Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3)
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation
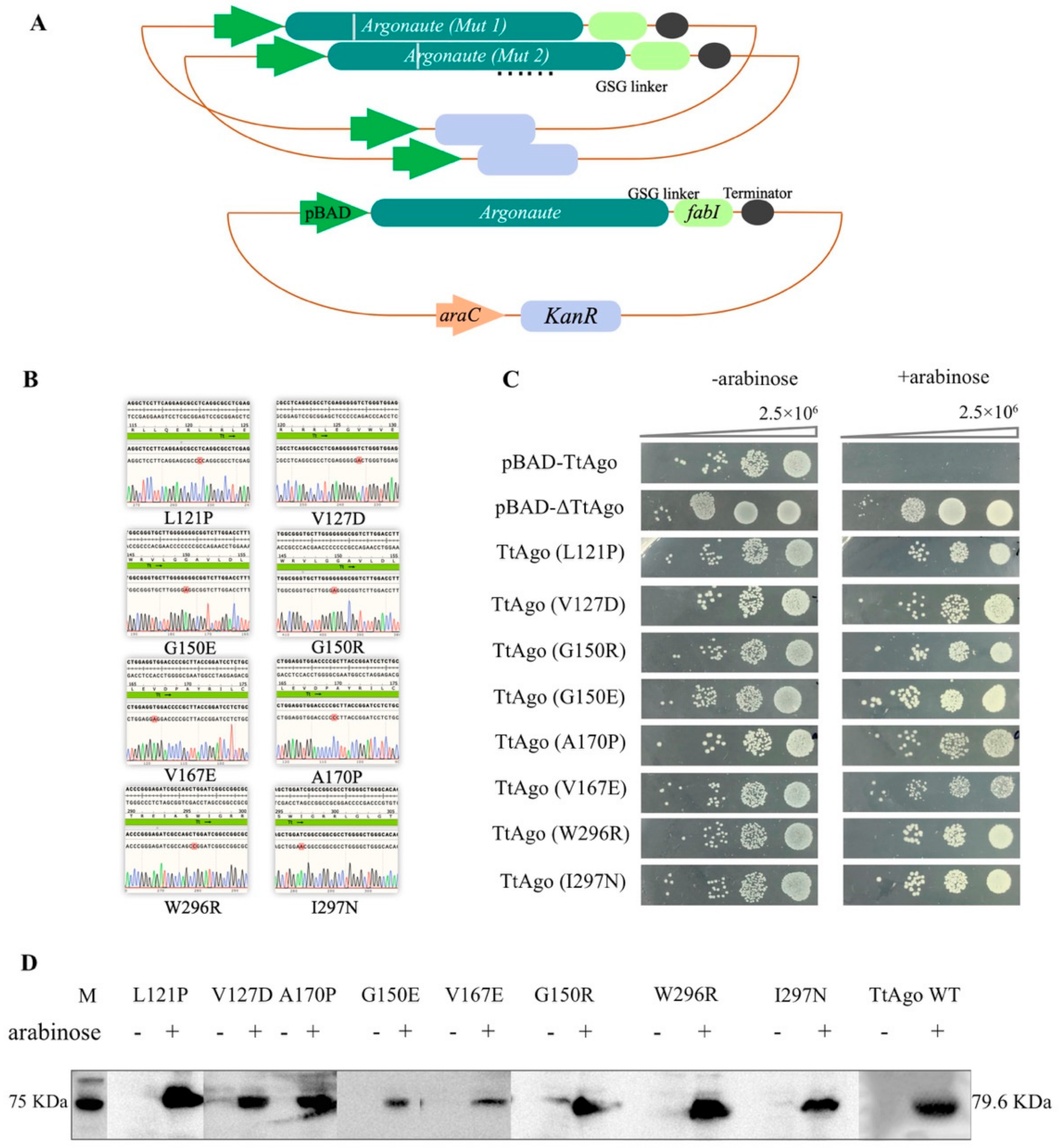
2.2. Construction of TtAgo Expression Vector pET28a-TtAgo and pET28a-pBAD-TtAgo
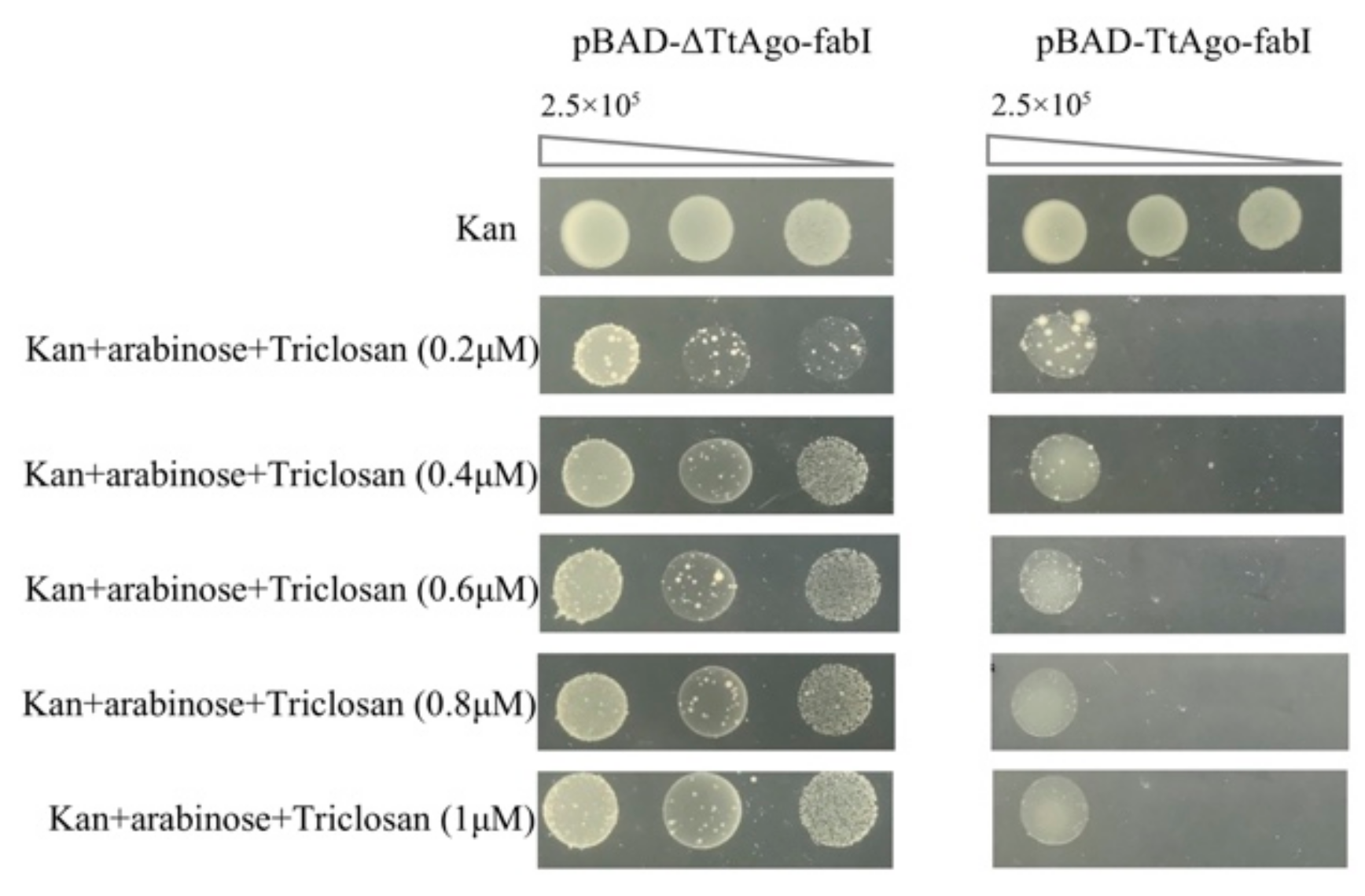
2.3. FabI-Fusion Construction
2.4. Construction of TtAgo Random Mutation Library by Error-Prone PCR (EP-PCR)
2.5. Construction and Selection for Loss of Enzyme Function Mutants
2.6. Construction of Single Point Mutations Isolated from Screening
2.7. Construction of the TtAgo DEDX Mutants by Site-Directed Mutagenesis
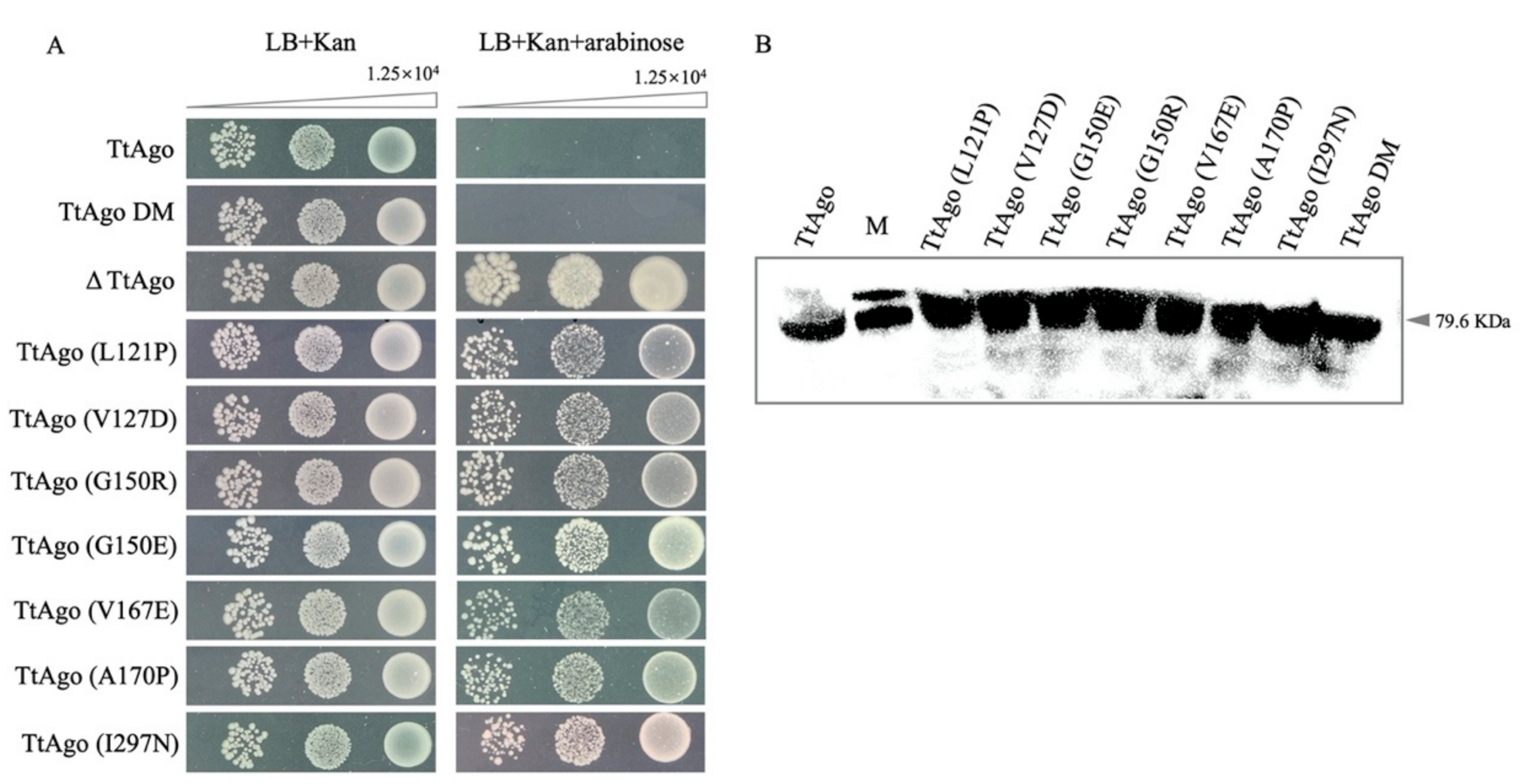
2.8. Spot Plating Assay
2.9. Protein Expression
2.10. Western Blotting
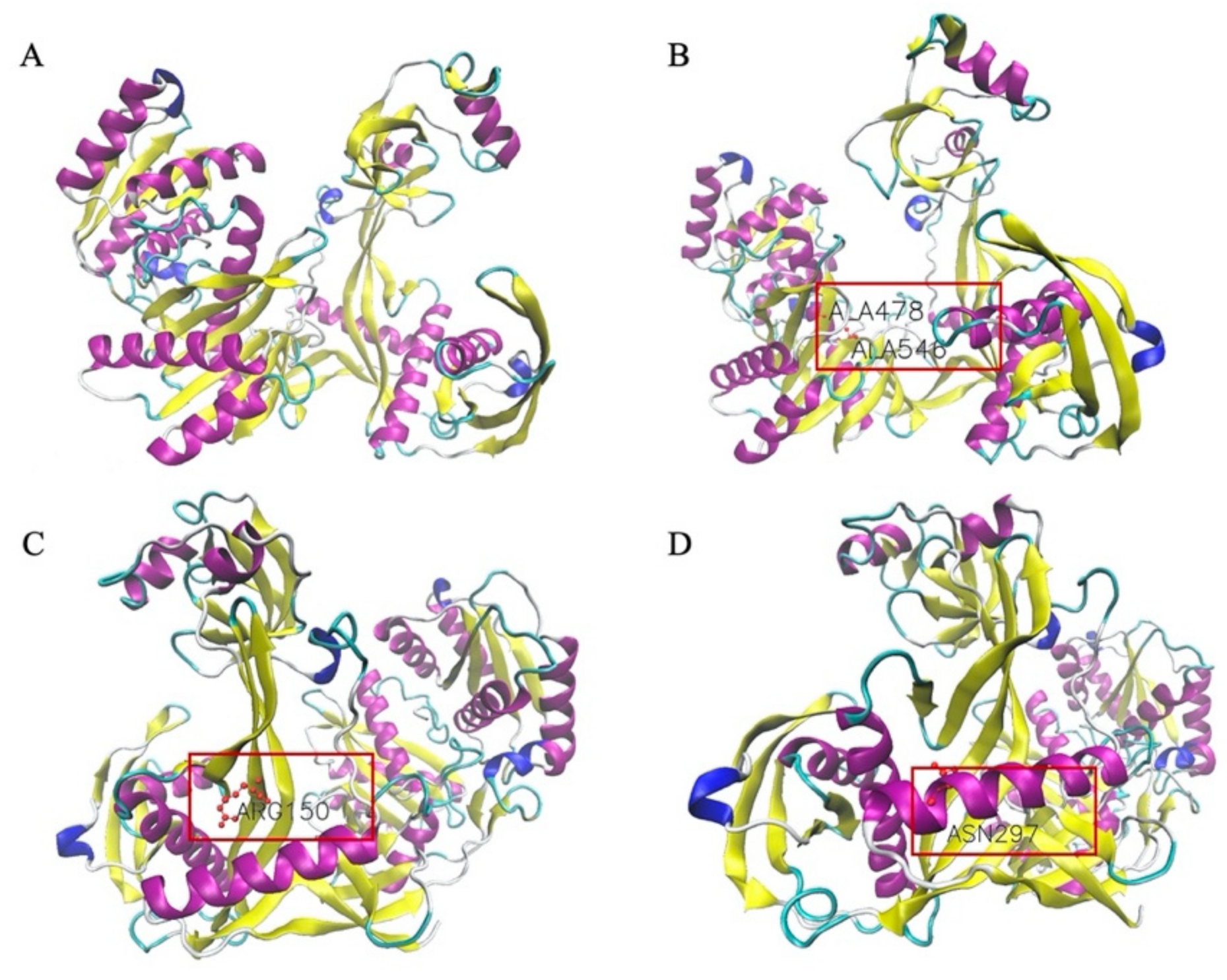
2.11. Protein Structure Simulation
3. Results
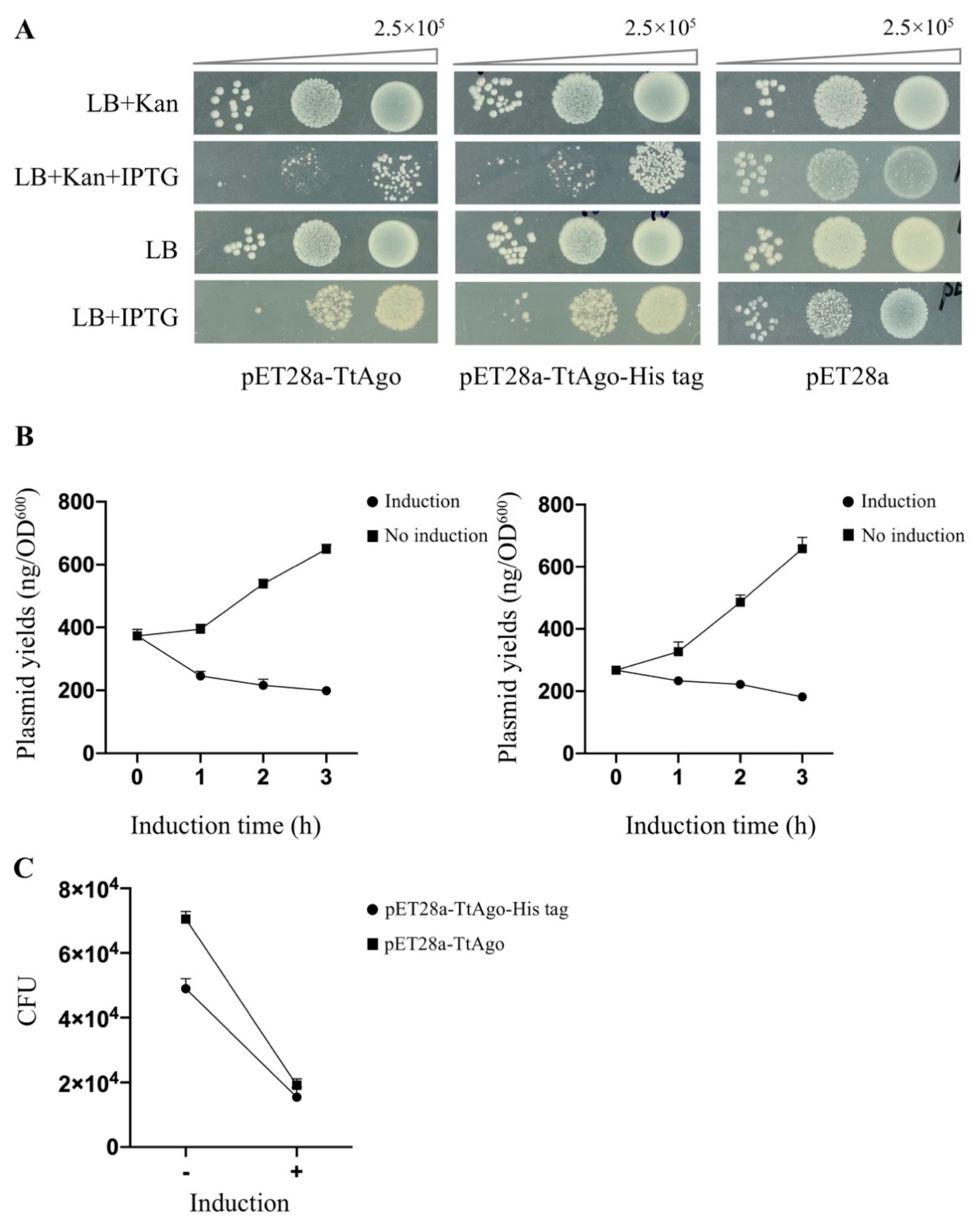
3.1. Expression of pAgo Protein in E. coli BL21 (DE3)
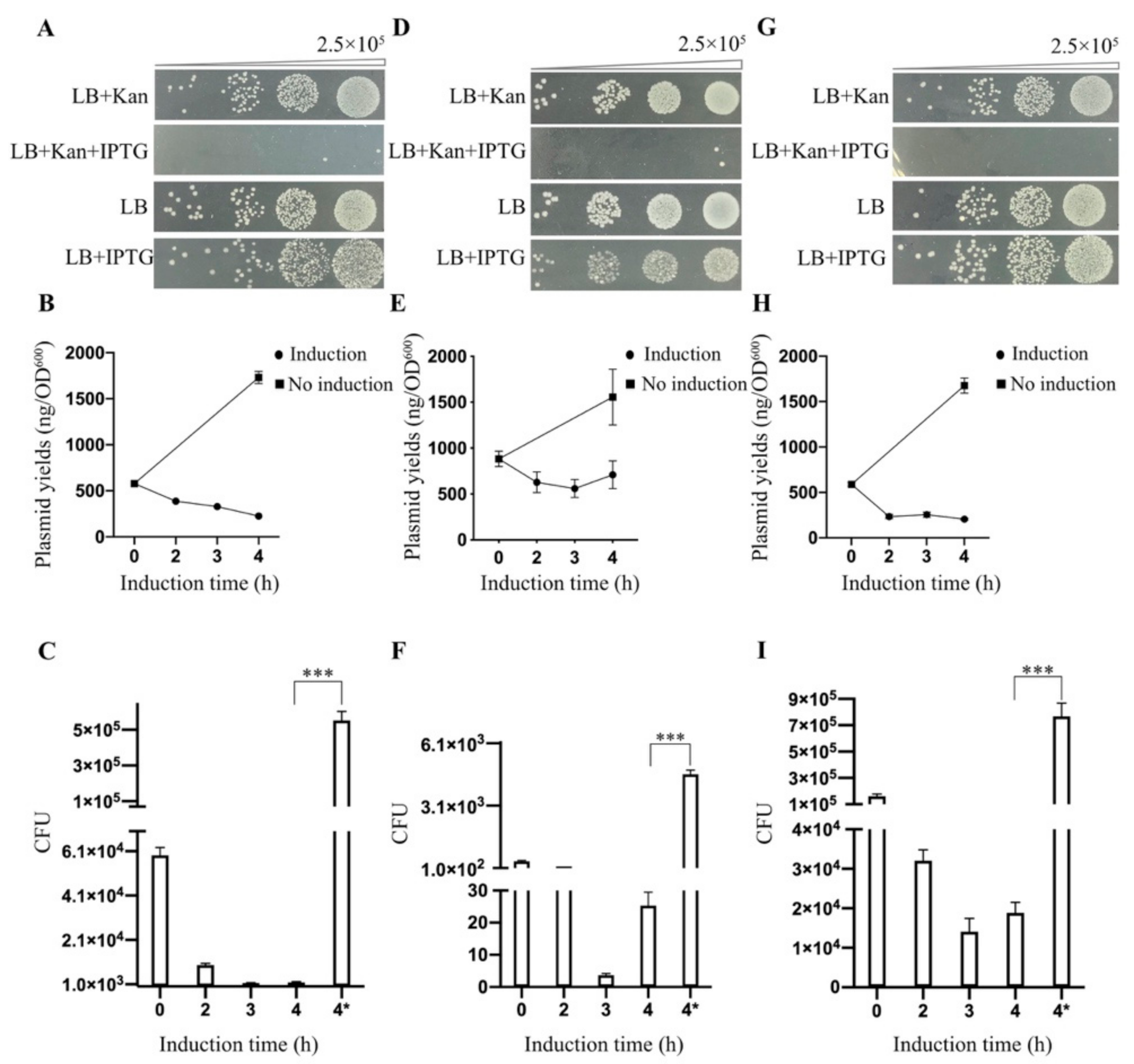
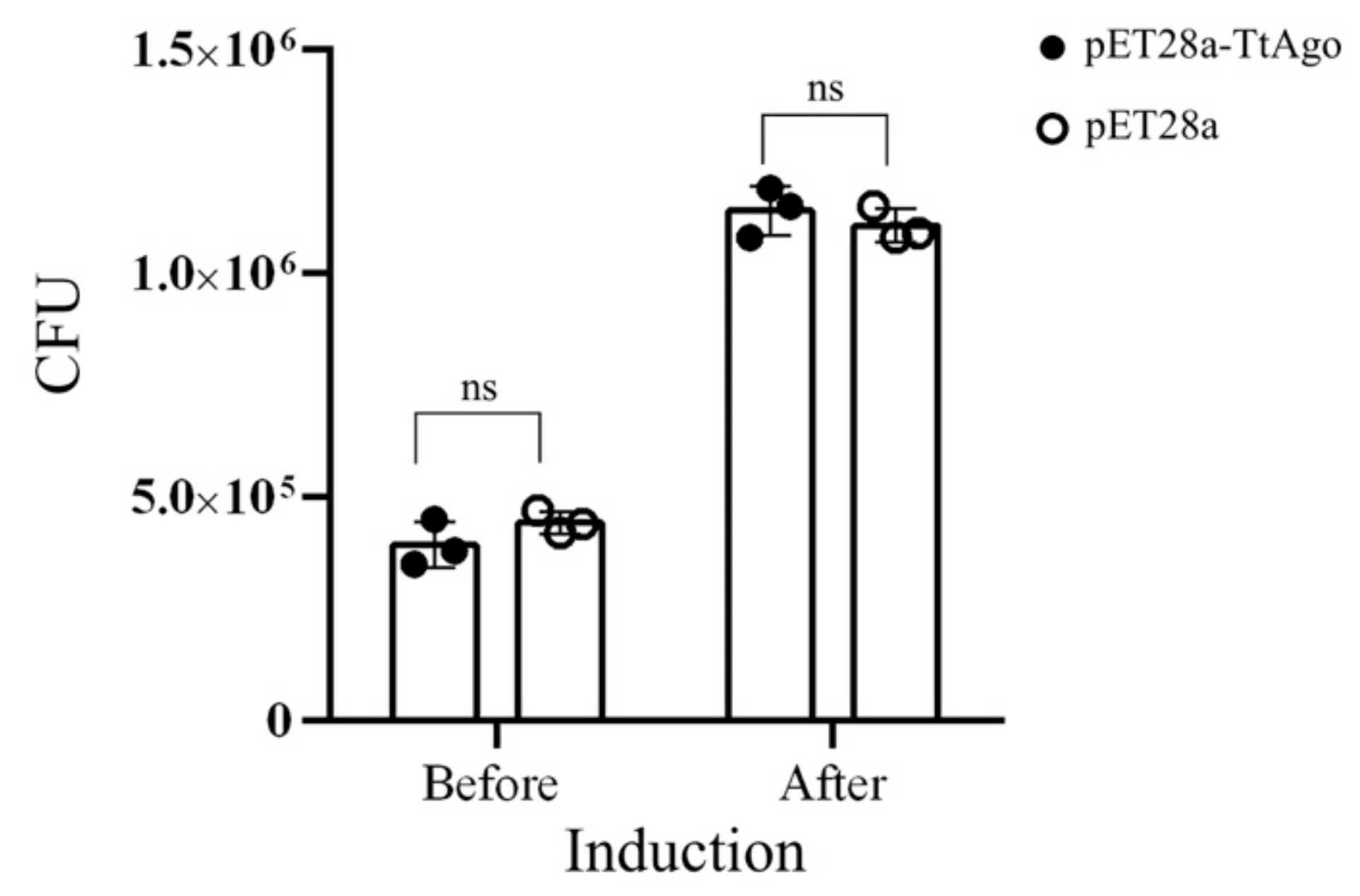
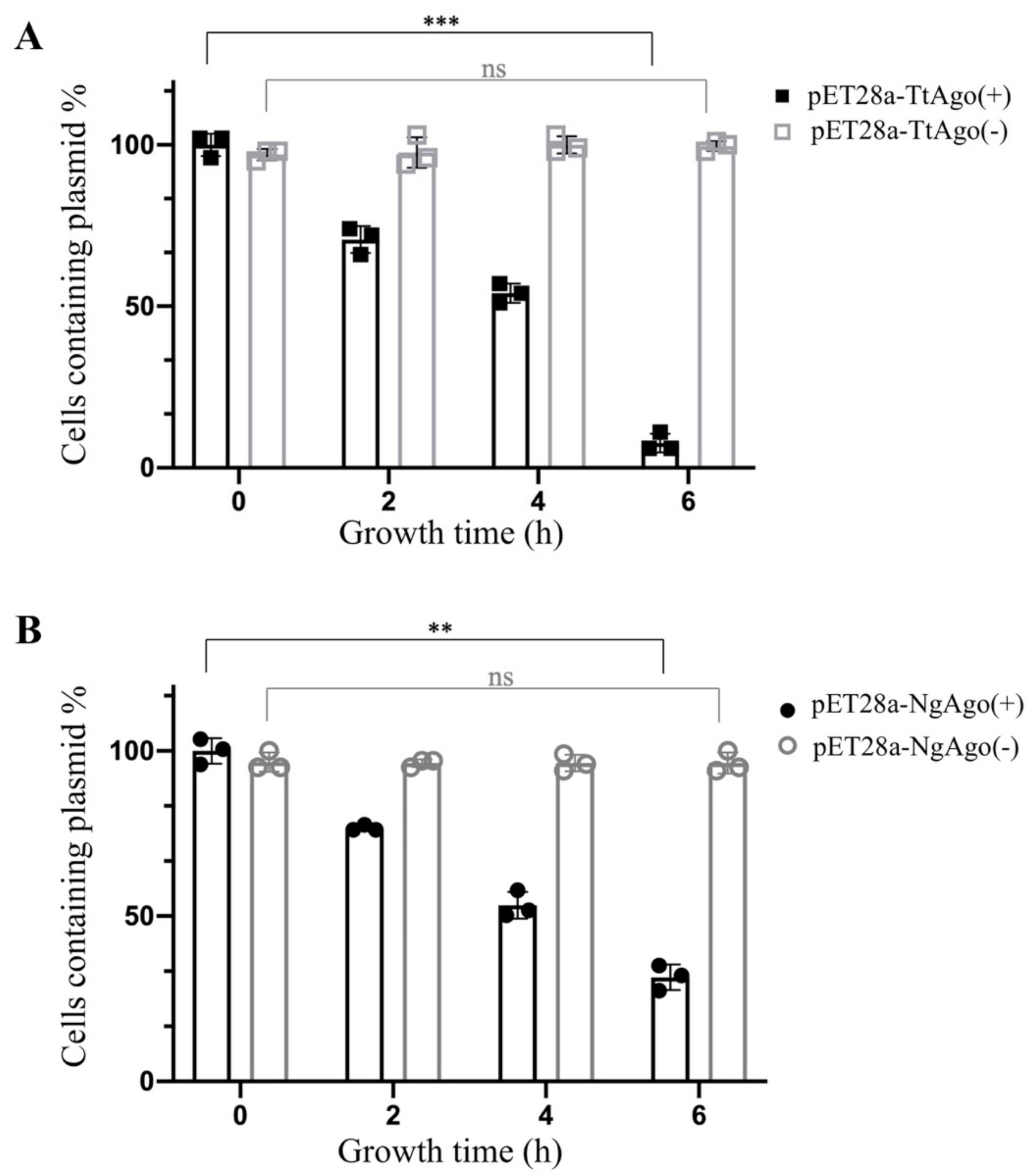
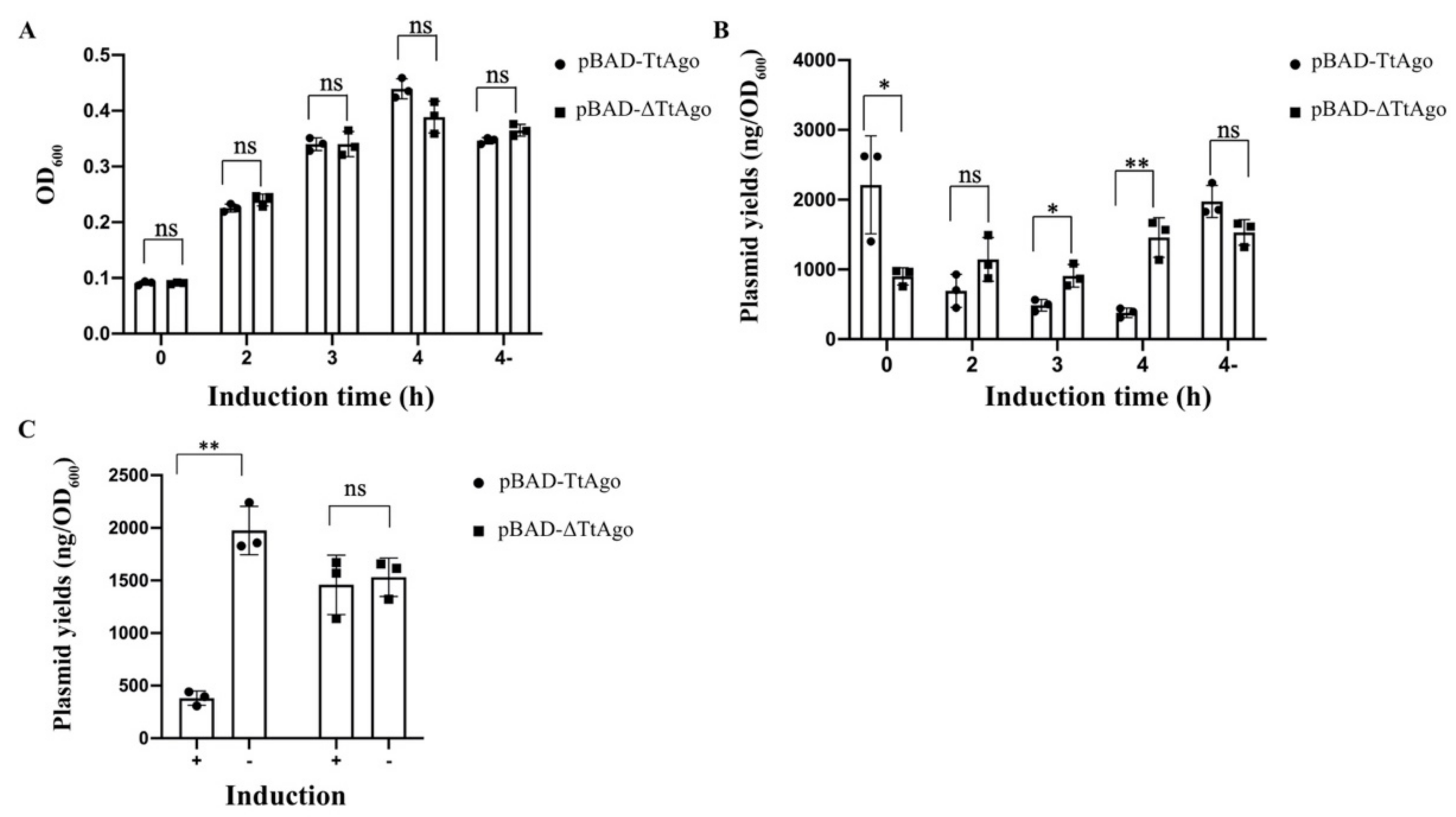
3.2. Effect of Deletion Mutant TtAgo (ΔTtAgo) Protein on Cell Growth and Plasmid Yields
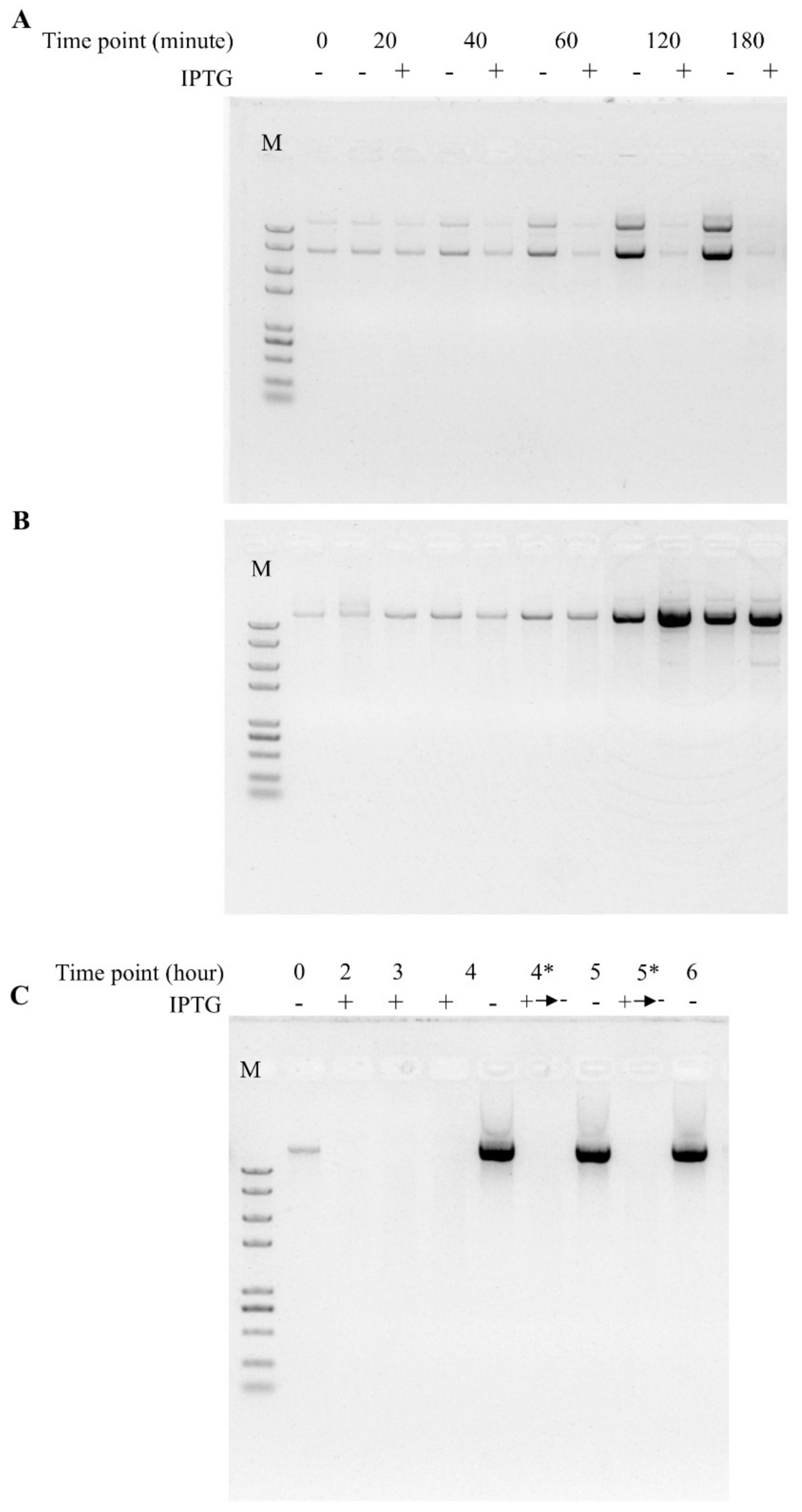
3.3. In Vivo Activity Comparison Between TtAgo Wild Type and 3′ Terminus Modification
3.4. Establishment of a High Throughput Screening System to Isolate Loss-of-Function Point Mutations of TtAgo in Vivo
3.5. Construction of a Random Mutation Library of TtAgo by EP-PCR
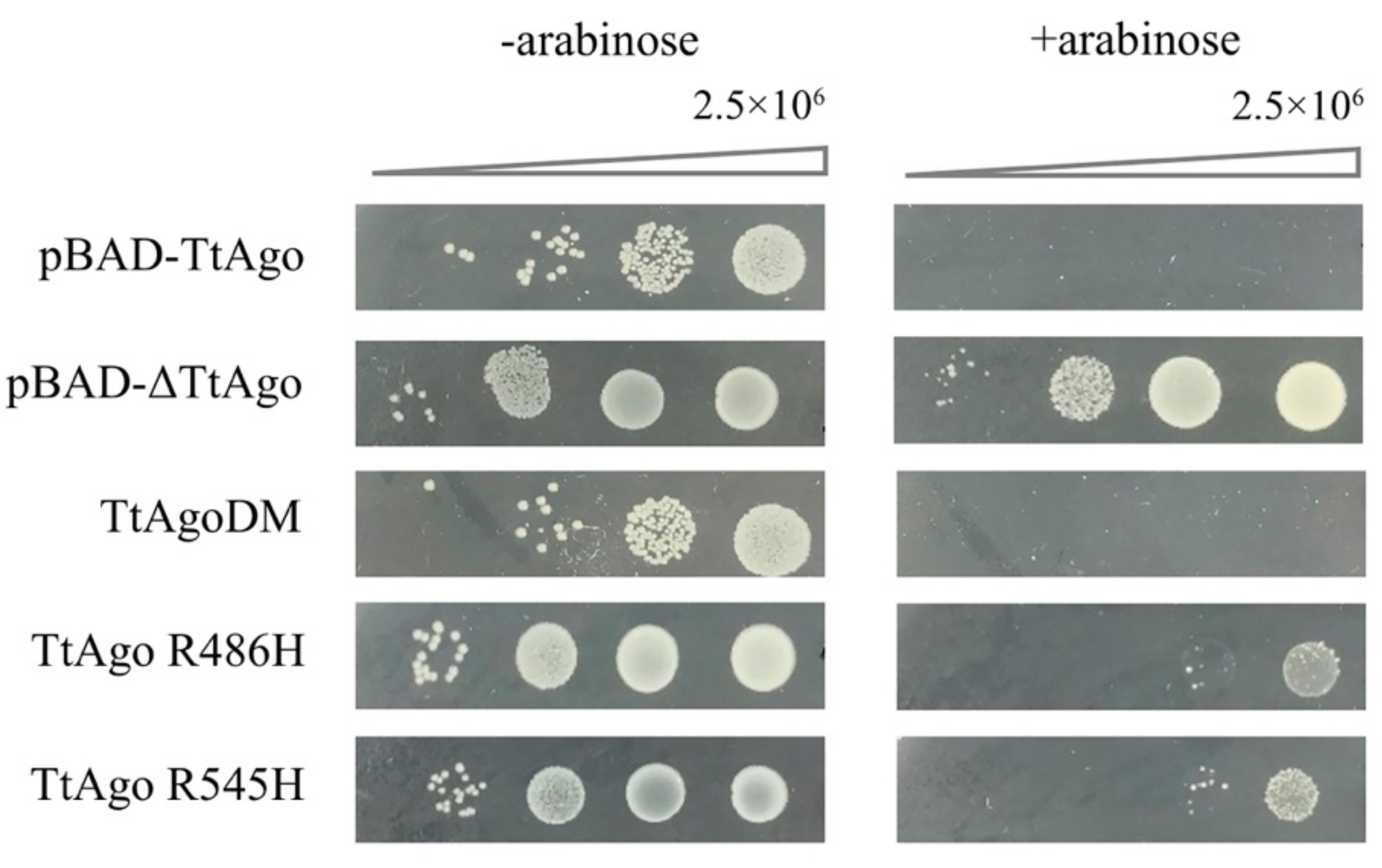
3.6. Function Analysis of TtAgo Point Mutations
