Impacts of Maternal Diet and Alcohol Consumption during Pregnancy on Maternal and Infant Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Statistical Methods
3. Results
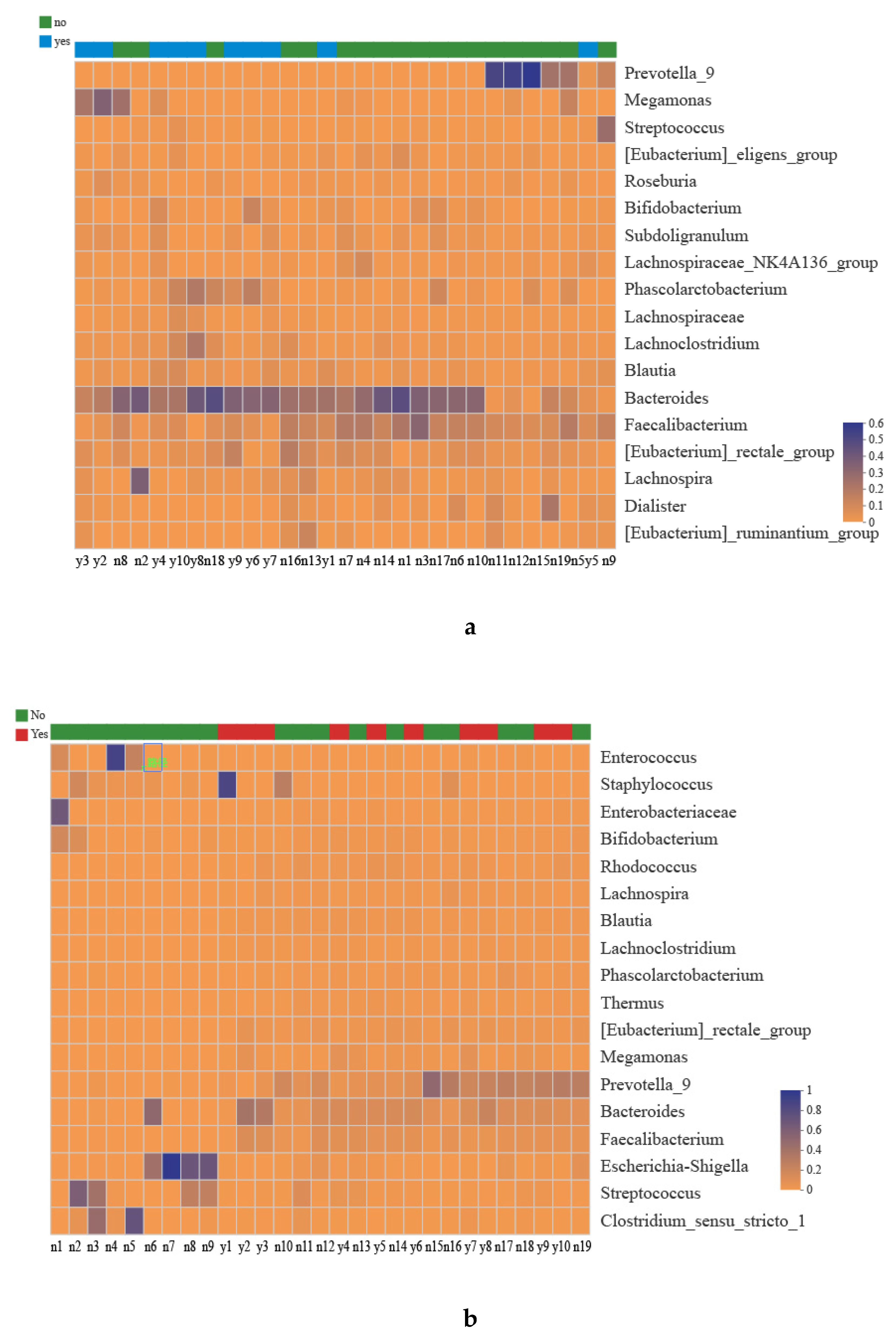
3.1. Taxonomies of the Gut Microbiota
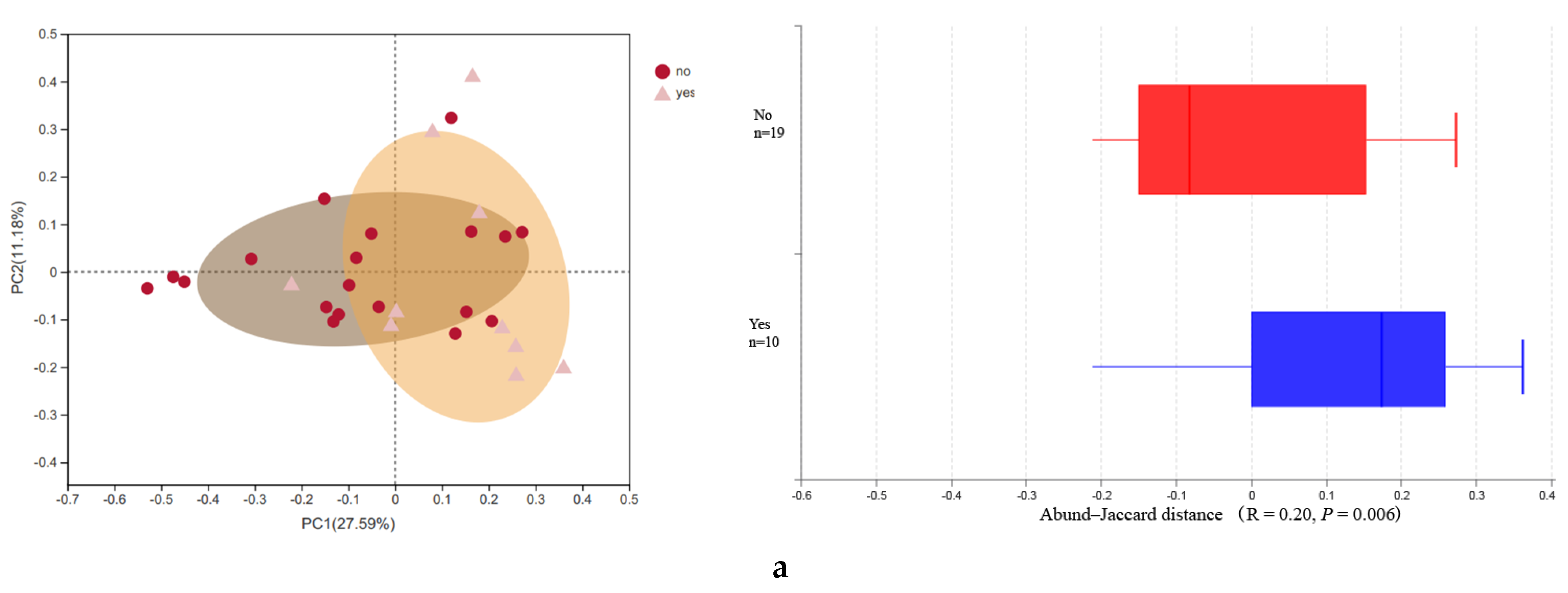
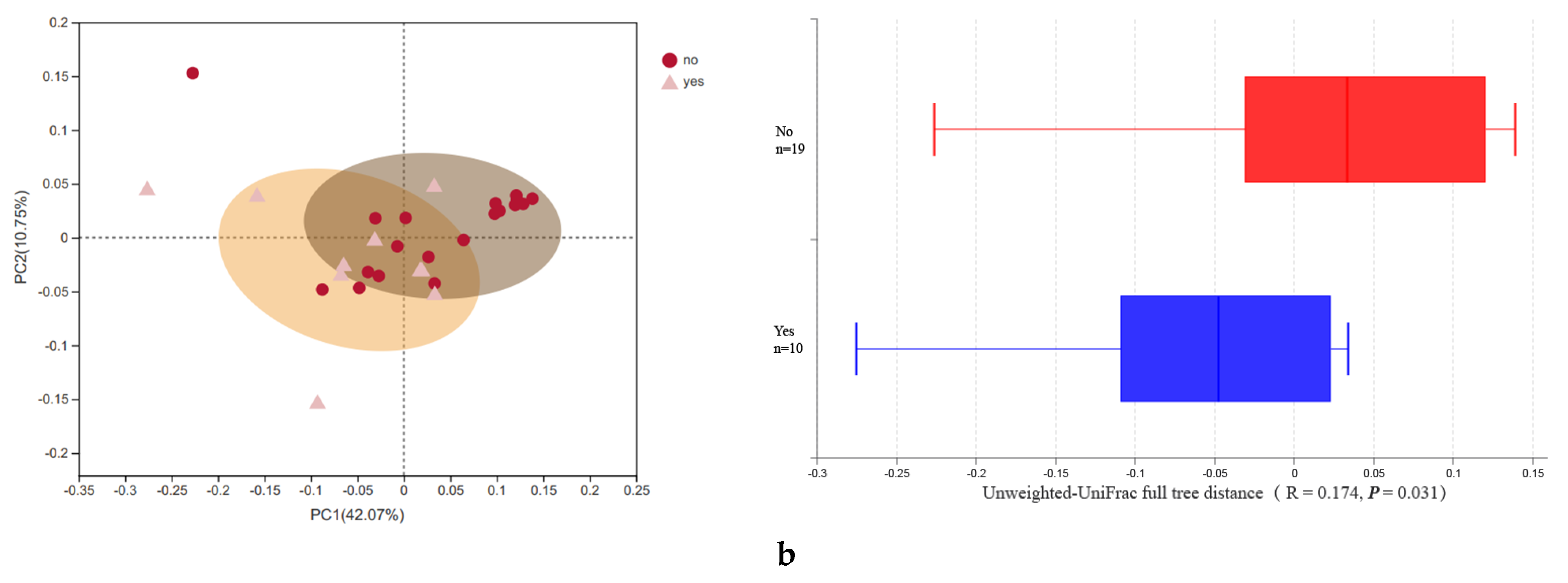
3.2. Microbial Diversity
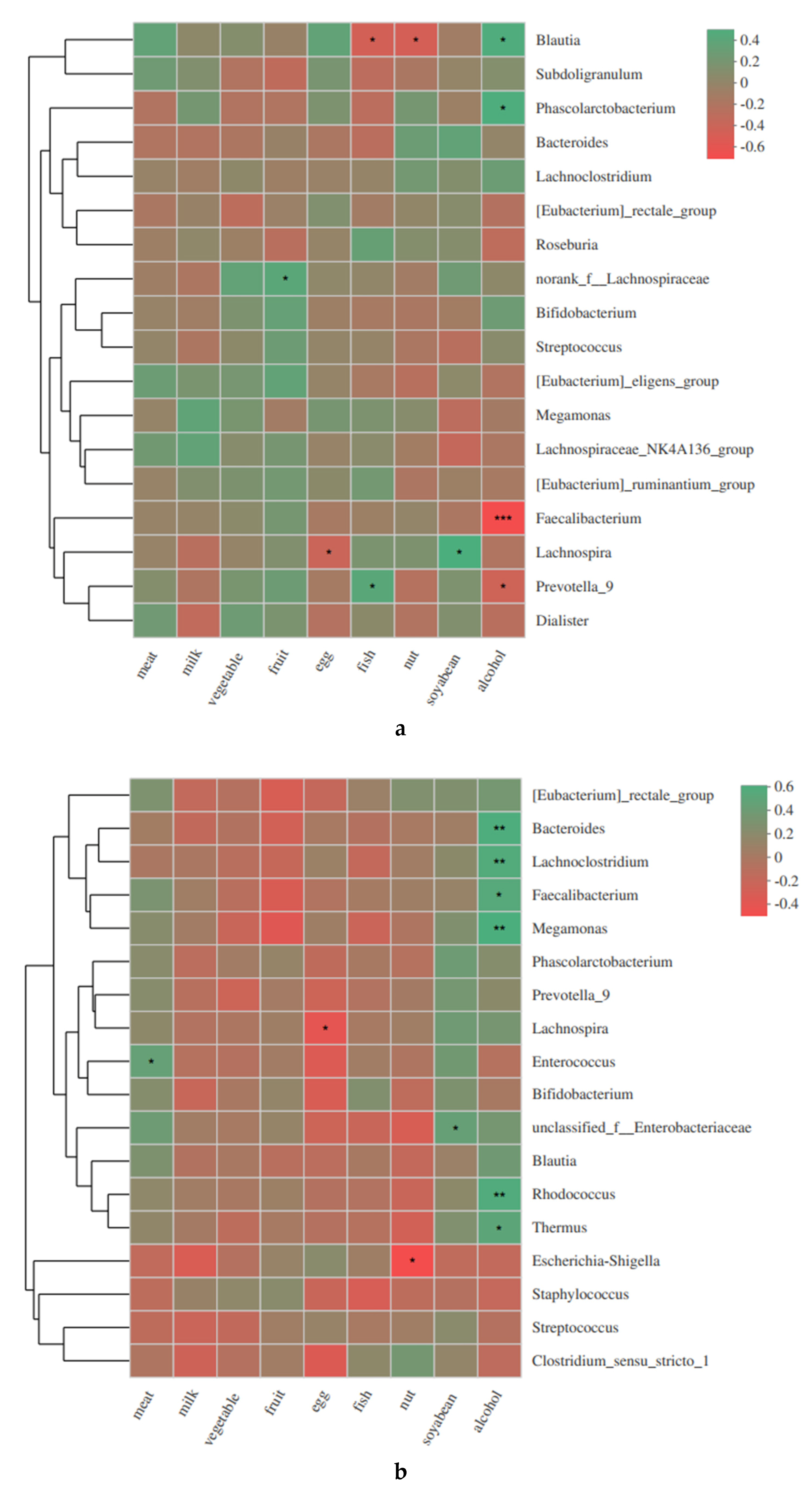
3.3. Associations between the Maternal Alcohol Consumption, Diet and Gut Microbiota Changes in Mothers and Infants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, J.; Hu, Y.; Bruner, D.W. Composition of gut microbiota and its association with body mass index and lifestyle factors in a cohort of 7-18 years old children from the American Gut Project. Pediatr. Obes. 2019, 14, e12480. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.L.; Yu, Y.; Liu, Y.Q.; Zhang, Q.; Bai, J.B. Association between Gut Microbiota and Infant’s Temperament in the First Year of Life in a Chinese Birth Cohort. Microorganisms 2020, 8, 753. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- McDonald, D.; Birmingham, A.; Knight, R. Context and the human microbiome. Microbiome 2015, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut microbes and the brain: Paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Chang, J.Y.; Shin, S.M.; Chun, J.; Lee, J.H.; Seo, J.K. Pyrosequencing-based molecular monitoring of the intestinal bacterial colonization in preterm infants. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 512–519. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Llop, S.; Valles, Y.; Moya, A.; Ballester, F.; Francino, M.P. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 2013, 43, 198–211. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Yang, K.; Yang, P.; Zhong, C.; Chen, C.; Wang, S.; Lu, Q.; Ning, K. Stratification of athletes’ gut microbiota: The multifaceted hubs associated with dietary factors, physical characteristics and performance. Gut Microbes 2020, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr 2013, 98, 111–120. