Blood Levels of Galectin-9, an Immuno-Regulating Molecule, Reflect the Severity for the Acute and Chronic Infectious Diseases
Abstract
1. Introduction
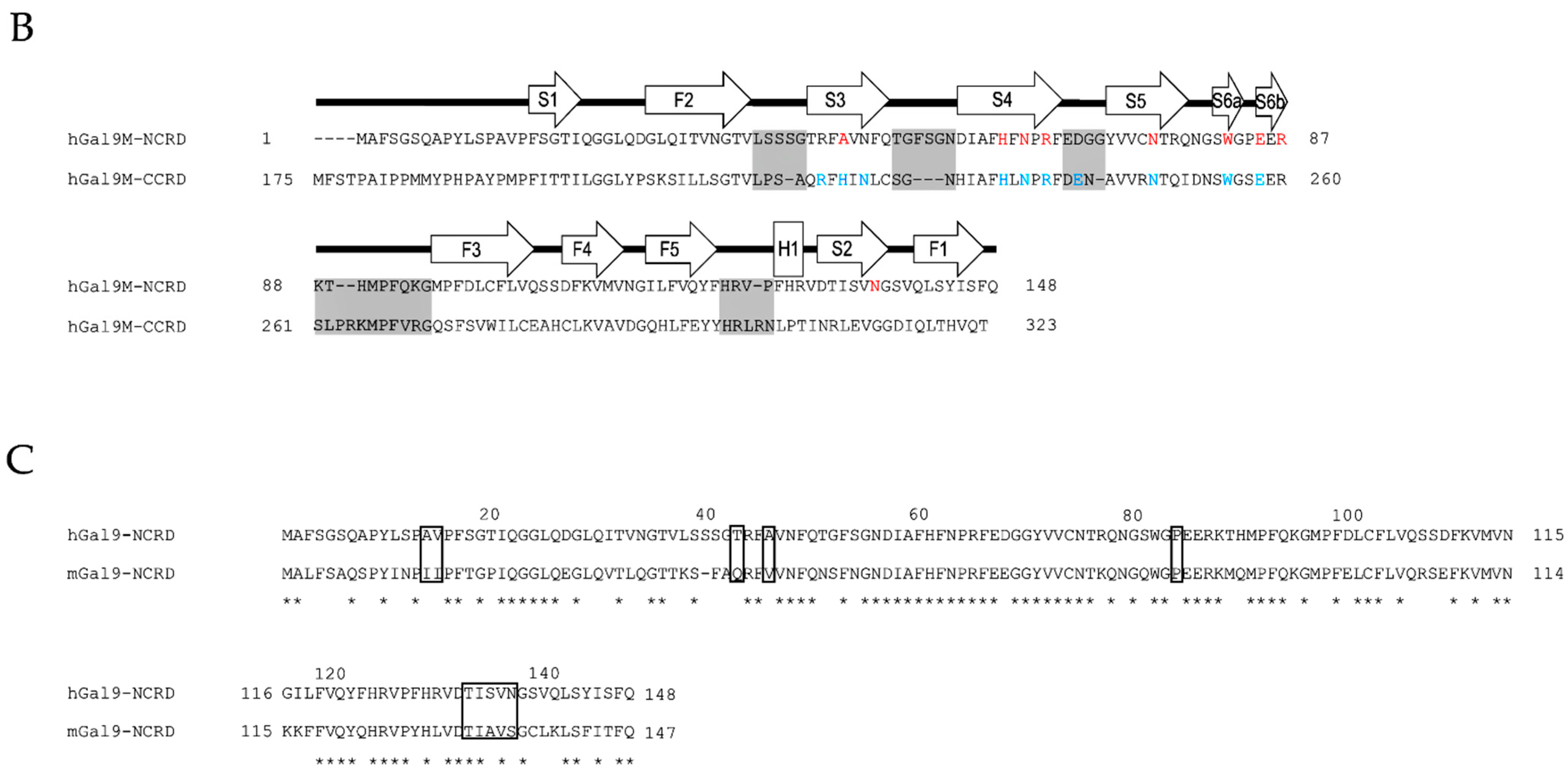
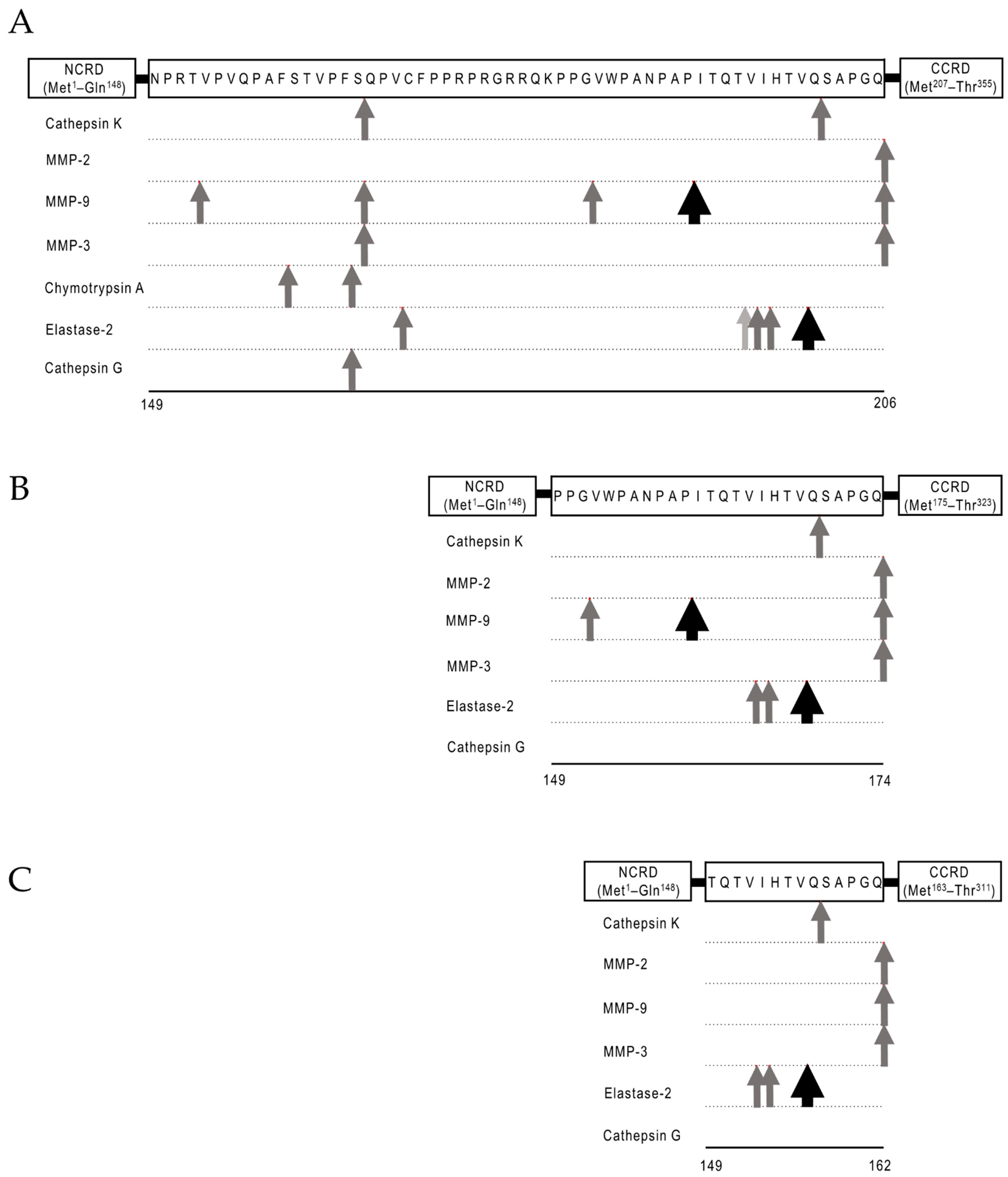
2. Galectin-9 Biochemistry
2.1. Structural Basis and Binding Properties of Gal-9
2.2. Protease-Susceptibility in the Linker-Peptides of Gal-9
3. Galectin-9 Molecular and Cellular Biology
3.1. Gal-9 Interaction with Multiple Receptors
3.1.1. Ig Superfamily
3.1.2. TNF Receptor Family
3.1.3. Adhesive Molecule
3.1.4. Enzyme
3.1.5. C-Type Lectin Receptor
3.2. Immune-Potentiating and -Suppressive Functions of Gal-9
4. Galectin-9 in a Variety of Infectious Diseases
4.1. Galectin-9 in Human Immunodeficiency Virus (HIV) Infection
4.2. Galectin-9 in Dengue Virus Infection
4.3. Galectin-9 in Malaria
4.4. Galectin-9 in Leptospirosis
4.5. Galectin-9 in Tuberculosis (TB)
4.6. Galectin-9 in Coronavirus Disease 2019 (COVID-19)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tureci, O.; Schmitt, H.; Fadle, N.; Pfreundschuh, M.; Sahin, U. Molecular definition of a novel human galectin which is immunogenic in patients with Hodgkin’s disease. J. Biol. Chem. 1997, 272, 6416–6422. [Google Scholar] [CrossRef]
- Hirashima, M.; Kashio, Y.; Nishi, N.; Yamauchi, A.; Imaizumi, T.A.; Kageshita, T.; Saita, N.; Nakamura, T. Galectin-9 in physiological and pathological conditions. Glycoconj. J. 2002, 19, 593–600. [Google Scholar] [CrossRef]
- Chagan-Yasutan, H.; Ndhlovu, L.C.; Lacuesta, T.L.; Kubo, T.; Leano, P.S.; Niki, T.; Oguma, S.; Morita, K.; Chew, G.M.; Barbour, J.D.; et al. Galectin-9 plasma levels reflect adverse hematological and immunological features in acute dengue virus infection. J. Clin. Virol. 2013, 58, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, B.; Zhao, J.; Okumura, M.; Chagan-Yasutan, H.; Yanai, H.; Mizuno, K.; Yoshiyama, T.; Idei, T.; Ashino, Y.; Nakajima, C.; et al. Immunological Roles of Elevated Plasma Levels of Matricellular Proteins in Japanese Patients with Pulmonary Tuberculosis. Int. J. Mol. Sci. 2016, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Dembele, B.P.; Chagan-Yasutan, H.; Niki, T.; Ashino, Y.; Tangpukdee, N.; Shinichi, E.; Krudsood, S.; Kano, S.; Hattori, T. Plasma levels of Galectin-9 reflect disease severity in malaria infection. Malar. J. 2016, 15, 403. [Google Scholar] [CrossRef] [PubMed]
- Merani, S.; Chen, W.; Elahi, S. The bitter side of sweet: The role of Galectin-9 in immunopathogenesis of viral infections. Rev. Med. Virol. 2015, 25, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Wada, J.; Kanwar, Y.S. Identification and characterization of galectin-9, a novel beta-galactoside-binding mammalian lectin. J. Biol. Chem. 1997, 272, 6078–6086. [Google Scholar] [CrossRef]
- Sato, M.; Nishi, N.; Shoji, H.; Seki, M.; Hashidate, T.; Hirabayashi, J.; Kasai Ki, K.; Hata, Y.; Suzuki, S.; Hirashima, M.; et al. Functional analysis of the carbohydrate recognition domains and a linker peptide of galectin-9 as to eosinophil chemoattractant activity. Glycobiology 2002, 12, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Heusschen, R.; Schulkens, I.A.; van Beijnum, J.; Griffioen, A.W.; Thijssen, V.L. Endothelial LGALS9 splice variant expression in endothelial cell biology and angiogenesis. Biochim. Biophys. Acta 2014, 1842, 284–292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heusschen, R.; Freitag, N.; Tirado-Gonzalez, I.; Barrientos, G.; Moschansky, P.; Munoz-Fernandez, R.; Leno-Duran, E.; Klapp, B.F.; Thijssen, V.L.; Blois, S.M. Profiling Lgals9 splice variant expression at the fetal-maternal interface: Implications in normal and pathological human pregnancy. Biol. Reprod. 2013, 88, 22. [Google Scholar] [CrossRef]
- Nagae, M.; Nishi, N.; Nakamura-Tsuruta, S.; Hirabayashi, J.; Wakatsuki, S.; Kato, R. Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue. J. Mol. Biol. 2008, 375, 119–135. [Google Scholar] [CrossRef]
- Yoshida, H.; Teraoka, M.; Nishi, N.; Nakakita, S.; Nakamura, T.; Hirashima, M.; Kamitori, S. X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose. J. Biol. Chem. 2010, 285, 36969–36976. [Google Scholar] [CrossRef] [PubMed]
- Leffler, H.; Carlsson, S.; Hedlund, M.; Qian, Y.; Poirier, F. Introduction to galectins. Glycoconj. J. 2002, 19, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.; et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim. Biophys. Acta 2002, 1572, 232–254. [Google Scholar] [CrossRef]
- Miyanishi, N.; Nishi, N.; Abe, H.; Kashio, Y.; Shinonaga, R.; Nakakita, S.; Sumiyoshi, W.; Yamauchi, A.; Nakamura, T.; Hirashima, M.; et al. Carbohydrate-recognition domains of galectin-9 are involved in intermolecular interaction with galectin-9 itself and other members of the galectin family. Glycobiology 2007, 17, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Nishi, N.; Murata, T.; Usui, T.; Nakamura, T.; Wakatsuki, S.; Kato, R. Crystal structure of the galectin-9 N-terminal carbohydrate recognition domain from Mus musculus reveals the basic mechanism of carbohydrate recognition. J. Biol. Chem. 2006, 281, 35884–35893. [Google Scholar] [CrossRef] [PubMed]
