Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genes, Strains and Reagents
2.2. Recombinant Plasmid Construction
2.3. Protein Expression and Purification
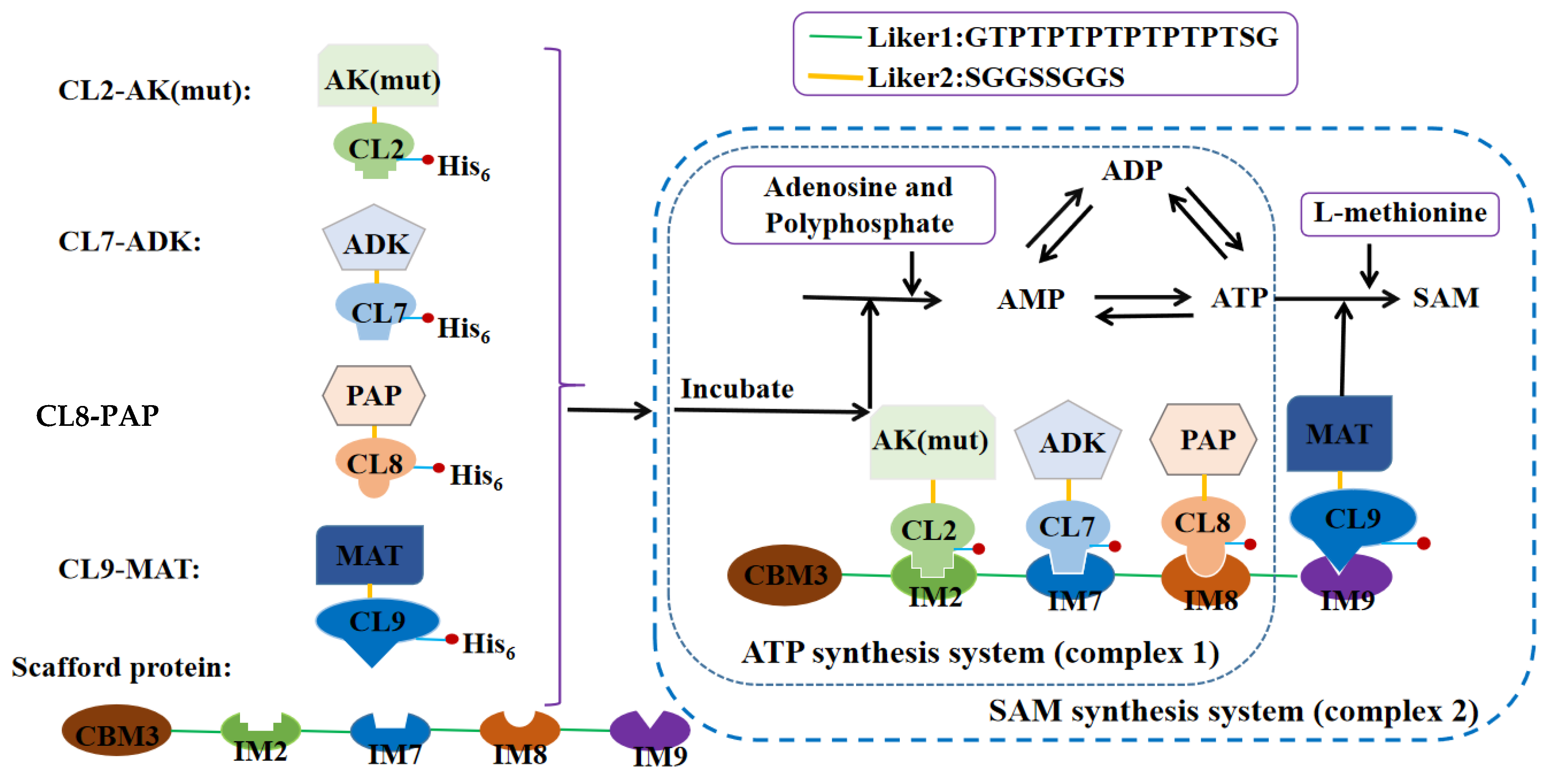
2.4. The Synthesis of ATP and SAM by Multidomain Scaffold Proteins (Un-Published in Our Lab)
2.5. Assembly of Enzyme and Scaffold Protein
2.6. Detection and Analysis by High-Performance Liquid Chromatography (HPLC)
2.7. Effect of Temperature, pH, Metal Ion Concentration and Reaction Time on ATP Synthesis
2.8. Effect of Temperature, pH, Metal Ion Concentration and Substrate Concentration on SAM Synthesis
3. Results
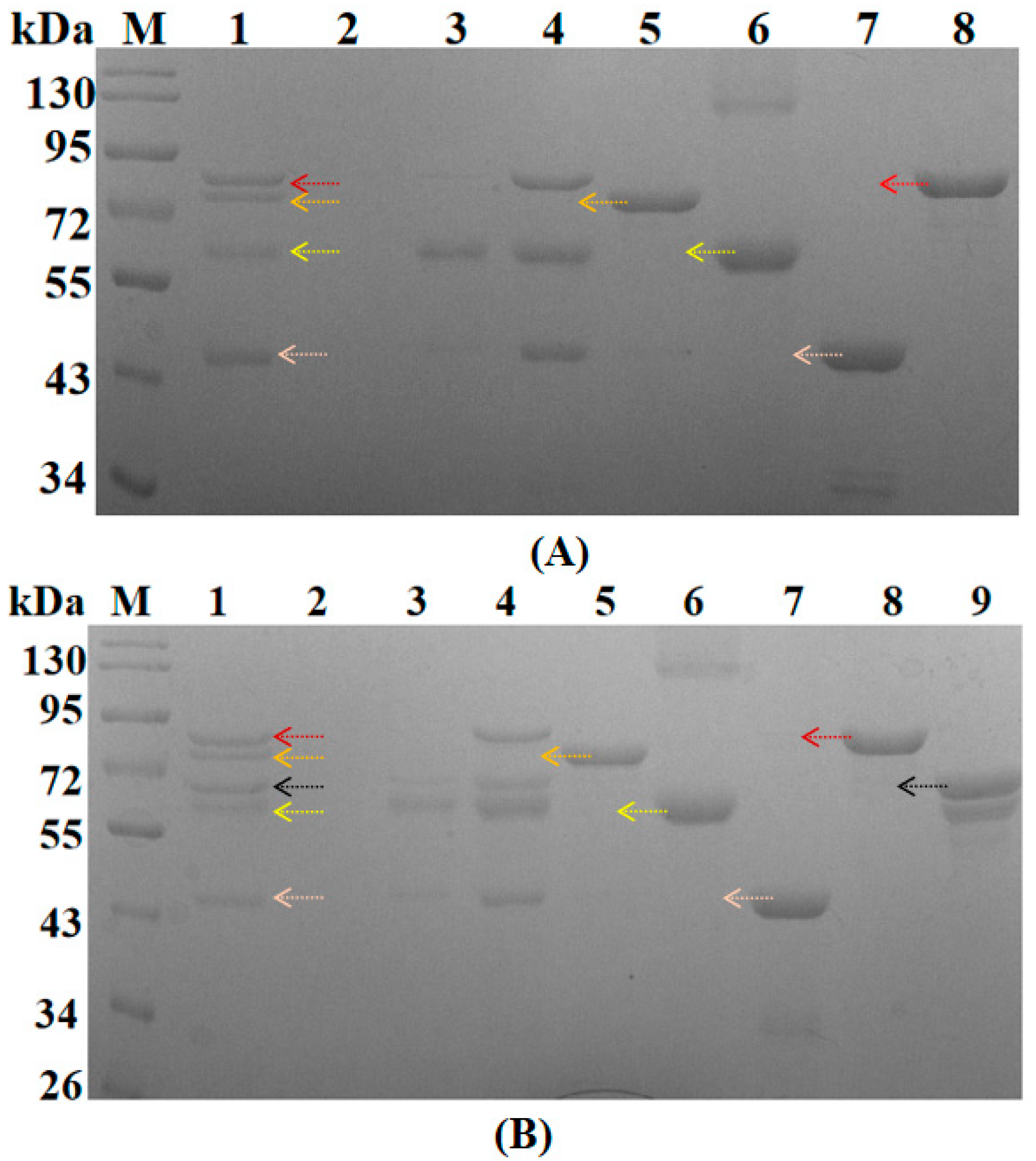
3.1. Expression and Purification of Components of the Engineered Multidomain Scaffold Proteins
3.2. The Assembly of Scaffold Proteins
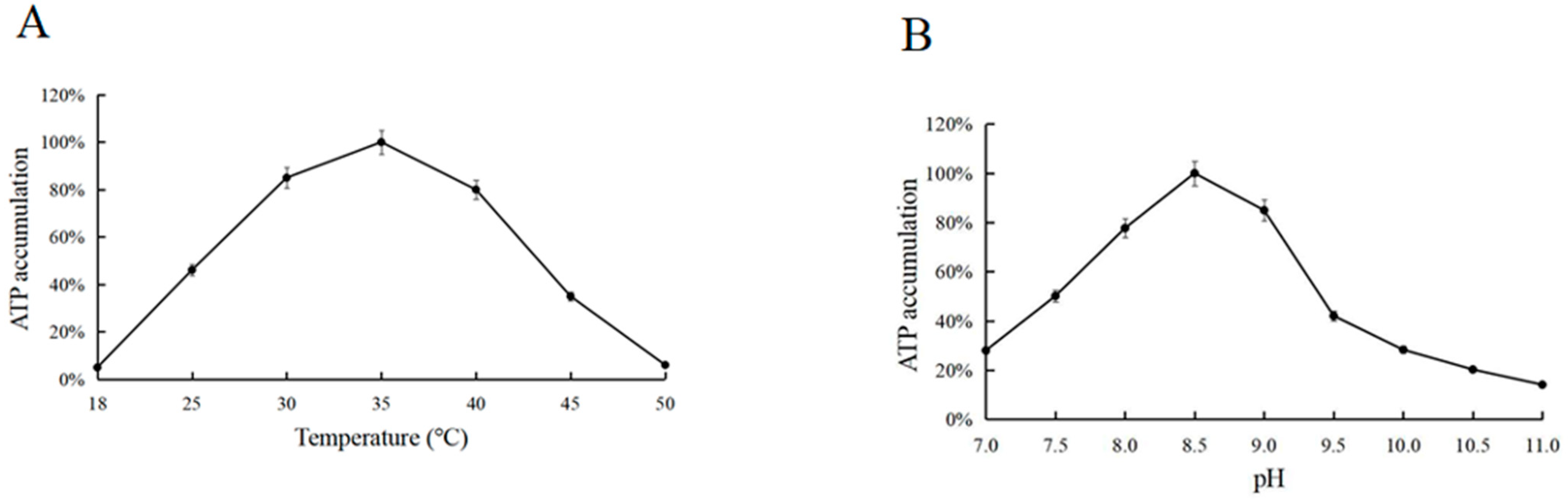
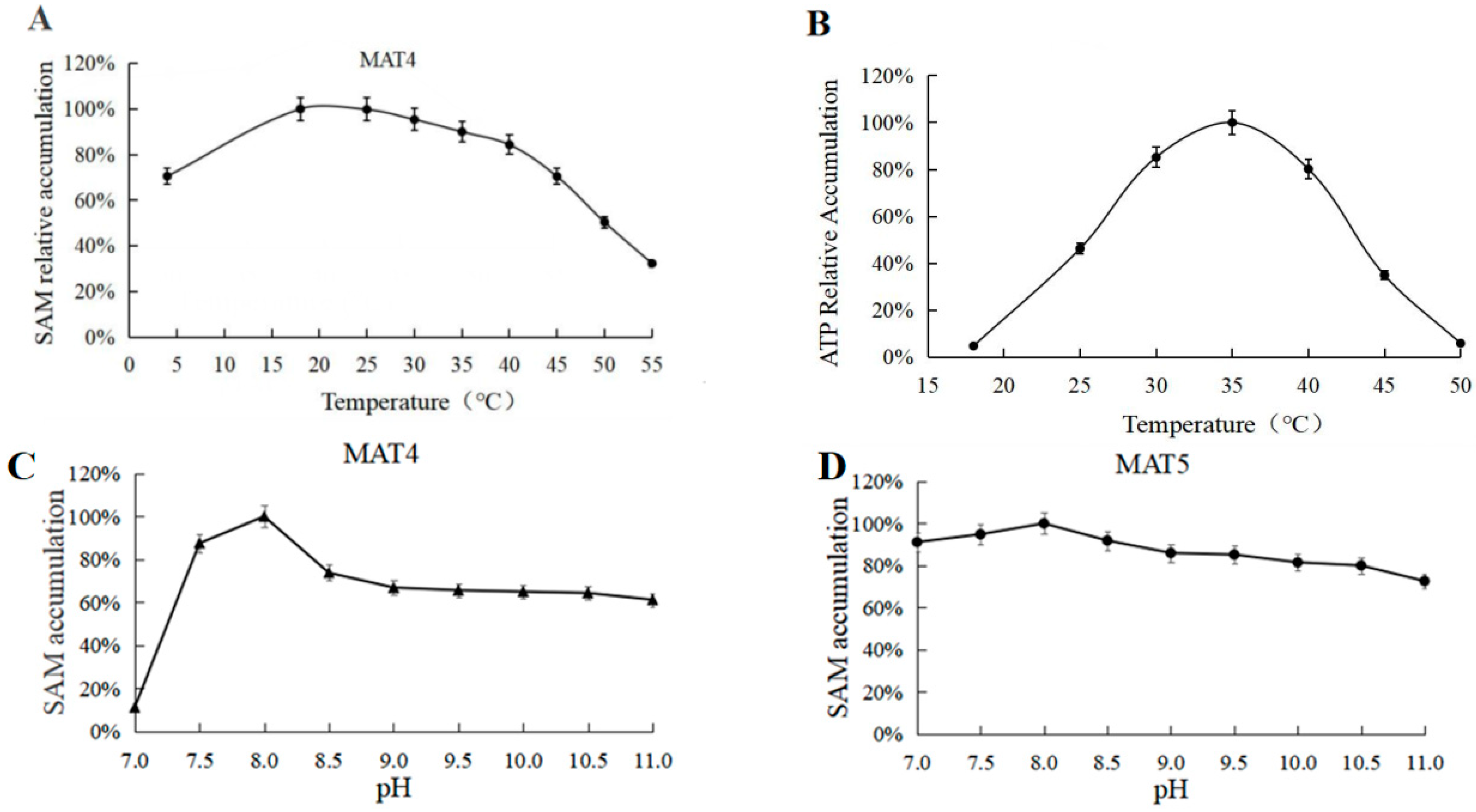
3.3. Optimum Reaction Temperature and pH for ATP Synthesis
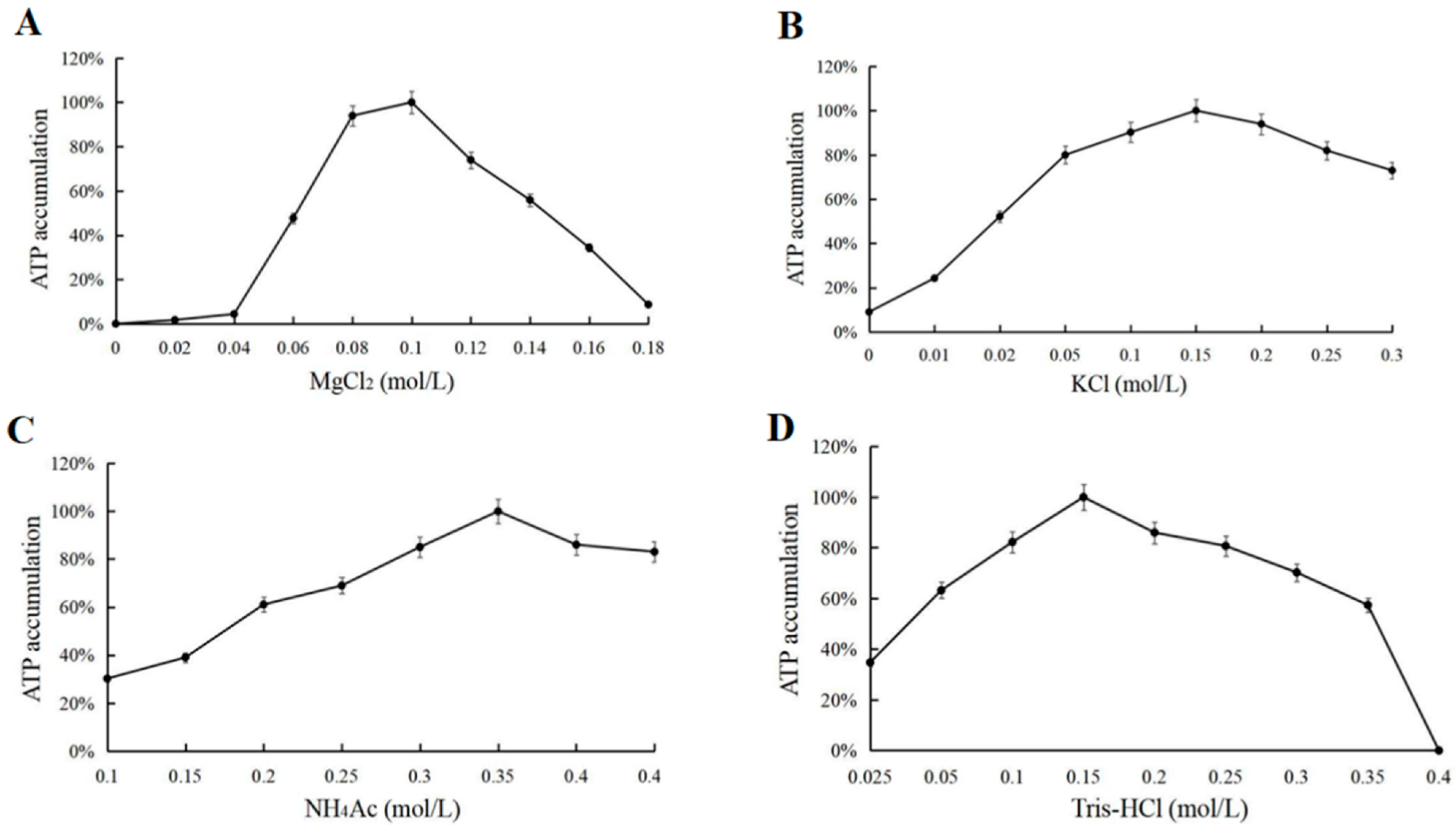
3.4. Effect of the Concentration of Mg2+, K+, NH4+, Na+, Mn2+ and Tris-HCl on ATP Synthesis
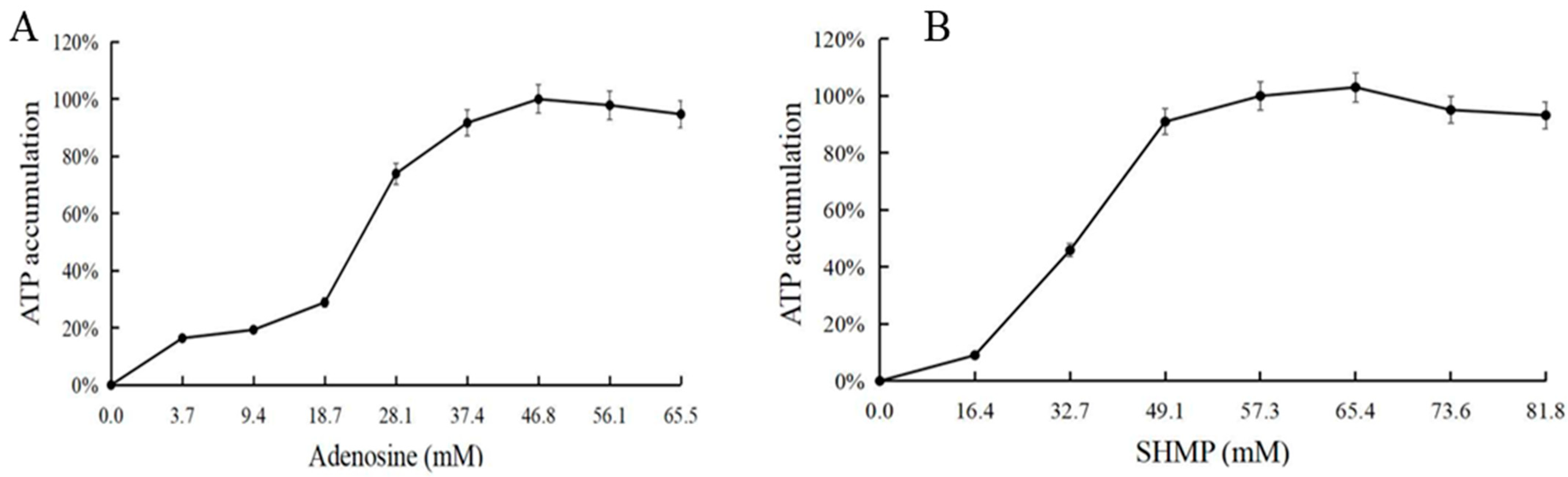
3.5. Effect of Adenosine and Sodium Hexametaphosphate on ATP Synthesis