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, J.E.; Zhou, Y.J.; McGeachie, M.J.; Ziniti, J.; Lange, N.; Laranjo, N.; Savage, J.R.; Carey, V.; O’Connor, G.; Sandel, M.; et al. Factors influencing the infant gut microbiome at age 3-6 months: Findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART). J. Allergy Clin. Immun. 2017, 139, 482–491.e14. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, S.N.; Madan, J.C.; Emond, J.A.; Morrison, H.G.; Christensen, B.C.; Karagas, M.R.; Hoen, A.G. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Selma-Royo, M.; Garcia-Mantrana, I.; Calatayud, M.; Parra-Llorca, A.; Martinez-Costa, C.; Collado, M.C. Maternal diet during pregnancy and intestinal markers are associated with early gut microbiota. Eur. J. Nutr. 2020. [Google Scholar] [CrossRef]
- Ma, J.; Prince, A.L.; Bader, D.; Hu, M.; Ganu, R.; Baquero, K.; Blundell, P.; Harris, R.A.; Frias, A.E.; Grove, K.L.; et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.S.; Jayaprakasha, G.K.; Vikram, A. Indigenous Crops of Asia and Southeast Asia: Exploring Health-promoting Properties. Hortscience 2012, 47, 821–827. [Google Scholar] [CrossRef]
- Diep, C.S.; Leung, R.; Thompson, D.I.; Gor, B.J.; Baranowski, T. Culture and Diet Among Chinese American Children Aged 9-13 Years: A Qualitative Study. J. Nutr. Educ. Behav. 2017, 49, 275–284. [Google Scholar] [CrossRef][Green Version]
- Rodriguez, L.M.; Litt, D.M.; Stewart, S.H. Drinking to cope with the pandemic: The unique associations of COVID-19-related perceived threat and psychological distress to drinking behaviors in American men and women. Addict. Behav. 2020, 110, 106532. [Google Scholar] [CrossRef]