- Nishi, N.; Itoh, A.; Fujiyama, A.; Yoshida, N.; Araya, S.; Hirashima, M.; Shoji, H.; Nakamura, T. Development of highly stable galectins: Truncation of the linker peptide confers protease-resistance on tandem-repeat type galectins. FEBS Lett. 2005, 579, 2058–2064. [Google Scholar] [CrossRef]
- Nishi, N.; Itoh, A.; Shoji, H.; Miyanaka, H.; Nakamura, T. Galectin-8 and galectin-9 are novel substrates for thrombin. Glycobiology 2006, 16, 15C–20C. [Google Scholar] [CrossRef]
- Song, J.; Tan, H.; Perry, A.J.; Akutsu, T.; Webb, G.I.; Whisstock, J.C.; Pike, R.N. PROSPER: An integrated feature-based tool for predicting protease substrate cleavage sites. PLoS ONE 2012, 7, e50300. [Google Scholar] [CrossRef]
- Itoh, A.; Fukata, Y.; Miyanaka, H.; Nonaka, Y.; Ogawa, T.; Nakamura, T.; Nishi, N. Optimization of the inter-domain structure of galectin-9 for recombinant production. Glycobiology 2013, 23, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Su, E.W.; Zhu, C.; Hainline, S.; Phuah, J.; Moroco, J.A.; Smithgall, T.E.; Kuchroo, V.K.; Kane, L.P. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol. Cell. Biol. 2011, 31, 3963–3974. [Google Scholar] [CrossRef] [PubMed]
- Van de Weyer, P.S.; Muehlfeit, M.; Klose, C.; Bonventre, J.V.; Walz, G.; Kuehn, E.W. A highly conserved tyrosine of Tim-3 is phosphorylated upon stimulation by its ligand galectin-9. Biochem. Biophys. Res. Commun. 2006, 351, 571–576. [Google Scholar] [CrossRef]
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- Niki, T.; Tsutsui, S.; Hirose, S.; Aradono, S.; Sugimoto, Y.; Takeshita, K.; Nishi, N.; Hirashima, M. Galectin-9 is a high affinity IgE-binding lectin with anti-allergic effect by blocking IgE-antigen complex formation. J. Biol. Chem. 2009, 284, 32344–32352. [Google Scholar] [CrossRef] [PubMed]
- Madireddi, S.; Eun, S.Y.; Lee, S.W.; Nemcovicova, I.; Mehta, A.K.; Zajonc, D.M.; Nishi, N.; Niki, T.; Hirashima, M.; Croft, M. Galectin-9 controls the therapeutic activity of 4-1BB-targeting antibodies. J. Exp. Med. 2014, 211, 1433–1448. [Google Scholar] [CrossRef] [PubMed]
- Cuadros, C.; Dominguez, A.L.; Lollini, P.L.; Croft, M.; Mittler, R.S.; Borgstrom, P.; Lustgarten, J. Vaccination with dendritic cells pulsed with apoptotic tumors in combination with anti-OX40 and anti-4-1BB monoclonal antibodies induces T cell-mediated protective immunity in Her-2/neu transgenic mice. Int. J. Cancer 2005, 116, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Salek-Ardakani, S.; Croft, M. Tumor necrosis factor receptor/tumor necrosis factor family members in antiviral CD8 T-cell immunity. J. Interferon Cytokine Res. 2010, 30, 205–218. [Google Scholar] [CrossRef]
- Snell, L.M.; Lin, G.H.; McPherson, A.J.; Moraes, T.J.; Watts, T.H. T-cell intrinsic effects of GITR and 4-1BB during viral infection and cancer immunotherapy. Immunol. Rev. 2011, 244, 197–217. [Google Scholar] [CrossRef]
- Madireddi, S.; Eun, S.Y.; Mehta, A.K.; Birta, A.; Zajonc, D.M.; Niki, T.; Hirashima, M.; Podack, E.R.; Schreiber, T.H.; Croft, M. Regulatory T Cell-Mediated Suppression of Inflammation Induced by DR3 Signaling Is Dependent on Galectin-9. J. Immunol. 2017, 199, 2721–2728. [Google Scholar] [CrossRef] [PubMed]
- Katoh, S.; Ishii, N.; Nobumoto, A.; Takeshita, K.; Dai, S.Y.; Shinonaga, R.; Niki, T.; Nishi, N.; Tominaga, A.; Yamauchi, A.; et al. Galectin-9 inhibits CD44-hyaluronan interaction and suppresses a murine model of allergic asthma. Am. J. Respir. Crit. Care Med. 2007, 176, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Nobumoto, A.; Nagahara, K.; Oomizu, S.; Katoh, S.; Nishi, N.; Takeshita, K.; Niki, T.; Tominaga, A.; Yamauchi, A.; Hirashima, M. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology 2008, 18, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Thalhamer, T.; Franca, R.F.; Xiao, S.; Wang, C.; Hotta, C.; Zhu, C.; Hirashima, M.; Anderson, A.C.; Kuchroo, V.K. Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity 2014, 41, 270–282. [Google Scholar] [CrossRef]
- Bi, S.; Hong, P.W.; Lee, B.; Baum, L.G. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. USA 2011, 108, 10650–10655. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, K.; Webb, N.E.; Pang, M.; Hernandez-Davies, J.E.; Lee, K.P.; Gonzalez, P.; Douglass, M.V.; Lee, B.; Baum, L.G. Galectin-9 binds to O-glycans on protein disulfide isomerase. Glycobiology 2017, 27, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Querol Cano, L.; Tagit, O.; Dolen, Y.; van Duffelen, A.; Dieltjes, S.; Buschow, S.I.; Niki, T.; Hirashima, M.; Joosten, B.; van den Dries, K.; et al. Intracellular Galectin-9 Controls Dendritic Cell Function by Maintaining Plasma Membrane Rigidity. iScience 2019, 22, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Kashio, Y.; Nakamura, K.; Abedin, M.J.; Seki, M.; Nishi, N.; Yoshida, N.; Nakamura, T.; Hirashima, M. Galectin-9 induces apoptosis through the calcium-calpain-caspase-1 pathway. J. Immunol. 2003, 170, 3631–3636. [Google Scholar] [CrossRef] [PubMed]