3.7. Structural Simulation of Isolated TtAgo Point Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swarts, D.C.; Hegge, J.W.; Hinojo, I.; Shiimori, M.; Ellis, M.A.; Dumrongkulraksa, J.; Terns, R.M.; Terns, M.P.; van der Oost, J. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015, 43, 5120–5129. [Google Scholar] [CrossRef] [PubMed]
- Hegge, J.W.; Swarts, D.C.; Chandradoss, S.D.; Cui, T.J.; Kneppers, J.; Jinek, M.; Joo, C.; van der Oost, J. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019, 47, 5809–5821. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Sun, W.; Wang, J.; Sheng, G.; Xiang, G.; Zhang, T.; Shi, W.; Li, C.; Wang, Y.; Zhao, F.; et al. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 degrees C. Cell Discov. 2019, 5, 38. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Jore, M.M.; Westra, E.R.; Zhu, Y.; Janssen, J.H.; Snijders, A.P.; Wang, Y.; Patel, D.J.; Berenguer, J.; Brouns, S.J.J.; et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nat. Cell Biol. 2014, 507, 258–261. [Google Scholar] [CrossRef]
- Zander, A.; Willkomm, S.; Ofer, S.; van Wolferen, M.; Egert, L.; Buchmeier, S.; Stockl, S.; Tinnefeld, P.; Schneider, S.; Klingl, A.; et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017, 2, 17034. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Xie, C.; Jin, Z.; Tu, Z.; Han, L.; Jin, M.; Xiang, Y.; Zhang, A. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019, 47, 3568–3579. [Google Scholar] [CrossRef]
- Jolly, S.M.; Gainetdinov, I.; Jouravleva, K.; Zhang, H.; Strittmatter, L.; Bailey, S.M.; Hendricks, G.M.; Dhabaria, A.; Ueberheide, B.; Zamore, P.D. Thermus thermophilus Argonaute Functions in the Completion of DNA Replication. Cell 2020, 182, 1545–1559.e18. [Google Scholar] [CrossRef]