- Peltier, M.R.; Verplaetse, T.L.; Mineur, Y.S.; Petrakis, I.L.; Cosgrove, K.P.; Picciotto, M.R.; McKee, S.A. Sex differences in stress-related alcohol use. Neurobiol. Stress 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Akison, L.K.; Kuo, J.; Reid, N.; Boyd, R.N.; Moritz, K.M. Effect of Choline Supplementation on Neurological, Cognitive, and Behavioral Outcomes in Offspring Arising from Alcohol Exposure During Development: A Quantitative Systematic Review of Clinical and Preclinical Studies. Alcohol. Clin. Exp. Res. 2018, 42, 1591–1611. [Google Scholar] [CrossRef] [PubMed]
- Muggl, E.; O’Leary, C.; Donath, S.; Orsini, F.; Forster, D.; Anderson, P.J.; Lewis, S.; Nagle, C.; Craig, J.M.; Elliott, E.J.B.P.H. “Did you ever drink more?” A detailed description of pregnant women’s drinking patterns. BMC Public Health, 2016; 16, 1–13. [Google Scholar]
- Isaacs, D. Maternal alcohol use and sudden infant death syndrome. J. Paediatr. Child. H 2016, 16, 871. [Google Scholar] [CrossRef][Green Version]
- Sundermann, A.C.; Velez Edwards, D.R.; Slaughter, J.C.; Wu, P.; Jones, S.H.; Torstenson, E.S.; Hartmann, K.E. Week-by-week alcohol consumption in early pregnancy and spontaneous abortion risk: A prospective cohort study. Am. J. Obstet Gynecol. 2020. [Google Scholar] [CrossRef]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Bull-Otterson, L.; Feng, W.; Kirpich, I.; Wang, Y.; Qin, X.; Liu, Y.; Gobejishvili, L.; Joshi-Barve, S.; Ayvaz, T.; Petrosino, J.; et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS ONE 2013, 8, e53028. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.K.; Seth, A.; Sheth, P. Recent Advances in Alcoholic Liver Disease I. Role of intestinal permeability and endotoxemia in alcoholic liver disease. Am. J. Physiol. Gastrointest Liver Physiol. 2004, 286, G881–G884. [Google Scholar] [CrossRef]
- Ames, N.J.; Barb, J.J.; Schuebel, K.; Mudra, S.; Meeks, B.K.; Tuason, R.T.S.; Brooks, A.T.; Kazmi, N.; Yang, S.; Ratteree, K.; et al. Longitudinal gut microbiome changes in alcohol use disorder are influenced by abstinence and drinking quantity. Gut Microbes 2020, 11, 1608–1631. [Google Scholar] [CrossRef]