- Oomizu, S.; Arikawa, T.; Niki, T.; Kadowaki, T.; Ueno, M.; Nishi, N.; Yamauchi, A.; Hirashima, M. Galectin-9 suppresses Th17 cell development in an IL-2-dependent but Tim-3-independent manner. Clin. Immunol. 2012, 143, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, J.; Liao, Y.; Wang, Y.; Liu, C.; Zhu, X.; Chen, Z.K.; Sun, Z. Tim-3 ligand galectin-9 reduces IL-17 level and accelerates Klebsiella pneumoniae infection. Cell. Immunol. 2011, 269, 22–28. [Google Scholar] [CrossRef]
- Gooden, M.J.; Wiersma, V.R.; Samplonius, D.F.; Gerssen, J.; van Ginkel, R.J.; Nijman, H.W.; Hirashima, M.; Niki, T.; Eggleton, P.; Helfrich, W.; et al. Galectin-9 activates and expands human T-helper 1 cells. PLoS ONE 2013, 8, e65616. [Google Scholar] [CrossRef]
- Seki, M.; Oomizu, S.; Sakata, K.M.; Sakata, A.; Arikawa, T.; Watanabe, K.; Ito, K.; Takeshita, K.; Niki, T.; Saita, N.; et al. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin. Immunol. 2008, 127, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Oomizu, S.; Arikawa, T.; Niki, T.; Kadowaki, T.; Ueno, M.; Nishi, N.; Yamauchi, A.; Hattori, T.; Masaki, T.; Hirashima, M. Cell surface galectin-9 expressing Th cells regulate Th17 and Foxp3+ Treg development by galectin-9 secretion. PLoS ONE 2012, 7, e48574. [Google Scholar] [CrossRef]
- Nagahara, K.; Arikawa, T.; Oomizu, S.; Kontani, K.; Nobumoto, A.; Tateno, H.; Watanabe, K.; Niki, T.; Katoh, S.; Miyake, M.; et al. Galectin-9 increases Tim-3+ dendritic cells and CD8+ T cells and enhances antitumor immunity via galectin-9-Tim-3 interactions. J. Immunol. 2008, 181, 7660–7669. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Facchinetti, V.; Voynova, E.; Hanabuchi, S.; Karnell, J.L.; Hanna, R.N.; Kolbeck, R.; Sanjuan, M.A.; Ettinger, R.; Liu, Y.J. Galectin-9 inhibits TLR7-mediated autoimmunity in murine lupus models. J. Clin. Investig. 2018, 128, 1873–1887. [Google Scholar] [CrossRef] [PubMed]
- Giovannone, N.; Liang, J.; Antonopoulos, A.; Geddes Sweeney, J.; King, S.L.; Pochebit, S.M.; Bhattacharyya, N.; Lee, G.S.; Dell, A.; Widlund, H.R.; et al. Galectin-9 suppresses B cell receptor signaling and is regulated by I-branching of N-glycans. Nat. Commun. 2018, 9, 3287. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, J.; Chen, F.; Liu, G.; Weng, Z.; Chen, J. Elevated Galectin-9 Suppresses Th1 Effector Function and Induces Apoptosis of Activated CD4(+) T Cells in Osteoarthritis. Inflammation 2017, 40, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- He, X.W.; Li, W.L.; Li, C.; Liu, P.; Shen, Y.G.; Zhu, M.; Jin, X.P. Serum levels of galectin-1, galectin-3, and galectin-9 are associated with large artery atherosclerotic stroke. Sci. Rep. 2017, 7, 40994. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Z.; Fu, Q.; Wang, Z.; Fu, H.; Liu, W.; Wang, Y.; Xu, J. Galectin-9 as a prognostic and predictive biomarker in bladder urothelial carcinoma. Urol. Oncol. 2017, 35, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Enninga, E.A.L.; Harrington, S.M.; Creedon, D.J.; Ruano, R.; Markovic, S.N.; Dong, H.; Dronca, R.S. Immune checkpoint molecules soluble program death ligand 1 and galectin-9 are increased in pregnancy. Am. J. Reprod. Immunol. 2018, 79, e12795. [Google Scholar] [CrossRef] [PubMed]
- Niki, T.; Fujita, K.; Rosen, H.; Hirashima, M.; Masaki, T.; Hattori, T.; Hoshino, K. Plasma Galectin-9 Concentrations in Normal and Diseased Condition. Cell. Physiol. Biochem. 2018, 50, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Ashino, Y.; Chagan-Yasutan, H.; Niki, T.; Hirashima, M.; Hattori, T. Rapid decrease of plasma galectin-9 levels in patients with acute HIV infection after therapy. Tohoku J. Exp. Med. 2012, 228, 157–161. [Google Scholar] [CrossRef]
- Tandon, R.; Chew, G.M.; Byron, M.M.; Borrow, P.; Niki, T.; Hirashima, M.; Barbour, J.D.; Norris, P.J.; Lanteri, M.C.; Martin, J.N.; et al. Galectin-9 is rapidly released during acute HIV-1 infection and remains sustained at high levels despite viral suppression even in elite controllers. Aids Res. Hum. Retrovir. 2014, 30, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.T.; Niki, T.; Furushima, D.; Bai, G.; Chagan-Yasutan, H.; Telan, E.F.; Tactacan-Abrenica, R.J.; Maeda, Y.; Solante, R.; Hattori, T. Plasma Levels of a Cleaved Form of Galectin-9 Are the Most Sensitive Biomarkers of Acquired Immune Deficiency Syndrome and Tuberculosis Coinfection. Biomolecules 2020, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Jost, S.; Moreno-Nieves, U.Y.; Garcia-Beltran, W.F.; Rands, K.; Reardon, J.; Toth, I.; Piechocka-Trocha, A.; Altfeld, M.; Addo, M.M. Dysregulated Tim-3 expression on natural killer cells is associated with increased Galectin-9 levels in HIV-1 infection. Retrovirology 2013, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.T.; Liu, Y.H.; Chen, Y.H.; Lin, C.Y.; Huang, C.H.; Yen, M.C.; Kuo, P.L. Serum Galectin-9 and Galectin-3-Binding Protein in Acute Dengue Virus Infection. Int. J. Mol. Sci. 2016, 17, 832. [Google Scholar] [CrossRef] [PubMed]