- Ketting, R.F. The many faces of RNAi. Dev. Cell 2011, 20, 148–161. [Google Scholar] [CrossRef]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Lapinaite, A.; Doudna, J.A.; Cate, J.H.D. Programmable RNA recognition using a CRISPR-associated Argonaute. Proc. Natl. Acad. Sci. USA 2018, 115, 3368–3373. [Google Scholar] [CrossRef]
- Swarts, D.C.; Koehorst, J.J.; Westra, E.R.; Schaap, P.J.; van der Oost, J. Effects of Argonaute on Gene Expression in Thermus thermophilus. PLoS ONE 2015, 10, e0124880. [Google Scholar]
- Kuzmenko, A.; Oguienko, A.; Esyunina, D.; Yudin, D.; Petrova, M.; Kudinova, A.; Maslova, O.; Ninova, M.; Ryazansky, S.; Leach, D.; et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nat. Cell Biol. 2020, 587, 632–637. [Google Scholar] [CrossRef]
- Garcia-Quintans, N.; Bowden, L.; Berenguer, J.; Mencia, M. DNA interference by a mesophilic Argonaute protein, CbcAgo. F1000Research 2019, 8, 321. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Dong, Z.; Shi, Y.; Wang, X.; Qin, Y.; Wang, Y.; Liu, D. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 2016, 26, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Tan, S.; Xu, L.; Gao, L.; Zhu, H.; Ma, C.; Liang, X. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017, 145, 20–23. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jung, Y.; Lim, D. Argonaute system of Kordia jejudonensis is a heterodimeric nucleic acid-guided nuclease. Biochem. Biophys. Res. Commun. 2020, 525, 755–758. [Google Scholar] [CrossRef]