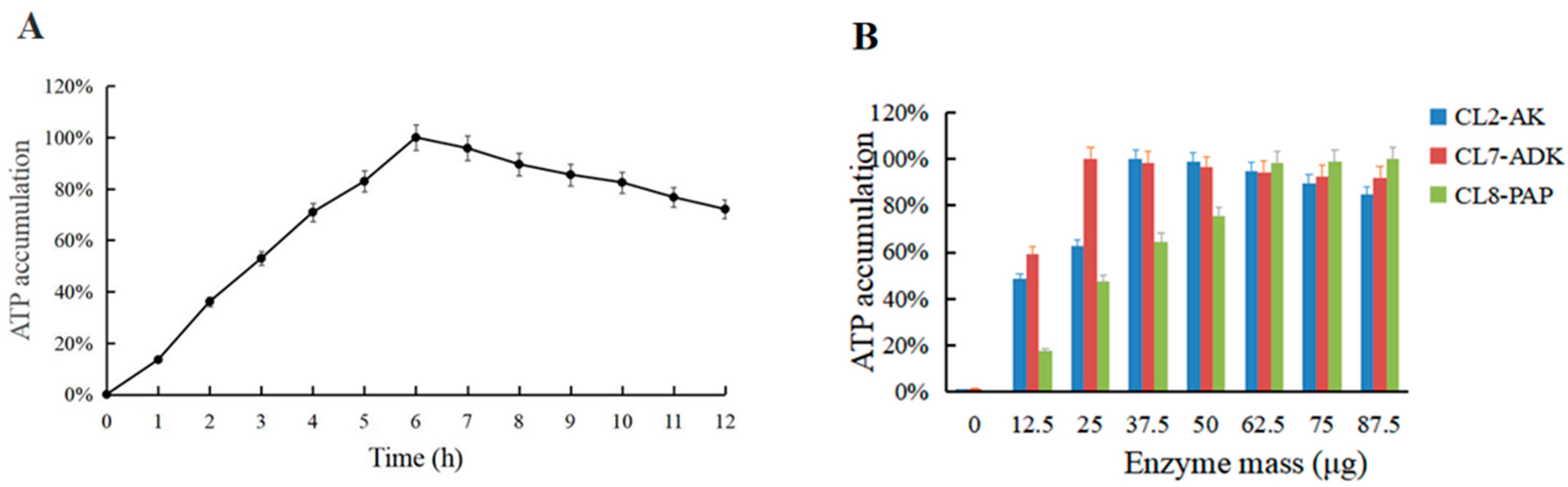
3.6. Effect of Reaction Time and Dosage Ratio of Multi-Enzyme Components on ATP Synthesis
3.7. Optimum Reaction Temperature and pH for MAT4 and MAT5
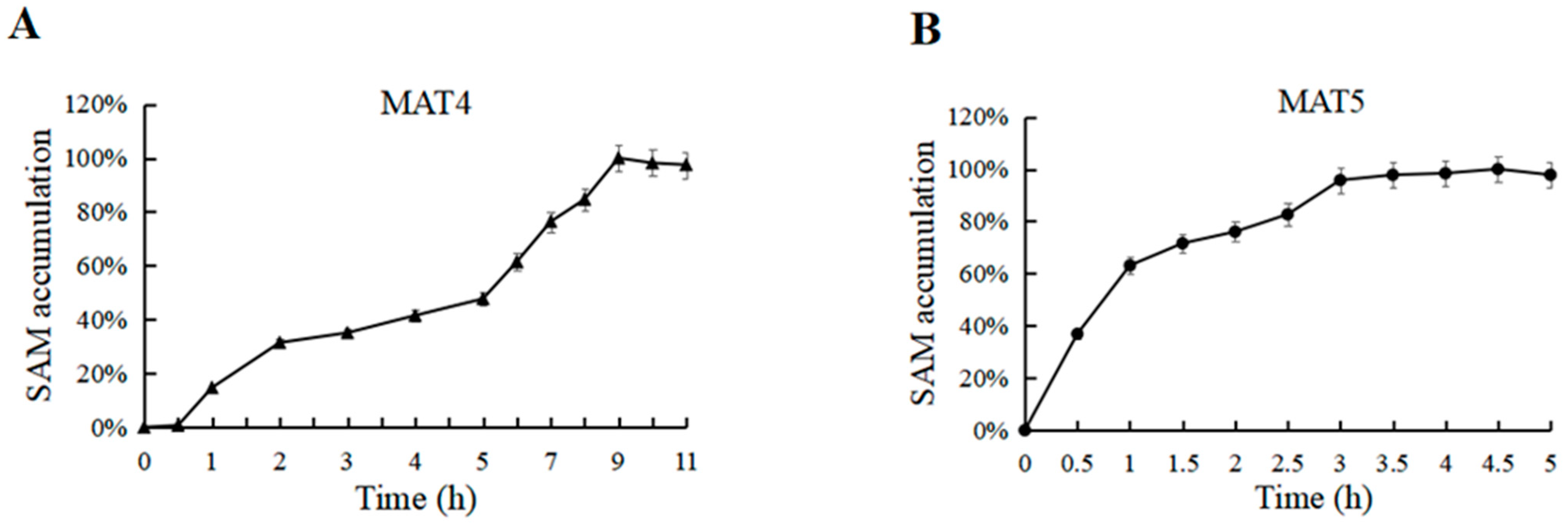
3.8. Effect of Synchronous and Asynchronous Reactions on SAM Synthesis
3.9. Effect of Catalytic Reaction Time on SAM Synthesis
3.10. Effect of l-Met and MAT on SAM Synthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, S.; Kalra, H.; Akundi, R.S.J. Extracellular ATP Mediates Cancer Cell Migration and Invasion Through Increased Expression of Cyclooxygenase 2. Front. Pharmacol. 2021, 11, 617211. [Google Scholar] [CrossRef] [PubMed]
- Jrgensen, P.E.; Eriksen, T.; Jensen, B.K. Estimation of viable biomass in wastewater and activated sludge by determination of ATP, oxygen utilization rate and FDA hydrolysis. Water. Res. 1992, 26, 1495–1501. [Google Scholar] [CrossRef]
- Fujio, T.; Furuya, A. Production of ATP from adenine by Brevibacterium ammoniagenes. J. Ferment. Technol. 1983, 61, 261–267. [Google Scholar]