- Mengshol, J.A.; Golden-Mason, L.; Arikawa, T.; Smith, M.; Niki, T.; McWilliams, R.; Randall, J.A.; McMahan, R.; Zimmerman, M.A.; Rangachari, M.; et al. A crucial role for Kupffer cell-derived galectin-9 in regulation of T cell immunity in hepatitis C infection. PLoS ONE 2010, 5, e9504. [Google Scholar] [CrossRef]
- Nishio, A.; Tatsumi, T.; Nawa, T.; Suda, T.; Yoshioka, T.; Onishi, Y.; Aono, S.; Shigekawa, M.; Hikita, H.; Sakamori, R.; et al. CD14(+) monocyte-derived galectin-9 induces natural killer cell cytotoxicity in chronic hepatitis C. Hepatology 2017, 65, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Nebbia, G.; Peppa, D.; Schurich, A.; Khanna, P.; Singh, H.D.; Cheng, Y.; Rosenberg, W.; Dusheiko, G.; Gilson, R.; ChinAleong, J.; et al. Upregulation of the Tim-3/galectin-9 pathway of T cell exhaustion in chronic hepatitis B virus infection. PLoS ONE 2012, 7, e47648. [Google Scholar] [CrossRef] [PubMed]
- Katoh, S.; Ikeda, M.; Shimizu, H.; Mouri, K.; Obase, Y.; Kobashi, Y.; Fukushima, K.; Hirashima, M.; Oka, M. Increased levels of plasma galectin-9 in patients with influenza virus infection. Tohoku J. Exp. Med. 2014, 232, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Machala, E.A.; Avdic, S.; Stern, L.; Zajonc, D.M.; Benedict, C.A.; Blyth, E.; Gottlieb, D.J.; Abendroth, A.; McSharry, B.P.; Slobedman, B. Restriction of Human Cytomegalovirus Infection by Galectin-9. J. Virol. 2019, 93, e01746-18. [Google Scholar] [CrossRef] [PubMed]
- Gualberto Cavalcanti, N.; Melo Vilar, K.; Branco Pinto Duarte, A.L.; Barreto de Melo Rego, M.J.; Pereira, M.C.; da Rocha Pitta, I.; Diniz Lopes Marques, C.; Galdino da Rocha Pitta, M. Increased serum levels of galectin-9 in patients with chikungunya fever. Virus Res. 2020, 286, 198062. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef] [PubMed]
- Bonifacius, A.; Tischer-Zimmermann, S.; Dragon, A.C.; Gussarow, D.; Vogel, A.; Krettek, U.; Godecke, N.; Yilmaz, M.; Kraft, A.R.M.; Hoeper, M.M.; et al. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity 2021, 54, 340–354 e346. [Google Scholar] [CrossRef] [PubMed]
- Chagan-Yasutan, H.; Hanan, F.; Niki, T.; Bai, G.; Ashino, Y.; Egawa, S.; Telan, E.F.O.; Hattori, T. Plasma Osteopontin Levels is Associated with Biochemical Markers of Kidney Injury in Patients with Leptospirosis. Diagnostics 2020, 10, 439. [Google Scholar] [CrossRef] [PubMed]
- UNAIDS. UNAIDS Data 2020; UNAIDS: Geneva, Switzerland, 2020. [Google Scholar]
- Finzi, D.; Hermankova, M.; Pierson, T.; Carruth, L.M.; Buck, C.; Chaisson, R.E.; Quinn, T.C.; Chadwick, K.; Margolick, J.; Brookmeyer, R.; et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.K.; Hezareh, M.; Gunthard, H.F.; Havlir, D.V.; Ignacio, C.C.; Spina, C.A.; Richman, D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997, 278, 1291–1295. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.R.; Smurzynski, M.; Parsons, T.D.; Wu, K.; Bosch, R.J.; Wu, J.; McArthur, J.C.; Collier, A.C.; Evans, S.R.; Ellis, R.J. The prevalence and incidence of neurocognitive impairment in the HAART era. AIDS 2007, 21, 1915–1921. [Google Scholar] [CrossRef] [PubMed]
- Currier, J.S.; Lundgren, J.D.; Carr, A.; Klein, D.; Sabin, C.A.; Sax, P.E.; Schouten, J.T.; Smieja, M.; Working, G. Epidemiological evidence for cardiovascular disease in HIV-infected patients and relationship to highly active antiretroviral therapy. Circulation 2008, 118, e29–e35. [Google Scholar] [CrossRef]
- Lambotte, O.; Delfraissy, J.F. [HIV controllers: A homogeneous group of HIV-1 infected patients with a spontaneous control of viral replication]. Pathol. Biol. 2006, 54, 566–571. [Google Scholar] [CrossRef]
- Sajadi, M.M.; Constantine, N.T.; Mann, D.L.; Charurat, M.; Dadzan, E.; Kadlecik, P.; Redfield, R.R. Epidemiologic characteristics and natural history of HIV-1 natural viral suppressors. J. Acquir. Immune Defic. Syndr. 2009, 50, 403–408. [Google Scholar] [CrossRef]
- Andrade, A.; Bailey, J.R.; Xu, J.; Philp, F.H.; Quinn, T.C.; Williams, T.M.; Ray, S.C.; Thomas, D.L.; Blankson, J.N. CD4+ T cell depletion in an untreated HIV type 1-infected human leukocyte antigen-B*5801-positive patient with an undetectable viral load. Clin. Infect. Dis. 2008, 46, e78–e82. [Google Scholar] [CrossRef]
- Hunt, P.W.; Brenchley, J.; Sinclair, E.; McCune, J.M.; Roland, M.; Page-Shafer, K.; Hsue, P.; Emu, B.; Krone, M.; Lampiris, H.; et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J. Infect. Dis. 2008, 197, 126–133. [Google Scholar] [CrossRef]
- Okulicz, J.F.; Lambotte, O. Epidemiology and clinical characteristics of elite controllers. Curr. Opin. Hiv Aids 2011, 6, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Chagan-Yasutan, H.; Saitoh, H.; Ashino, Y.; Arikawa, T.; Hirashima, M.; Li, S.; Usuzawa, M.; Oguma, S.; EF, O.T.; Obi, C.L.; et al. Persistent elevation of plasma osteopontin levels in HIV patients despite highly active antiretroviral therapy. Tohoku J. Exp. Med. 2009, 218, 285–292. [Google Scholar] [CrossRef]
