Cyclic Tetra-Adenylate (cA4) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus
Abstract
1. Introduction
1.1. CARF Domains
1.2. Cyclic Oligo-Adenylate Second Messengers
1.3. Transcriptional Regulation by Csa3
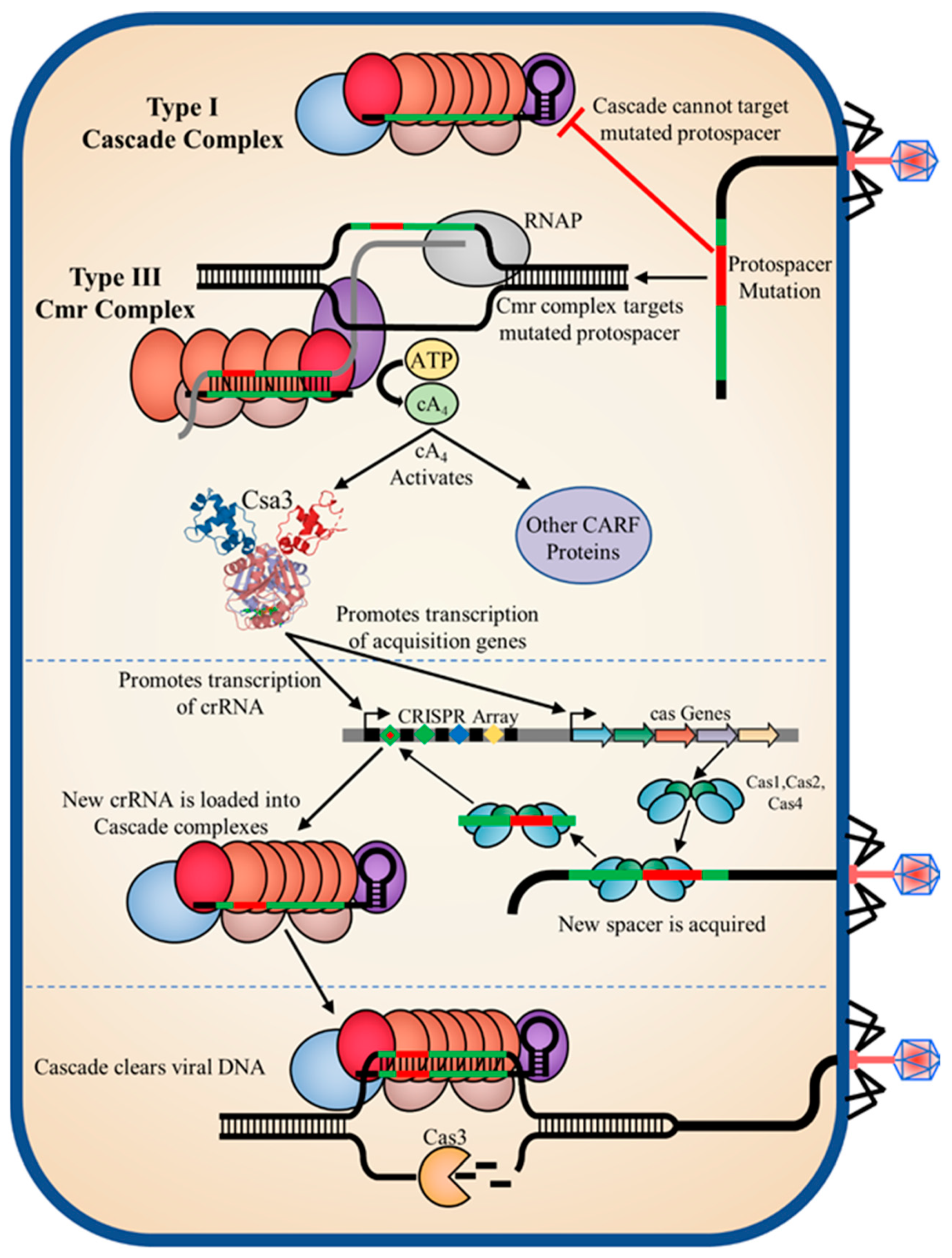
1.4. Integration of Class 1 Systems
2. Materials and Methods
2.1. Bioinformatics
2.2. Csa3a Expression and Purification
2.3. Isothermal Titration Calorimetry
2.4. Cyclic Oligonucleotide Phosphodiesterase (Ring Nuclease) Assays
