.; Yamashita, M.; Choi, I. Molecular mechanism underlying muscle mass retention in hibernating bats: Role of periodic arousal. J. Cell Physiol. 2010, 222, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Storey, K.B. Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J. Exp. Biol. 2012, 215, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Szereszewski, K.E.; Storey, K.B. Translational regulation in the anoxic turtle, Trachemys scripta elegans. Mol. Cell Biochem. 2018, 242, 110653. [Google Scholar] [CrossRef] [PubMed]
- Al-attar, R.; Childers, C.L.; Nguyen, V.C.; Pamenter, M.E.; Storey, K.B. Differential protein phosphorylation is responsible for hypoxia-induced regulation of the Akt/mTOR pathway in naked mole rats. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2020, 242, 110653. [Google Scholar] [CrossRef]
- Wu, C.W.; Tessier, S.N.; Storey, K.B. Regulation of the insulin-Akt signaling pathway and glycolysis during dehydration stress in the African clawed frog Xenopus laevis. Biochem. Cell Biol. 2017, 95, 663–671. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Brugarolas, J.; Lei, K.; Hurley, R.L.; Manning, B.D.; Reiling, J.H.; Hafen, E.; Witters, L.A.; Ellisen, L.W.; Kaelin, W.G. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004, 18, 2893–2904. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Hu, W.; De Stanchina, E.; Teresky, A.K.; Jin, S.; Lowe, S.; Levine, A.J. The regulation of AMPK β1, TSC2, and PTEN expression by p53, Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007, 67, 3043–3053. [Google Scholar] [CrossRef] [PubMed]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [PubMed]
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jiang, X.; Li, B.; Yang, H.J.; Miller, M.; Yang, A.; Dhar, A.; Pavletich, N.P. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 2017, 552, 368–373. [Google Scholar] [CrossRef]
- Wu, C.W.; Storey, K.B. Life in the cold: Links between mammalian hibernation and longevity. Biomol. Concepts. 2016, 7, 41–52. [Google Scholar] [CrossRef]
- Storey, K.B. Out cold: Biochemical regulation of mammalian hibernation-A mini-review. Gerontology 2010, 56, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Frerichs, K.U.; Smith, C.B.; Brenner, M.; Degracia, D.J.; Krause, G.S.; Marrone, L.; Dever, T.E.; Hallenbeck, J.M. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc. Natl. Acad. Sci. USA 1998, 95, 14511–14516. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Manning, B.D.; Tee, A.R.; Logsdon, M.N.; Blenis, J.; Cantley, L.C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol. Cell 2002, 10, 151–162. [Google Scholar] [CrossRef]
- Garami, A.; Zwartkruis, F.J.T.; Nobukuni, T.; Joaquin, M.; Roccio, M.; Stocker, H.; Kozma, S.C.; Hafen, E.; Bos, J.L.; Thomas, G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 2003, 11, 1457–1466. [Google Scholar] [CrossRef]
- Long, X.; Lin, Y.; Ortiz-Vega, S.; Yonezawa, K.; Avruch, J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005, 15, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; McCarron, R.M.; Yu, E.Z.; Li, Y.; Hallenbeck, J. Akt phosphorylation and kinase activity are down-regulated during hibernation in the 13-lined ground squirrel. Brain Res. 2004, 1014, 14–21. [Google Scholar] [CrossRef]
- McMullen, D.C.; Hallenbeck, J.M. Regulation of Akt during torpor in the hibernating ground squirrel, Ictidomys tridecemlineatus. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2010. [Google Scholar] [CrossRef]
- Hoehn, K.L.; Hudachek, S.F.; Summers, S.A.; Florant, G.L. Seasonal, tissue-specific regulation of Akt/protein kinase B and glycogen synthase in hibernators. Am. J. Physiol-Regul. Integr. Comp. Physiol. 2004, 286, 498–504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, H.; Peng, X.; Yan, X.; Zhang, J.; Xu, S.; Wang, H.; Wang, Z.; Ma, X.; Gao, Y. Autophagy and Akt-mTOR signaling display periodic oscillations during torpor-arousal cycles in oxidative skeletal muscle of Daurian ground squirrels (Spermophilus dauricus). J Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2020, 190, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Abnous, K.; Dieni, C.A.; Storey, K.B. Regulation of Akt during hibernation in Richardson’s ground squirrels. Biochim. Biophys. Acta-Gen. Subj. 2008, 1780, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Copp, J.; Manning, G.; Hunter, T. TORC-Specific phosphorylation of mammalian target of rapamycin (mTOR): Phospho-Ser 2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009, 69, 1821–1827. [Google Scholar] [CrossRef]
- Chiang, G.G.; Abraham, R.T. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 2005, 280, 25485–25490. [Google Scholar] [CrossRef]