- Elahi, S.; Dinges, W.L.; Lejarcegui, N.; Laing, K.J.; Collier, A.C.; Koelle, D.M.; McElrath, M.J.; Horton, H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat. Med. 2011, 17, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Ndhlovu, L.C.; Barbour, J.D.; Sheth, P.M.; Jha, A.R.; Long, B.R.; Wong, J.C.; Satkunarajah, M.; Schweneker, M.; Chapman, J.M.; et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J. Exp. Med. 2008, 205, 2763–2779. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, S.; Reddy, P.B.; Rajasagi, N.; Suryawanshi, A.; Hirashima, M.; Rouse, B.T. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS Pathog 2010, 6, e1000882. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, M.; Shahbaz, S.; Fu, L.; Dunsmore, G.; Xu, L.; Harrington, R.; Houston, S.; Elahi, S. Galectin-9 Expression Defines a Subpopulation of NK Cells with Impaired Cytotoxic Effector Molecules but Enhanced IFN-gamma Production, Dichotomous to TIGIT, in HIV-1 Infection. Immunohorizons 2019, 3, 531–546. [Google Scholar] [CrossRef]
- Golden-Mason, L.; McMahan, R.H.; Strong, M.; Reisdorph, R.; Mahaffey, S.; Palmer, B.E.; Cheng, L.; Kulesza, C.; Hirashima, M.; Niki, T.; et al. Galectin-9 functionally impairs natural killer cells in humans and mice. J. Virol. 2013, 87, 4835–4845. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, S.; Dunsmore, G.; Koleva, P.; Xu, L.; Houston, S.; Elahi, S. Galectin-9 and VISTA Expression Define Terminally Exhausted T Cells in HIV-1 Infection. J. Immunol. 2020, 204, 2474–2491. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Schacker, T.W.; Ruff, L.E.; Price, D.A.; Taylor, J.H.; Beilman, G.J.; Nguyen, P.L.; Khoruts, A.; Larson, M.; Haase, A.T.; et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 2004, 200, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Stacey, A.R.; Norris, P.J.; Qin, L.; Haygreen, E.A.; Taylor, E.; Heitman, J.; Lebedeva, M.; DeCamp, A.; Li, D.; Grove, D.; et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 2009, 83, 3719–3733. [Google Scholar] [CrossRef] [PubMed]
- Herbeuval, J.P.; Hardy, A.W.; Boasso, A.; Anderson, S.A.; Dolan, M.J.; Dy, M.; Shearer, G.M. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: Role of type I IFN-producing plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA 2005, 102, 13974–13979. [Google Scholar] [CrossRef] [PubMed]
- Doitsh, G.; Cavrois, M.; Lassen, K.G.; Zepeda, O.; Yang, Z.; Santiago, M.L.; Hebbeler, A.M.; Greene, W.C. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 2010, 143, 789–801. [Google Scholar] [CrossRef]
- Doitsh, G.; Galloway, N.L.; Geng, X.; Yang, Z.; Monroe, K.M.; Zepeda, O.; Hunt, P.W.; Hatano, H.; Sowinski, S.; Munoz-Arias, I.; et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 2014, 505, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Elahi, S.; Niki, T.; Hirashima, M.; Horton, H. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood 2012, 119, 4192–4204. [Google Scholar] [CrossRef] [PubMed]
- Lhuillier, C.; Barjon, C.; Niki, T.; Gelin, A.; Praz, F.; Morales, O.; Souquere, S.; Hirashima, M.; Wei, M.; Dellis, O.; et al. Impact of Exogenous Galectin-9 on Human T Cells: Contribution of the T cell receptor complex to antigen-independent activation but not to apoptosis induction. J. Biol. Chem. 2015, 290, 16797–16811. [Google Scholar] [CrossRef]
- Abdel-Mohsen, M.; Chavez, L.; Tandon, R.; Chew, G.M.; Deng, X.; Danesh, A.; Keating, S.; Lanteri, M.; Samuels, M.L.; Hoh, R.; et al. Human Galectin-9 Is a Potent Mediator of HIV Transcription and Reactivation. PLoS Pathog 2016, 12, e1005677. [Google Scholar] [CrossRef]
- Colomb, F.; Giron, L.B.; Premeaux, T.A.; Mitchell, B.I.; Niki, T.; Papasavvas, E.; Montaner, L.J.; Ndhlovu, L.C.; Abdel-Mohsen, M. Galectin-9 Mediates HIV Transcription by Inducing TCR-Dependent ERK Signaling. Front. Immunol. 2019, 10, 267. [Google Scholar] [CrossRef]
- Shete, A.; Dhayarkar, S.; Dhamanage, A.; Kulkarni, S.; Ghate, M.; Sangle, S.; Medhe, U.; Verma, V.; Rajan, S.; Hattori, T.; et al. Possible role of plasma Galectin-9 levels as a surrogate marker of viremia in HIV infected patients on antiretroviral therapy in resource-limited settings. Aids Res. Ther. 2020, 17, 43. [Google Scholar] [CrossRef]
- Glaziou, P.; Floyd, K.; Raviglione, M.C. Global Epidemiology of Tuberculosis. Semin. Respir. Crit. Care Med. 2018, 39, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Elahi, S.; Weiss, R.H.; Merani, S. Atorvastatin restricts HIV replication in CD4+ T cells by upregulation of p21. AIDS 2016, 30, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; Margolick, J.B. Understanding frailty, aging, and inflammation in HIV infection. Curr. Hiv Aids Rep. 2015, 12, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Aberg, J.A. Aging, inflammation, and HIV infection. Top. Antivir. Med. 2012, 20, 101–105. [Google Scholar] [PubMed]
- Sharan, R.; Bucsan, A.N.; Ganatra, S.; Paiardini, M.; Mohan, M.; Mehra, S.; Khader, S.A.; Kaushal, D. Chronic Immune Activation in TB/HIV Co-infection. Trends Microbiol. 2020, 28, 699. [Google Scholar] [CrossRef] [PubMed]