- Kim, M.J.; Whitesides, G.M. Enzyme-catalyzed synthesis of nucleoside triphosphates from nucleoside monophosphates. Appl. Biochem. Biotechnol. 1987, 16, 95–108. [Google Scholar] [CrossRef]
- Akihiko, M.; Tatsuro, F. ATP Production from Adenine by a Self-coupling Enzymatic Process: High-level Accumulation under Ammonium-limited Conditions. Biosci. Biotechnol. Biochem. 2001, 65, 644–650. [Google Scholar]
- Chen, H.; Wang, Z.; Cai, H.; Zhou, C. Progress in the microbial production of S-adenosyl-L-methionine. World. J. Microb. Biot. 2016, 32, 153. [Google Scholar] [CrossRef]
- Chu, J.; Qian, J.; Zhuang, Y.; Zhang, S.; Li, Y. Progress in the research of S-adenosyl-L-methionine production. Appl. Microbiol. Biot. 2013, 97, 41–49. [Google Scholar] [CrossRef]
- Lu, S.C. S-Adenosylmethionine. Int. J. Biochem. Cell. Biol. 2000, 32, 391–395. [Google Scholar] [CrossRef]
- Galizia, I.; Oldani, L.; Macritchie, K.; Amari, E.; Young, A.H. S-adenosyl methionine (SAMe) for depression in adults. Cochrane Database Syst. Rev. 2016, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, A.L.; Girard, C.; Jui, D.; Sabina, A.; Katz, D.L. S-adenosylmethionine (SAMe) as treatment for depression: A systematic review. Clin. Investig. Med. 2005, 28, 132–139. [Google Scholar]