- Lowe, P.P.; Gyongyosi, B.; Satishchandran, A.; Iracheta-Vellve, A.; Ambade, A.; Kodys, K.; Catalano, D.; Ward, D.V.; Szabo, G. Alcohol-related changes in the intestinal microbiome influence neutrophil infiltration, inflammation and steatosis in early alcoholic hepatitis in mice. PLoS ONE 2017, 12, e0174544. [Google Scholar] [CrossRef]
- Guo, Z. Interpretation of “Dietary Guidelines for Pregnant Women (2016)” by The Chinese Nutrition Society. J. Pract. Obstetr. Gynecol. 2018, 34. [Google Scholar]
- Wang, Y.; Liu, Y.; Bai, J.; Chen, X. The Effect of Maternal Postpartum Practices on Infant Gut Microbiota: A Chinese Cohort Study. Microorganisms 2019, 7, 511. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 22 February 2021).
- Yan, A.W.; Fouts, D.E.; Brandl, J.; Starkel, P.; Torralba, M.; Schott, E.; Tsukamoto, H.; Nelson, K.E.; Brenner, D.A.; Schnabl, B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 2011, 53, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Kumbhare, S.V.; Patangia, D.V.V.; Patil, R.H.; Shouche, Y.S.; Patil, N.P. Factors influencing the gut microbiome in children: From infancy to childhood. J. Biosci. 2019, 44. [Google Scholar] [CrossRef]
- Labrecque, M.T.; Malone, D.; Caldwell, K.E.; Allan, A.M. Impact of Ethanol and Saccharin on Fecal Microbiome in Pregnant and Non-Pregnant Mice. J. Pregnancy Child. Health 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, Y.; Carnielli, V.P.; Ksiazyk, J.; Luna, M.S.; Migacheva, N.; Mosselmans, J.M.; Picaud, J.C.; Possner, M.; Singhal, A.; Wabitsch, M. Factors affecting early-life intestinal microbiota development. Nutrition 2020, 78. [Google Scholar] [CrossRef]