- Shete, A.; Bichare, S.; Pujari, V.; Virkar, R.; Thakar, M.; Ghate, M.; Patil, S.; Vyakarnam, A.; Gangakhedkar, R.; Bai, G.; et al. Elevated Levels of Galectin-9 but Not Osteopontin in HIV and Tuberculosis Infections Indicate Their Roles in Detecting MTB Infection in HIV Infected Individuals. Front. Microbiol. 2020, 11, 1685. [Google Scholar] [CrossRef] [PubMed]
- Kathamuthu, G.R.; Kumar, N.P.; Moideen, K.; Nair, D.; Banurekha, V.V.; Sridhar, R.; Baskaran, D.; Babu, S. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases Are Potential Biomarkers of Pulmonary and Extra-Pulmonary Tuberculosis. Front. Immunol. 2020, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- WHO. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- WHO. Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control, 2nd ed.; World Health Organization: Geneva, Switzerland, 1997. [Google Scholar]
- Ho, L.J.; Wang, J.J.; Shaio, M.F.; Kao, C.L.; Chang, D.M.; Han, S.W.; Lai, J.H. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J. Immunol. 2001, 166, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Malasit, P.; Seliger, B.; Bhakdi, S.; Husmann, M. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J. Immunol. 1998, 161, 6338–6346. [Google Scholar]
- Warke, R.V.; Xhaja, K.; Martin, K.J.; Fournier, M.F.; Shaw, S.K.; Brizuela, N.; de Bosch, N.; Lapointe, D.; Ennis, F.A.; Rothman, A.L.; et al. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J. Virol. 2003, 77, 11822–11832. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Roberts, T.G.; Edgil, D.; Lu, B.; Ernst, J.; Harris, E. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 2000, 74, 4957–4966. [Google Scholar] [CrossRef] [PubMed]
- Appanna, R.; Wang, S.M.; Ponnampalavanar, S.A.; Lum, L.C.; Sekaran, S.D. Cytokine factors present in dengue patient sera induces alterations of junctional proteins in human endothelial cells. Am. J. Trop. Med. Hyg. 2012, 87, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Becquart, P.; Wauquier, N.; Nkoghe, D.; Ndjoyi-Mbiguino, A.; Padilla, C.; Souris, M.; Leroy, E.M. Acute dengue virus 2 infection in Gabonese patients is associated with an early innate immune response, including strong interferon alpha production. BMC Infect. Dis. 2010, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, C.D.; Arrieta, A.F.; Ramirez, N.D.; Rodriguez, L.S.; Vega, R.; Bosch, I.; Rodriguez, J.A.; Narvaez, C.F.; Salgado, D.M. High plasma levels of soluble ST2 but not its ligand IL-33 is associated with severe forms of pediatric dengue. Cytokine 2013, 61, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Dapat, I.C.; Pascapurnama, D.N.; Iwasaki, H.; Labayo, H.K.; Chagan-Yasutan, H.; Egawa, S.; Hattori, T. Secretion of Galectin-9 as a DAMP during Dengue Virus Infection in THP-1 Cells. Int. J. Mol. Sci. 2017, 18, 1644. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Wang, M.Y.; Ho, L.J.; Huang, C.Y.; Lai, J.H. Up-regulation of galectin-9 induces cell migration in human dendritic cells infected with dengue virus. J. Cell. Mol. Med. 2015, 19, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Malaria Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Kern, P.; Hemmer, C.J.; Van Damme, J.; Gruss, H.J.; Dietrich, M. Elevated tumor necrosis factor alpha and interleukin-6 serum levels as markers for complicated Plasmodium falciparum malaria. Am. J. Med. 1989, 87, 139–143. [Google Scholar] [CrossRef]
- Kwiatkowski, D.; Hill, A.V.; Sambou, I.; Twumasi, P.; Castracane, J.; Manogue, K.R.; Cerami, A.; Brewster, D.R.; Greenwood, B.M. TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet 1990, 336, 1201–1204. [Google Scholar] [CrossRef]
- Hojo-Souza, N.S.; Pereira, D.B.; Passos, L.S.; Gazzinelli-Guimaraes, P.H.; Cardoso, M.S.; Tada, M.S.; Zanini, G.M.; Bartholomeu, D.C.; Fujiwara, R.T.; Bueno, L.L. Phenotypic profiling of CD8(+) T cells during Plasmodium vivax blood-stage infection. BMC Infect. Dis. 2015, 15, 35. [Google Scholar] [CrossRef]
- Horne-Debets, J.M.; Karunarathne, D.S.; Faleiro, R.J.; Poh, C.M.; Renia, L.; Wykes, M.N. Mice lacking Programmed cell death-1 show a role for CD8(+) T cells in long-term immunity against blood-stage malaria. Sci. Rep. 2016, 6, 26210. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, J.; Huang, S.; Lu, F. Increased Gal-9 and Tim-3 expressions during liver damage in a murine malarial model. Parasitol. Res. 2016, 115, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, S.; Huang, S.; Pei, F.; Lu, F. Upregulated Tim-3/galectin-9 expressions in acute lung injury in a murine malarial model. Parasitol. Res. 2016, 115, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Zhao, S.; Xiang, J.; Ju, C.; Chen, X.; Gramaglia, I.; Yan, X. Targeting the CD146/Galectin-9 axis protects the integrity of the blood-brain barrier in experimental cerebral malaria. Cell. Mol. Immunol. 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Teglia, O.F.; Battagliotti, C.; Villavicencio, R.L.; Cunha, B.A. Leptospiral pneumonia. Chest 1995, 108, 874–875. [Google Scholar] [CrossRef]
- McBride, A.J.; Athanazio, D.A.; Reis, M.G.; Ko, A.I. Leptospirosis. Curr. Opin. Infect. Dis. 2005, 18, 376–386. [Google Scholar] [CrossRef]