- Willkomm, S.; Makarova, K.S.; Grohmann, D. DNA silencing by prokaryotic Argonaute proteins adds a new layer of defense against invading nucleic acids. FEMS Microbiol. Rev. 2018, 42, 376–387. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; van der Oost, J.; Koonin, E.V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 2009, 4, 1–15. [Google Scholar] [CrossRef]
- Hegge, J.W.; Swarts, D.C.; van der Oost, J. Prokaryotic Argonaute proteins: Novel genome-editing tools? Nat. Rev. Genet. 2018, 16, 5–11. [Google Scholar] [CrossRef]
- Blesa, A.; Cesar, C.E.; Averhoff, B.; Berenguer, J. Noncanonical cell-to-cell DNA transfer in Thermus spp. is insensitive to argonaute-mediated interference. J Bacteriol. 2015, 197, 138–146. [Google Scholar] [CrossRef]
- He, R.; Wang, L.; Wang, F.; Li, W.; Liu, Y.; Li, A.; Wang, Y.; Mao, W.; Zhai, C.; Ma, L. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. 2019, 55, 13219–13222. [Google Scholar] [CrossRef]
- Zander, A.; Holzmeister, P.; Klose, D.; Tinnefeld, P.; Grohmann, D. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014, 11, 45–56. [Google Scholar] [CrossRef]
- Willkomm, S.; Oellig, C.A.; Zander, A.; Restle, T.; Keegan, R.; Grohmann, D.; Schneider, S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol. 2017, 2, 17035. [Google Scholar] [CrossRef] [PubMed]
- Doxzen, K.W.; Doudna, J.A. DNA recognition by an RNA-guided bacterial Argonaute. PLoS ONE 2017, 12, e0177097. [Google Scholar] [CrossRef] [PubMed]
- Kaya, E.; Doxzen, K.W.; Knoll, K.R.; Wilson, R.C.; Strutt, S.C.; Kranzusch, P.J.; Doudna, J.A. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. USA 2016, 113, 4057–4062. [Google Scholar] [CrossRef]