- Guruswamy, S.; Swamy, M.V.; Choi, C.I.; Steele, V.E.; Rao, C.V. S-adenosyl L-methionine inhibits azoxymethane-induced colonic aberrant crypt foci in F344 rats and suppresses human colon cancer Caco-2 cell growth in 3D culture. Int. J. Cancer. 2010, 122, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Barve, S.; Chen, T.; Nelson, W.; Uriarte, S.; Hill, D.; Mcclain, C. S-adenosylmethionine (AdoMet) modulates endotoxin stimulated interleukin-10 production in monocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 284, G949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, C.S. S-adenosyl-L-methionine: Its role in the treatment of liver disorders. Am. J. Clin. Nutr. 2002, 5, 1183S–1187S. [Google Scholar]
- Rountree, C.B.; Senadheera, S.; Mato, J.M.; Crooks, G.M.; Lu, S.C. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology 2010, 47, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Gl, A.; Hui, L.A.; Yt, A.; Ning, H.A.; Xy, B.; Kc, A.; Po, A. Improved S-adenosyl-l-methionine production in Saccharomyces cerevisiae using tofu yellow serofluid. J. Biotechnol. 2020, 309, 100–106. [Google Scholar]
- Gross, A.; Geresh, S.; Whitesides, G.M. Enzymatic synthesis of S-adenosyl-L-methionine from L-methionine and ATP. Appl. Biochem. Biotechnol. 1983, 8, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Markham, G.D.; Hafner, E.W.; Tabor, C.W.; Tabor, H. S-Adenosylmethionine synthetase from Escherichia coli. J. Biol. Inorg. Chem. 1980, 255, 9082–9092. [Google Scholar] [CrossRef]
- Yu, P.; Zhu, P. Improving the production of S-adenosyl-L-methionine in Escherichia coli by overexpressing metk. Prep. Biochem. Biotechnol. 2017, 47, 1350976. [Google Scholar] [CrossRef] [PubMed]
- Hui, H.; Qian, J.C.; Chu, J.; Wang, Y.H.; Zhuang, Y.P.; Zhang, S.L. Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Appl. Microbiol. Biotechnol. 2009, 83, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Fehlau, M.; Kaspar, F.; Hellendahl, K.F.; Schollmeyer, J.; Neubauer, P.; Wagner, A. Modular Enzymatic Cascade Synthesis of Nucleotides Using a (d)ATP Regeneration System. Front. Bioeng. Biotechnol. 2020, 8, 854. [Google Scholar] [CrossRef]