- Logan, S.M.; Wu, C.W.; Storey, K.B. The squirrel with the lagging eIF2, Global suppression of protein synthesis during torpor. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2019, 227, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.P.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904. [Google Scholar] [CrossRef]
- Wickler, S.J.; Hoyt, D.F.; Van Breukelen, F. Disuse atrophy in the hibernating golden-mantled ground squirrel, Spermophilus lateralis. Am. J. Physiol-Regul. Integr. Comp. Physiol. 1991, 261, R1214–R1217. [Google Scholar] [CrossRef]
- Zatzman, M.L. Renal and cardiovascular effects of hibernation and hypothermia. Cryobiology 1984, 21, 539–614. [Google Scholar] [CrossRef]
- Carey, H.V.; Andrews, M.T.; Martin, S.L. Mammalian hibernation: Cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev. 2003, 83, 1153–1181. [Google Scholar] [CrossRef]
- Rigano, K.S.; Gehring, J.L.; Evans Hutzenbiler, B.D.; Chen, A.V.; Nelson, O.L.; Vella, C.A.; Robbins, C.T.; Jansen, H.T. Life in the fat lane: Seasonal regulation of insulin sensitivity, food intake, and adipose biology in brown bears. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2017, 187, 649–676. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Shimozuru, M.; Tsubota, T. Skeletal muscles of hibernating black bears show minimal atrophy and phenotype shifting despite prolonged physical inactivity and starvation. PLoS ONE 2019, 14, e0215489. [Google Scholar]
- Schmid, J. Daily torpor in the gray mouse lemur (Microcebus murinus) in Madagascar: Energetic consequences and biological significance. Oecologia 2000, 123, 175–183. [Google Scholar] [CrossRef]
- Bozinovic, F.; Ruiz, G.; Rosenmann, M. Energetics and torpor of a South American “living fossil”, the microbiotheriid Dromiciops gliroides. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2004, 174, 293–297. [Google Scholar] [CrossRef]
- Giroud, S.; Habold, C.; Nespolo, R.F.; Mejías, C.; Terrien, J.; Logan, S.M.; Henning, R.H.; Storey, K.B. The Torpid State: Recent Advances in Metabolic Adaptations and Protective Mechanisms†. Front. Physiol. 2021, 20, 623665. [Google Scholar] [CrossRef]
- Tessier, S.N.; Zhang, J.; Biggar, K.K.; Wu, C.W.; Pifferi, F.; Perret, M.; Storey, K.B. Regulation of the PI3K/AKT Pathway and Fuel Utilization During Primate Torpor in the Gray Mouse Lemur, Microcebus murinus. Genom. Proteom. Bioinforma 2015, 13, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Luu, B.E.; Wijenayake, S.; Zhang, J.; Tessier, S.N.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. Strategies of biochemical adaptation for hibernation in a South American marsupial, Dromiciops gliroides: 2. Control of the Akt pathway and protein translation machinery. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 224, 19–25. [Google Scholar] [CrossRef]
- Schmid, J.; Ganzhorn, J.U. Optional strategies for reduced metabolism in gray mouse lemurs. Naturwissenschaften 2009, 96, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Halsall, A.L.; Boyles, J.G.; Whitaker, J.O. Body temperature patterns of big brown bats during winter in a building hibernaculum. J. Mammal. 2012, 93, 497–503. [Google Scholar] [CrossRef]
- Williams, C.T.; Goropashnaya, A.V.; Buck, C.L.; Fedorov, V.B.; Kohl, F.; Lee, T.N.; Barnes, B.M. Hibernating above the permafrost: Effects of ambient temperature and season on expression of metabolic genes in liver and brown adipose tissue of arctic ground squirrels. J. Exp. Biol. 2011, 214, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Oppliger, W.; Albert, V.; Robitaille, A.M.; Trapani, F.; Quagliata, L.; Fuhrer, T.; Sauer, U.; Terracciano, L.; Hall, M.N. Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc. Natl. Acad. Sci. USA 2014, 111, 11592–11599. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Moussa, H.; Moggridge, J.A.; Luu, B.E.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. The hibernating South American marsupial, Dromiciops gliroides, displays torpor-sensitive microRNA expression patterns. Sci. Rep. 2016, 6, 24627. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Biggar, K.K.; Luu, B.E.; Szereszewski, K.E.; Storey, K.B. Analysis of microRNA expression during the torpor-arousal cycle of a mammalian hibernator, the 13-lined ground squirrel. Physiol. Genom. 2016, 48, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Li, J.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Sun, L.; Zhang, Y.; Cui, Y.; Zhang, F.; et al. MicroRNA-193a-3p and-5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 2015, 34, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Arsham, A.M.; Howell, J.J.; Simon, M.C. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 2003, 278, 29655–29660. [Google Scholar] [CrossRef]
- Wouters, B.G.; Koritzinsky, M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer 2008, 8, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Buffenstein, R. The naked mole-rat: A new long-living model for human aging research. J. Gerontol-Ser. A Biol. Sci. Med. Sci. 2005, 60, 1369–1377. [Google Scholar] [CrossRef]