- Kuzmenko, A.; Yudin, D.; Ryazansky, S.; Kulbachinskiy, A.; Aravin, A.A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019, 47, 5822–5836. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, I.; Chan, K.; Sachidanandam, R.; Newman, D.K.; Aravin, A.A. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell 2013, 51, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, T.; Ito, K.; Murakami, R.; Uchiumi, T. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016, 7, 11846. [Google Scholar] [CrossRef]
- Lisitskaya, L.; Aravin, A.A.; Kulbachinskiy, A. DNA interference and beyond: Structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Swarts, D.C.; Szczepaniak, M.; Sheng, G.; Chandradoss, S.D.; Zhu, Y.; Timmers, E.M.; Zhang, Y.; Zhao, H.; Lou, J.; Wang, Y.; et al. Autonomous Generation and Loading of DNA Guides by Bacterial Argonaute. Mol. Cell 2017, 65, 985–998 e986. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Tuschl, T.; Patel, D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nat. Cell Biol. 2008, 456, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nat. Cell Biol. 2008, 456, 209–213. [Google Scholar] [CrossRef]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Wardle, G.S.; Tuschl, T.; Patel, D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nat. Cell Biol. 2009, 461, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.; Zhao, H.; Wang, J.; Rao, Y.; Tian, W.; Swarts, D.C.; van der Oost, J.; Patel, D.J.; Wang, Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 2013, 111, 652–657. [Google Scholar] [CrossRef]