- Frisch, J.; Maršić, T.; Loderer, C. A Novel One-Pot Enzyme Cascade for the Biosynthesis of Cladribine Triphosphate. Biomolecules 2021, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lucas, J. Multienzymatic synthesis of nucleic acid derivatives: A general perspective. Appl. Biochem. Biotechnol. 2015, 99, 4615–4627. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, L.; Ottmann, C.; Brunsveld, L. Conjugated protein domains as engineered scaffold proteins. Bioconjug. Chem. 2020, 31, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qiao, J.; Lin, S.; Liu, Y.; Ma, L. A Highly Efficient Indirect P. pastoris Surface Display Method Based on the CL7/Im7 Ultra-High-Affinity System. Molecules 2019, 24, 1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Li, Y.; He, H.; Liu, L.; Wang, Y. A Novel Adenosine Kinase from Bombyx mori: Enzymatic Activity, Structure, and Biological Function. Int. J. Mol. Sci. 2019, 20, 3732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Tsai, S.L.; Chen, W. Artificial scaffolds for enhanced biocatalysis. Method Enzymol. 2019, 617, 363–383. [Google Scholar]
- Beales, P.A.; Ciani, B.; Cleasby, A.J. Nature’s lessons in design: Nanomachines to scaffold, remodel and shape membrane compartments. Phys. Chem. Chem. Phys. 2015, 17, 15489–15507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damien, G.; Anthony, B. The surprising dynamics of scaffolding proteins. Mol. Biol. Cell. 2014, 25, 2315–2319. [Google Scholar]