- Chu, D.M.; Meyer, K.M.; Prince, A.L.; Aagaard, K.M. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function (vol 7, pg 459, 2016). Gut Microbes 2017, 8. [Google Scholar] [CrossRef]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pract. Res. Cl Ga 2017, 31, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, J.; Ran, Z.H. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroent Res. Pract. 2014, 2014. [Google Scholar] [CrossRef]
- Eppinga, H.; Weiland, C.J.S.; Thio, H.B.; van der Woude, C.J.; Nijsten, T.E.C.; Peppelenbosch, M.P.; Konstantinov, S.R. Similar Depletion of Protective Faecalibacterium prausnitzii in Psoriasis and Inflammatory Bowel Disease, but not in Hidradenitis Suppurativa. J. Crohns Colitis 2016, 10, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Banan, A.; Shaikh, M.; Zhang, L.J.; Forsythe, C.; Choudhary, S.; Farhadi, A.; Fields, J.Z.; Keshavarzian, A. NF-kappaB activation as a key mechanism in ethanol-induced disruption of the F-actin cytoskeleton and barrier integrity of intestinal epithelial monolayers. Alcohol Clin. Exp. Res. 2005, 29, 139a. [Google Scholar]
- Leclercq, S.; Matamoros, S.; Cani, P.D.; Neyrinck, A.M.; Jamar, F.; Starkel, P.; Windey, K.; Tremaroli, V.; Backhed, F.; Verbeke, K.; et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA 2014, 111, E4485–E4493. [Google Scholar] [CrossRef] [PubMed]
- Sobhonslidsuk, A.; Chanprasertyothin, S.; Pongrujikorn, T.; Kaewduang, P.; Promson, K.; Petraksa, S.; Ongphiphadhanakul, B. The Association of Gut Microbiota with Nonalcoholic Steatohepatitis in Thais. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Sterbini, F.P.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Canesso, M.C.C.; Queiroz, N.L.; Marcantonio, C.; Lauar, J.; Almeida, D.; Gamba, C.; Cassali, G.; Pedroso, S.H.; Moreira, C.; Martins, F.S.; et al. Comparing the effects of acute alcohol consumption in germ-free and conventional mice: The role of the gut microbiota. BMC Microbiol. 2014, 14, 240. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Ciafrè, S.; Ferraguti, G.; Greco, A.; Polimeni, A.; Ralli, M.; Ceci, F.M.; Ceccanti, M.; Fiore, M. Alcohol as an early life stressor: Epigenetics, metabolic, neuroendocrine and neurobehavioral implications. Neuroscience Biobehav. Rev. 2020, 118, 654–668. [Google Scholar] [CrossRef]