- Rodricks, J.V.; Swenberg, J.A.; Borzelleca, J.F.; Maronpot, R.R.; Shipp, A.M. Triclosan: A critical review of the experimental data and development of margins of safety for consumer products. Crit. Rev. Toxicol. 2010, 40, 422–484. [Google Scholar] [CrossRef]
- Dann, A.B.; Hontela, A. Triclosan: Environmental exposure, toxicity and mechanisms of action. J. Appl. Toxicol. 2010, 31, 285–311. [Google Scholar] [CrossRef]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef]
- Balzer, S.; Kucharova, V.; Megerle, J.; Lale, R.; Brautaset, T.; Valla, S. A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli. Microb. Cell Fact. 2013, 12, 26. [Google Scholar] [CrossRef]
- Jang, C.W.; Magnuson, T. A novel selection marker for efficient DNA cloning and recombineering in E. coli. PLoS ONE 2013, 8, e57075. [Google Scholar] [CrossRef]
- Yu, B.J.; Kim, J.A.; Pan, J.G. Signature gene expression profile of triclosan-resistant Escherichia coli. J. Antimicrob. Chemother. 2010, 65, 1171–1177. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. 1), S162–S173. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, Z.; Zhu, J.; Wang, S.; Xu, D.; Han, W. Why Is a High Temperature Needed by Thermus thermophilus Argonaute During mRNA Silencing: A Theoretical Study. Front. Chem. 2018, 6, 223. [Google Scholar] [CrossRef]
- Lei, J.; Sheng, G.; Cheung, P.P.; Wang, S.; Li, Y.; Gao, X.; Zhang, Y.; Wang, Y.; Huang, X. Two symmetric arginine residues play distinct roles in Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 2019, 116, 845–853. [Google Scholar] [CrossRef]
- Zhu, L.; Jiang, H.; Sheong, F.K.; Cui, X.; Gao, X.; Wang, Y.; Huang, X. A Flexible Domain-Domain Hinge Promotes an Induced-fit Dominant Mechanism for the Loading of Guide-DNA into Argonaute Protein in Thermus thermophilus. J. Phys. Chem. B 2016, 120, 2709–2720. [Google Scholar] [CrossRef]