- Matsuura, A.; Tsukada, J.; Mizobe, T.; Higashi, T.; Mouri, F.; Tanikawa, R.; Yamauchi, A.; Hirashima, M.; Tanaka, Y. Intracellular galectin-9 activates inflammatory cytokines in monocytes. Genes Cells 2009, 14, 511–521. [Google Scholar] [CrossRef]
- Faisal, S.M.; Varma, V.P.; Subathra, M.; Azam, S.; Sunkara, A.K.; Akif, M.; Baig, M.S.; Chang, Y.F. Leptospira surface adhesin (Lsa21) induces Toll like receptor 2 and 4 mediated inflammatory responses in macrophages. Sci. Rep. 2016, 6, 39530. [Google Scholar] [CrossRef]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Compagno, D.; Tiraboschi, C.; Garcia, J.D.; Rondon, Y.; Corapi, E.; Velazquez, C.; Laderach, D.J. Galectins as Checkpoints of the Immune System in Cancers, Their Clinical Relevance, and Implication in Clinical Trials. Biomolecules 2020, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Jayaraman, P.; Sada-Ovalle, I.; Beladi, S.; Anderson, A.C.; Dardalhon, V.; Hotta, C.; Kuchroo, V.K.; Behar, S.M. Tim3 binding to galectin-9 stimulates antimicrobial immunity. J. Exp. Med. 2010, 207, 2343–2354. [Google Scholar] [CrossRef]
- Jayaraman, P.; Sada-Ovalle, I.; Nishimura, T.; Anderson, A.C.; Kuchroo, V.K.; Remold, H.G.; Behar, S.M. IL-1beta promotes antimicrobial immunity in macrophages by regulating TNFR signaling and caspase-3 activation. J. Immunol. 2013, 190, 4196–4204. [Google Scholar] [CrossRef] [PubMed]
- Zajac, A.J.; Blattman, J.N.; Murali-Krishna, K.; Sourdive, D.J.; Suresh, M.; Altman, J.D.; Ahmed, R. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 1998, 188, 2205–2213. [Google Scholar] [CrossRef] [PubMed]
- Gallimore, A.; Glithero, A.; Godkin, A.; Tissot, A.C.; Pluckthun, A.; Elliott, T.; Hengartner, H.; Zinkernagel, R. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 1998, 187, 1383–1393. [Google Scholar] [CrossRef]
- Okoye, I.S.; Houghton, M.; Tyrrell, L.; Barakat, K.; Elahi, S. Coinhibitory Receptor Expression and Immune Checkpoint Blockade: Maintaining a Balance in CD8(+) T Cell Responses to Chronic Viral Infections and Cancer. Front. Immunol. 2017, 8, 1215. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, P.; Jacques, M.K.; Zhu, C.; Steblenko, K.M.; Stowell, B.L.; Madi, A.; Anderson, A.C.; Kuchroo, V.K.; Behar, S.M. TIM3 Mediates T Cell Exhaustion during Mycobacterium tuberculosis Infection. PLoS Pathog 2016, 12, e1005490. [Google Scholar] [CrossRef]
- Chavez-Galan, L.; Ramon-Luing, L.; Carranza, C.; Garcia, I.; Sada-Ovalle, I. Lipoarabinomannan Decreases Galectin-9 Expression and Tumor Necrosis Factor Pathway in Macrophages Favoring Mycobacterium tuberculosis Intracellular Growth. Front. Immunol. 2017, 8, 1659. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; Hlh Across Speciality Collaboration, U.K. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Ueland, T.; Holter, J.C.; Holten, A.R.; Muller, K.E.; Lind, A.; Bekken, G.K.; Dudman, S.; Aukrust, P.; Dyrhol-Riise, A.M.; Heggelund, L. Distinct and early increase in circulating MMP-9 in COVID-19 patients with respiratory failure. J. Infect. 2020, 81, e41–e43. [Google Scholar] [CrossRef] [PubMed]
- Ashino, Y.; Chagan-Yasutan, H.; Hatta, M.; Shirato, Y.; Kyogoku, Y.; Komuro, H.; Hattori, T. Successful Treatment of a COVID-19 Case with Pneumonia and Renal Injury Using Tocilizumab. Reports 2020, 3, 29. [Google Scholar] [CrossRef]
- Mancuso, P.; Gidaro, A.; Gregato, G.; Raveane, A.; Cremonesi, P.; Quarna, J.; Caccia, S.; Gusso, L.; Rusconi, S.; Giacomelli, A.; et al. Circulating endothelial progenitors are increased in COVID-19 patients and correlate with SARS-CoV-2 RNA in severe cases. J. Thromb. Haemost. 2020, 18, 2744–2750. [Google Scholar] [CrossRef] [PubMed]

| Disease. | Gal-9 [pg/mL] | p Value | Stat. | Statistical Analysis | Sample Type | Immuno Assay | Description | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Patients | Control | ||||||||
| HIV infection | Acute HIV 4196 (n = 3) | HC <46 (n = 30) | Data not shown | Median | Data not shown | Plasma | ELISA (GalP) | [51] | |
| Chronic HIV 325.6 (n = 58) | HC 54 (n = 19) | <0.0001 | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | [52] | ||
| HIV/TB 567 (n = 33) | HC 55.5 (n = 30) | <0.0001 | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | [53] | ||
| HIV/OI* 650 (n = 24) | <0.0001 | ||||||||
| Early untreated HIV 1000 (n = 11) | HC 160 (n = 13) | NS | Median | Mann-Whiney t test | Plasma | ELISA (Uscn) | Values were read from the figure | [54] | |
| Dengue | 1525 (n = 65) | HC 196 (n = 30) | <0.001 | Mean | Mann-Whiney test | Plasma | ELISA (GalP) | [3] | |
| 10,287 (n = 187) | HC 5061 (n = 20) | <0.0001 | Median | Mann-Whiney U test | Serum | Multiplex (RDS) | [55] | ||
| HCV infection | Chronic HCV 1276 (n = 22) | HC 112 (n = 10) | 0.0005 | Mean | Two tailed Mann-Whiney test | Plasma | ELISA (GalP) | Patients: SD = 1503 HC: SD = 210 | [56] |
| Chronic HCV 146 (n = 50) | HC 0 (n = 39) | 0.05 | Median | Mann-Whiney test | Serum | ELISA (GalP) | [57] | ||