- Lees, B.; Mewton, L.; Jacobus, J.; Valadez, E.A.; Stapinski, L.A.; Teesson, M.; Tapert, S.F.; Squeglia, L.M. Association of Prenatal Alcohol Exposure With Psychological, Behavioral, and Neurodevelopmental Outcomes in Children From the Adolescent Brain Cognitive Development Study. Am. J. Psychiat 2020, 177, 1060–1072. [Google Scholar] [CrossRef]
- Wang, X.J.; Carlson, V.C.C.; Studholme, C.; Newman, N.; Ford, M.M.; Grant, K.A.; Kroenke, C.D. In utero MRI identifies consequences of early-gestation alcohol drinking on fetal brain development in rhesus macaques. Proc. Natl. Acad. Sci. USA 2020, 117, 10035–10044. [Google Scholar] [CrossRef]
- Tun, H.M.; Bridgman, S.L.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Sears, M.R.; et al. Roles of Birth Mode and Infant Gut Microbiota in Intergenerational Transmission of Overweight and Obesity From Mother to Offspring. JAMA Pediatr. 2018, 172, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Odenyo, A.A.; Bishop, R.; Asefa, G.; Jamnadass, R.; Odongo, D.; Osuji, P. Characterization of tannin-tolerant bacterial isolates from East African ruminants. Anaerobe 2001, 7, 5–15. [Google Scholar] [CrossRef]
- Wiseman, H.; Casey, K.; Bowey, E.A.; Duffy, R.; Davies, M.; Rowland, I.R.; Lloyd, A.S.; Murray, A.; Thompson, R.; Clarke, D.B. Influence of 10 wk of soy consumption on plasma concentrations and excretion of isoflavonoids and on gut microflora metabolism in healthy adults. Am. J. Clin. Nutr. 2004, 80, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Minamida, K.; Tanaka, M.; Abe, A.; Sone, T.; Tomita, F.; Hara, H.; Asano, K. Production of equol from daidzein by gram-positive rod-shaped bacterium isolated from rat intestine. J. Biosci. Bioeng. 