- Klum, S.M.; Chandradoss, S.D.; Schirle, N.T.; Joo, C.; MacRae, I.J. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2017, 37, 75–88. [Google Scholar] [CrossRef]
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef]
| Mutation | Location |
|---|---|
| L121P | Linker 1 |
| V127D | Linker 1 |
| G150R G150E V167E A170P W296R I297N | Linker 1 Linker 1 Linker 1 Linker 1 Linker 2 Linker 2 |
| Plasmid Name | Description |
|---|---|
| pET28a-pBAD-TtAgo (R486H) | Arginine 486 replaced with histidine in TtAgo |
| pET28a-pBAD-TtAgo (R545H) | Arginine 545 replaced with histidine in TtAgo |
| pET28a-pBAD-TtAgoDM (D478A, D546A) | Aspartic acid 478 and 546 replaced with alanine in TtAgo |
| pET28a-pBAD-ΔTtAgo | Amino acids 25 to 215 of TtAgo deleted |
| pET28a-pBAD-TtAgo (L121P) | Leucine 121 replaced with proline in TtAgo |
| pET28a-pBAD-TtAgo (V127D) | Valine 127 replaced with aspartic acid in TtAgo |
| pET28a-pBAD-TtAgo (G150R) | Glycine 150 replaced with arginine in TtAgo |
| pET28a-pBAD-TtAgo (G150E) | Glycine 150 replaced with glutamic acid in TtAgo |
| pET28a-pBAD-TtAgo (V167E) | Valine 167 replaced with glutamic acid in TtAgo |
| pET28a-pBAD-TtAgo (A170P) | Alanine 170 replaced with proline in TtAgo |
| pET28a-pBAD-TtAgo (W296R) | Tryptophan 296 replaced with arginine in TtAgo |
| pET28a-pBAD-TtAgo (I297N) | Isoleucine 297 replaced with asparagine in TtAgo |
| Colony Number | Mutation Position and Type |
|---|---|
| 1 | P97S, G150R, L189Q |
| 2 | V127D |