| HBV infection | CHB (ALT>100 IU/L) 14,000 (n = 9) | HC 5700 (n = 10) | 0.02 | Mean | Mann-Whiney test | Serum | ELISA (Uscn) | Values were read from the figure | [58] |
| Influenza infection | 184 (n = 43) | HC 14 (n = 20) | <0.05 | Mean | Two tailed Mann-Whiney U test | Plasma | ELISA (GalP) | Values were read from the figure | [59] |
| HCMV infection | HCMV+ reactivators 24,400 (n = 3) | HCMV- 8800 (n = 3) | <0.001 | Mean | Bonferroni Dunn test | Plasma | ELISA (RDS) | Day-53 post transplant | [60] |
| CHIKF | 2192 (n = 44) | HC 46.88 (n = 49) | <0.0001 | Median | Mann-Whiney U test | Serum | ELISA (RDS) | [61] | |
| COVID-19 | 24,770 (n = 23) | HC 6902 (n = 15) | <0.0001 | Mean | Two tailed Mann-Whiney test | Plasma | Multiplex (RDS) | Patients: SD = 7512.62 HC: SD = 1551.97 | [62] |
| Active COVID-19 2,250,000 (n = 65-92) | Recovered COVID-19 500,000 (n = 47-66) | <0.0001 | Mean | Krustal-Wallis test and Dunn’s test | Plasma | Multiplex (BioLegend) | Values were read from the figure | [63] | |
| HC 450,000 (n = 24-43) | <0.0001 | ||||||||
| Malaria | Day-0 illness 686.5 (n = 50) | Day-28 illness 243 (n = 50) | <0.0001 | Median | Mann-Whiney test | Plasma | ELISA (GalP) | [5] | |
| Leptospirosis | 613 (n = 111) | HC 196 (n = 30) | <0.0001 | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | [64] | |
| TB | 358 (n = 49) | HC 55.5 (n = 30) | <0.0001 | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | [53] | |
| 171,500 (n = 36) | HC 14,000 (n = 19) | 0.0002 | Median | Mann-Whiney U test | Plasma | ELISA (RDS) | Patients: active PTB | [4] | |
| Disease | Gal-9 [pg/mL] | p Value | Stat. | Statistical Analysis | Sample Type | Immuno Assay | Description | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| HIV infection | Non-contollers 555.6 (n = 20) | Elite-controllers 250 (n = 20) | NS | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | Values of Elite-cont. and ART-suppressed were read from the figure | [52] |
| ART-suppressed 200 (n = 20) | NS | ||||||||
| Aviremic 4000 (n = 62) | Viremic 11,000 (n = 43) | <0.0001 | Data not shown | One tailed Mann-Whiney test | Plasma | ELISA (RDS) | Values were read from the figure | [91] | |
| Untreated chronic HIV 398 (n = 47) | Early untreated HIV 1000 (n = 11) | <0.001 | Median | Mann-Whiney t test | Plasma | ELISA (Uscn) | Values were read from the figure | [54] | |
| HAART-treated HIV 398 (n = 15) | 0.0009 | ||||||||
| HIV/OI * | Alive 554 (n = 21) | Dead 1042 (n = 3) | 0.0301 | Median | Mann-Whiney U test | Plasma | ELISA (GalP) | [53] | |
| HIV/TB | HIV/LTBI 3500 (n = 17) | HIV/PTB 11,290 (n = 14) | ≤0.001 | Median | Krustal-Wallis test | Plasma | ELISA (RDS) | [97] | |
| Dengue | DF 1407 (n = 53) | DHF 2464 (n = 12) | Data not shown | Mean | Mann-Whiney test | Plasma | ELISA (GalP) | [3] | |
| HCV infection | HCV infection alone 715 (n = 15) | HCV infection with hepatocellular carcinoma 1376 (n = 7) | Data not shown | Median | Two tailed Mann-Whiney test | Plasma | ELISA (GalP) | [56] | |
| SVR after treatment 20.8 (n = 24) | chronic HCV infection 146 (n = 50) | 0.05 | Median | Mann-Whiney test | Serum | ELISA (GalP) | [57] | ||
| HBV infection | CHB (ALT<50 IU/L) 6000 (n = 16) | CHB (ALT>100 IU/L) 14,000 (n = 9) | 0.01 | Mean | Mann-Whiney test | Serum | ELISA (Uscn) | Values were read from the figure | [58] |
| HCMV infection | HCMV+ non- reactivators 14,100 (n = 3) | HCMV+ reactivators 24,400 (n = 3) | <0.01 | Mean | Bonferroni Dunn test | Plasma | ELISA (RDS) | Day-53 post transplant | [60] |
| Malaria | UM (n = 41) 617 348 | SM (n = 9) 923 659 | 0.03 0.02 | Median | Mann-Whiney test | Plasma | ELISA (GalP) | Day-0 illness Day-7 illness | [5] |
| BUN/creatinine <20 (mg/dL) 576.2 (n = 28) | BUN/creatinine ≥20 (mg/dL) 817.3 (n = 22) | 0.007 | Day-0 illness | ||||||
| TB | LTBI 1190 (n = 22) | EPTB 6800 (n = 33) | ≤0.001 | Median | Krustal-Wallis test with Dunn's test | Plasma | ELISA (RDS) | [97] | |
| PTB 5900 (n = 21) | ≤0.001 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwasaki-Hozumi, H.; Chagan-Yasutan, H.; Ashino, Y.; Hattori, T. Blood Levels of Galectin-9, an Immuno-Regulating Molecule, Reflect the Severity for the Acute and Chronic Infectious Diseases. Biomolecules 2021, 11, 430. https://doi.org/10.3390/biom11030430
Iwasaki-Hozumi H, Chagan-Yasutan H, Ashino Y, Hattori T. Blood Levels of Galectin-9, an Immuno-Regulating Molecule, Reflect the Severity for the Acute and Chronic Infectious Diseases. Biomolecules. 2021; 11(3):430. https://doi.org/10.3390/biom11030430
Chicago/Turabian StyleIwasaki-Hozumi, Hiroko, Haorile Chagan-Yasutan, Yugo Ashino, and Toshio Hattori. 2021. "Blood Levels of Galectin-9, an Immuno-Regulating Molecule, Reflect the Severity for the Acute and Chronic Infectious Diseases" Biomolecules 11, no. 3: 430. https://doi.org/10.3390/biom11030430
APA StyleIwasaki-Hozumi, H., Chagan-Yasutan, H., Ashino, Y., & Hattori, T. (2021). Blood Levels of Galectin-9, an Immuno-Regulating Molecule, Reflect the Severity for the Acute and Chronic Infectious Diseases. Biomolecules, 11(3), 430. https://doi.org/10.3390/biom11030430