2006, 102, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.M.; Wahala, K.; Adlercreutz, H. Identification of urinary metabolites of the red clover isoflavones formononetin and biochanin A in human subjects. J. Agric. Food Chem. 2004, 52, 6802–6809. [Google Scholar] [CrossRef]
- Coldham, N.G.; Darby, C.; Hows, M.; King, L.J.; Zhang, A.Q.; Sauer, M.J. Comparative metabolism of genistin by human and rat gut microflora: Detection and identification of the end-products of metabolism. Xenobiotica 2002, 32, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Paul, H.A.; Bomhof, M.R.; Vogel, H.J.; Reimer, R.A. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Sci. Rep. 2016, 6, 20683. [Google Scholar] [CrossRef] [PubMed]
- Parnell, J.A.; Reimer, R.A. Prebiotic fiber modulation of the gut microbiota improves risk factors for obesity and the metabolic syndrome. Gut Microbes 2012, 3, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Legarrea, P.; Fuller, N.R.; Zulet, M.A.; Martinez, J.A.; Caterson, I.D. The influence of Mediterranean, carbohydrate and high protein diets on gut microbiota composition in the treatment of obesity and associated inflammatory state. Asia Pac. J. Clin. Nutr. 2014, 23, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Lahtinen, S.J.; Boyle, R.J.; Kivivuori, S.; Oppedisano, F.; Smith, K.R.; Robins-Browne, R.; Salminen, S.J.; Tang, M.L.K. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. J. Allergy Clin. Immun. 2009, 123, 499–501. [Google Scholar] [CrossRef]
- Chu, D.M.; Antony, K.M.; Ma, J.; Prince, A.L.; Showalter, L.; Moller, M.; Aagaard, K.M. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef]