| 3 4 5 6 7 8 9 10 11 12 13 14 15 16, 40 17 18, 21, 22 19 20 23 24 25 26, 28 27, 30 29 31 32 33 34 35, 41 36 37 38 39 42 | A278T, W296R V127D, R651W A555V, E483D A555V, A659S A555V, L556P G150R A53E, R59H, G144R R427M, E483D A133V, V127D A151E, I297N L265P, A278T, W296R V127D, R286Q, R299Q G150E, L430H G150E P97S, G150R, L189Q, E210K, H637Y L265P, A278T, W296R V127D, R651W A113V, V127D, G388G F10Y, R123H, G150E, G220S, L638Q P37L, V127D, A133V V167E G150W, G467D L121P L174P, I293F, L430P, K599Q A170P E41A, G126E, R136Q, R141Q, A345T G150E A114V, V127D, A245V, F485L L75F, I293F R122G, G150R, S229P, R427S R155K, G150W, A555P G126E, G150E L148P L121P, R420H |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, J.; Ma, L.; Cheng, X.; Ma, J.; Wang, R.; Xu, K.; Mymryk, J.S.; Zhang, Z. Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3). Biomolecules 2021, 11, 524. https://doi.org/10.3390/biom11040524
Xing J, Ma L, Cheng X, Ma J, Wang R, Xu K, Mymryk JS, Zhang Z. Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3). Biomolecules. 2021; 11(4):524. https://doi.org/10.3390/biom11040524
Chicago/Turabian StyleXing, Jiani, Lixia Ma, Xinzhen Cheng, Jinrong Ma, Ruyu Wang, Kun Xu, Joe S. Mymryk, and Zhiying Zhang. 2021. "Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3)" Biomolecules 11, no. 4: 524. https://doi.org/10.3390/biom11040524
APA StyleXing, J., Ma, L., Cheng, X., Ma, J., Wang, R., Xu, K., Mymryk, J. S., & Zhang, Z. (2021). Expression and Functional Analysis of the Argonaute Protein of Thermus thermophilus (TtAgo) in E. coli BL21(DE3). Biomolecules, 11(4), 524. https://doi.org/10.3390/biom11040524