- Ilacqua, A.N.; Kirby, A.M.; Pamenter, M.E. Behavioural responses of naked mole rats to acute hypoxia and anoxia. Biol. Lett. 2017, 13, 20170545. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Reznick, J.; Peterson, B.L.; Blass, G.; Omerbašić, D.; Bennett, N.C.; Kuich, P.H.J.L.; Zasada, C.; Browe, B.M.; Hamann, W.; et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science (80-) 2017, 356, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. MTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, L.J.; Hadj-Moussa, H.; Nguyen, V.C.; Pamenter, M.E.; Storey, K.B. Naked mole rats activate neuroprotective proteins during hypoxia. J. Exp. Zool Part A Ecol. Integr. Physiol. 2019, 331, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Robin, E.D.; Vester, J.W.; Murdaugh, H.V.; Millen, J.E. Prolonged anaerobiosis in a vertebrate: Anaerobic metabolism in the freshwater turtle. J. Cell Comp. Physiol. 1964, 63, 287–297. [Google Scholar] [CrossRef]
- Fraser, K.P.P.; Houlihan, D.F.; Lutz, P.L.; Leone-Kabler, S.; Manuel, L.; Brechin, J.G. Complete suppression of protein synthesis during anoxia with no post-anoxia protein synthesis debt in the red-eared slider turtle Trachemys scripta elegans. J. Exp. Biol. 2001, 204, 4353–4360. [Google Scholar] [CrossRef]
- Brooks, S.P.J.; Storey, K.B. De novo protein synthesis and protein phosphorylation during anoxia and recovery in the red-eared turtle. Am. J. Physiol-Regul Integr. Comp. Physiol. 1993, 6, 1380–1386. [Google Scholar] [CrossRef]
- Biggar, K.K.; Zhang, J.; Storey, K.B. Navigating oxygen deprivation: Liver transcriptomic responses of the red eared slider turtle to environmental anoxia. PeerJ 2019, 7, e8144. [Google Scholar] [CrossRef]
- Warren, D.E.; Jackson, D.C. Lactate metabolism in anoxic turtles: An integrative review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2008, 178, 133–148. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Aestivation: Signaling and hypometabolism. J. Exp. Biol. 2012, 215, 1425–1433. [Google Scholar] [CrossRef]
- Romspert, A.P. Osmoregulation of the african clawed frog Xenopus laevis, in hypersaline media. Comp. Biochem. Physiol. Part A Physiol. 1976, 54, 1976. [Google Scholar]
- Meier, R.; Thelen, M.; Hemmings, B.A. Inactivation and dephosphorylation of protein kinase Bα (PKBα) promoted by hyperosmotic stress. EMBO J. 1998, 17, 7294–7303. [Google Scholar] [CrossRef]
- Lornejad-Schäfer, M.R.; Schäfer, C.; Graf, D.; Häussinger, D.; Schliess, F. Osmotic regulation of insulin-induced mitogen-activated protein kinase phosphatase (MKP-1) expression in H4IIE rat hepatoma cells. Biochem. J. 2003, 371, 609–619. [Google Scholar] [CrossRef][Green Version]
- Fielenbach, N.; Antebi, A.C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008, 22, 2149–2165. [Google Scholar] [CrossRef]
- Albert, P.S.; Riddle, D.L. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 1988, 126, 270–293. [Google Scholar] [CrossRef]
- Jia, K.; Chen, D.; Riddle, D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 2004, 131, 3897–3906. [Google Scholar] [CrossRef]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science (80-) 1997, 277, 942–946. [Google Scholar] [CrossRef]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef]
- Long, X.; Spycher, C.; Han, Z.S.; Rose, A.M.; Müller, F.; Avruch, J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002, 12, 1448–1461. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Kazyken, D.; Magnuson, B.; Bodur, C.; Acosta-Jaquez, H.A.; Zhang, D.; Tong, X.; Barnes, T.M.; Steinl, G.K.; Patterson, N.E.; Altheim, C.H.; et al. AMPK directly activates mTORC2 to promote cell survival during acute energetic stress. Sci. Signal. 2019, 12, eeav3249. [Google Scholar] [CrossRef] [PubMed]
- Logan, S.M.; Storey, K.B. Avoiding apoptosis during mammalian hibernation. Temperature 2017, 4, 15–17. [Google Scholar] [CrossRef]
- Logan, S.M.; Luu, B.E.; Storey, K.B. Turn down genes for WAT? Activation of anti-apoptosis pathways protects white adipose tissue in metabolically depressed thirteen-lined ground squirrels. Mol. Cell Biochem. 2016, 416, 47–62. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-W.; Storey, K.B. mTOR Signaling in Metabolic Stress Adaptation. Biomolecules 2021, 11, 681. https://doi.org/10.3390/biom11050681
Wu C-W, Storey KB. mTOR Signaling in Metabolic Stress Adaptation. Biomolecules. 2021; 11(5):681. https://doi.org/10.3390/biom11050681
Chicago/Turabian StyleWu, Cheng-Wei, and Kenneth B. Storey. 2021. "mTOR Signaling in Metabolic Stress Adaptation" Biomolecules 11, no. 5: 681. https://doi.org/10.3390/biom11050681
APA StyleWu, C.-W., & Storey, K. B. (2021). mTOR Signaling in Metabolic Stress Adaptation. Biomolecules, 11(5), 681. https://doi.org/10.3390/biom11050681