- Rgensen, K.; Rasmussen, A.V.; Morant, M.; Nielsen, A.H.; Bjarnholt, N.; Zagrobelny, M.; Bak, S.; Møller, B.L. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant. Biol. 2005, 8, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Vassylyeva, M.N.; Klyuyev, S.; Vassylyev, A.D.; Wesson, H.; Zhang, Z.; Renfrow, M.B.; Wang, H.B.; Higgins, N.P.; Chow, L.T.; Vassylyev, D.G. Efficient, ultra-high-affinity chromatography in a one-step purification of complex proteins. Proc. Natl. Acad. Sci. USA 2017, 114, E5138–E5147. [Google Scholar] [CrossRef] [Green Version]
- Mordhorst, S.; Siegrist, J.; Müller, M.; Richter, M.; Andexer, J.N. Catalytic Alkylation Using a Cyclic S -Adenosylmethionine Regeneration System. Angew. Chem. Int. Ed. 2017, 56, 4037–4041. [Google Scholar] [CrossRef] [PubMed]
- Popadić, D.; Mhaindarkar, D.; Dang Thai, M.H.N.; Hailes, H.C.; Mordhorst, S.; Andexer, J.N. A bicyclic S-adenosylmethionine regeneration system applicable with different nucleosides or nucleotides as cofactor building blocks. RSC Chem. Biol. 2021, 2, 883–891. [Google Scholar] [CrossRef]
- Becker, M.; Nikel, P.; Andexer, J.N.; Lütz, S.; Rosenthal, K. A Multi-Enzyme Cascade Reaction for the Production of 2′3′-cGAMP. Biomolecules 2021, 11, 590. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, G.; Li, X.; Yang, J.; Li, Z.; Hou, J.; Rao, B.; Hu, Y.; Ma, L.; Wang, Y. Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins. Biomolecules 2021, 11, 1706. https://doi.org/10.3390/biom11111706
Yan G, Li X, Yang J, Li Z, Hou J, Rao B, Hu Y, Ma L, Wang Y. Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins. Biomolecules. 2021; 11(11):1706. https://doi.org/10.3390/biom11111706
Chicago/Turabian StyleYan, Guangbo, Xia Li, Jun Yang, Zhongchen Li, Jia Hou, Ben Rao, Yong Hu, Lixin Ma, and Yaping Wang. 2021. "Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins" Biomolecules 11, no. 11: 1706. https://doi.org/10.3390/biom11111706
APA StyleYan, G., Li, X., Yang, J., Li, Z., Hou, J., Rao, B., Hu, Y., Ma, L., & Wang, Y. (2021). Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins. Biomolecules, 11(11), 1706. https://doi.org/10.3390/biom11111706