- Penney, N.; Barton, W.; Posma, J.M.; Darzi, A.; Frost, G.; Cotter, P.D.; Holmes, E.; Shanahan, F.; O’Sullivan, O.; Garcia-Perez, I. Investigating the Role of Diet and Exercise in Gut Microbe-Host Cometabolism. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Kaczmarek, J.L.; Musaad, S.M.A.; Holscher, H.D. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am. J. Clin. Nutr. 2017, 106, 1220–1231. [Google Scholar] [CrossRef]
| Variable | Maternal Alcohol Consumption | p Value | ||
|---|---|---|---|---|
| Yes (10, 34.38%) | No (19, 65.52%) | |||
| Maternal age | 29.8 ± 2.04 | 30.9 ± 4.15 | 0.44 | |
| Pre-BMI | 20.77 ± 3.02 | 21.15 ± 2.63 | 0.73 | |
| GWG | 15.95 ± 1.63 | 15.53 ± 4.52 | 0.82 | |
| Gestational week | 40.13 ± 1.08 | 39.55 ± 4.15 | 0.15 | |
| Mode of delivery | C-section | 7 (24.14%) | 8 (27.59%) | 0.16 |
| Vaginal delivery | 3 (10.34%) | 11 (37.93%) | ||
| Infant gender | Male | 5 (17.24%) | 5 (17.24%) | 0.24 |
| Female | 5 (17.24%) | 14 (48.28%) | ||
| Newborns’ alpha diversity | Shannon | 4.01 ± 1.07 | 2.62 ± 1.70 | 0.03 * |
| Maternal alpha diversity | Shannon | 3.66 ± 0.50 | 3.19 ± 0.49 | 0.02 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Xie, T.; Wu, Y.; Liu, Y.; Zou, Z.; Bai, J. Impacts of Maternal Diet and Alcohol Consumption during Pregnancy on Maternal and Infant Gut Microbiota. Biomolecules 2021, 11, 369. https://doi.org/10.3390/biom11030369
Wang Y, Xie T, Wu Y, Liu Y, Zou Z, Bai J. Impacts of Maternal Diet and Alcohol Consumption during Pregnancy on Maternal and Infant Gut Microbiota. Biomolecules. 2021; 11(3):369. https://doi.org/10.3390/biom11030369
Chicago/Turabian StyleWang, Ying, Tianqu Xie, Yinyin Wu, Yanqun Liu, Zhijie Zou, and Jinbing Bai. 2021. "Impacts of Maternal Diet and Alcohol Consumption during Pregnancy on Maternal and Infant Gut Microbiota" Biomolecules 11, no. 3: 369. https://doi.org/10.3390/biom11030369
APA StyleWang, Y., Xie, T., Wu, Y., Liu, Y., Zou, Z., & Bai, J. (2021). Impacts of Maternal Diet and Alcohol Consumption during Pregnancy on Maternal and Infant Gut Microbiota. Biomolecules, 11(3), 369. https://doi.org/10.3390/biom11030369