2.5. Crystallization and Data Collection
2.6. Structure Determination and Refinement
2.7. Electromobility Shift Assays (EMSAs)
2.8. Fluorescence Polarization (FP) Assays
2.9. Sulfolobus Strains, Growth Conditions
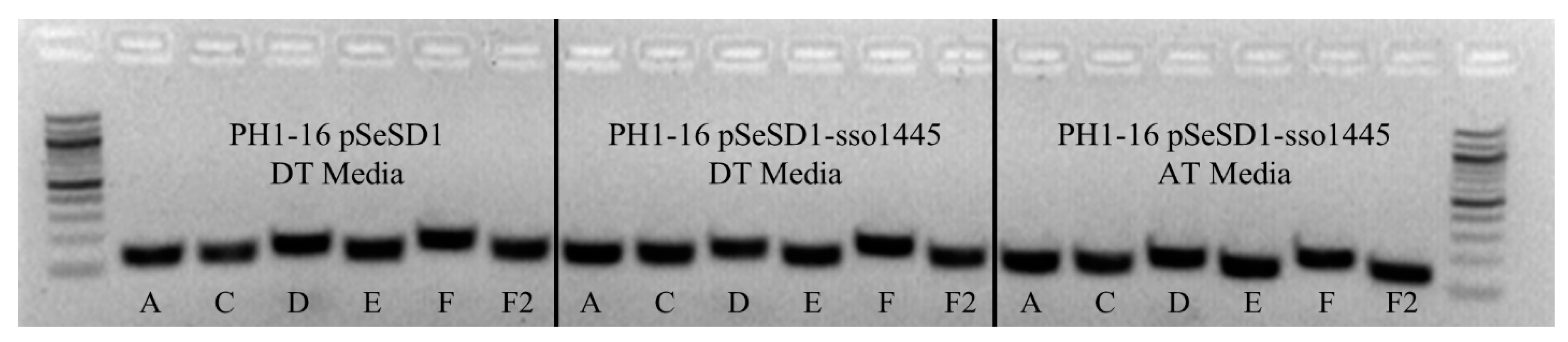
2.10. Acquisition Assay
3. Results
3.1. CRISPR/Cas Systems in S. solfataricus
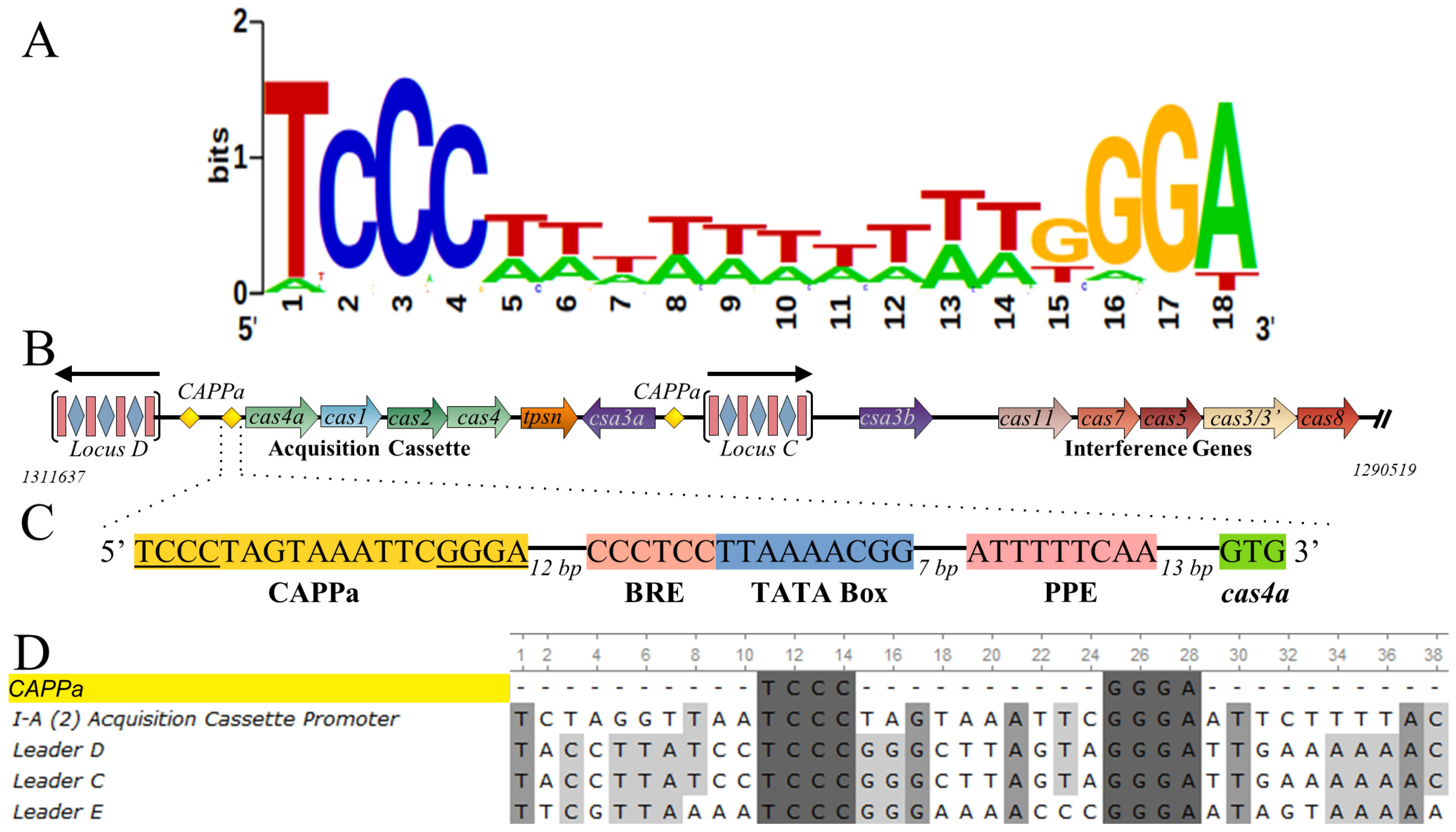
3.2. CRISPR-Associated Palindromes
3.3. Csa3a Binds cA4
3.4. Csa3a Lacks Ring Nuclease Activity
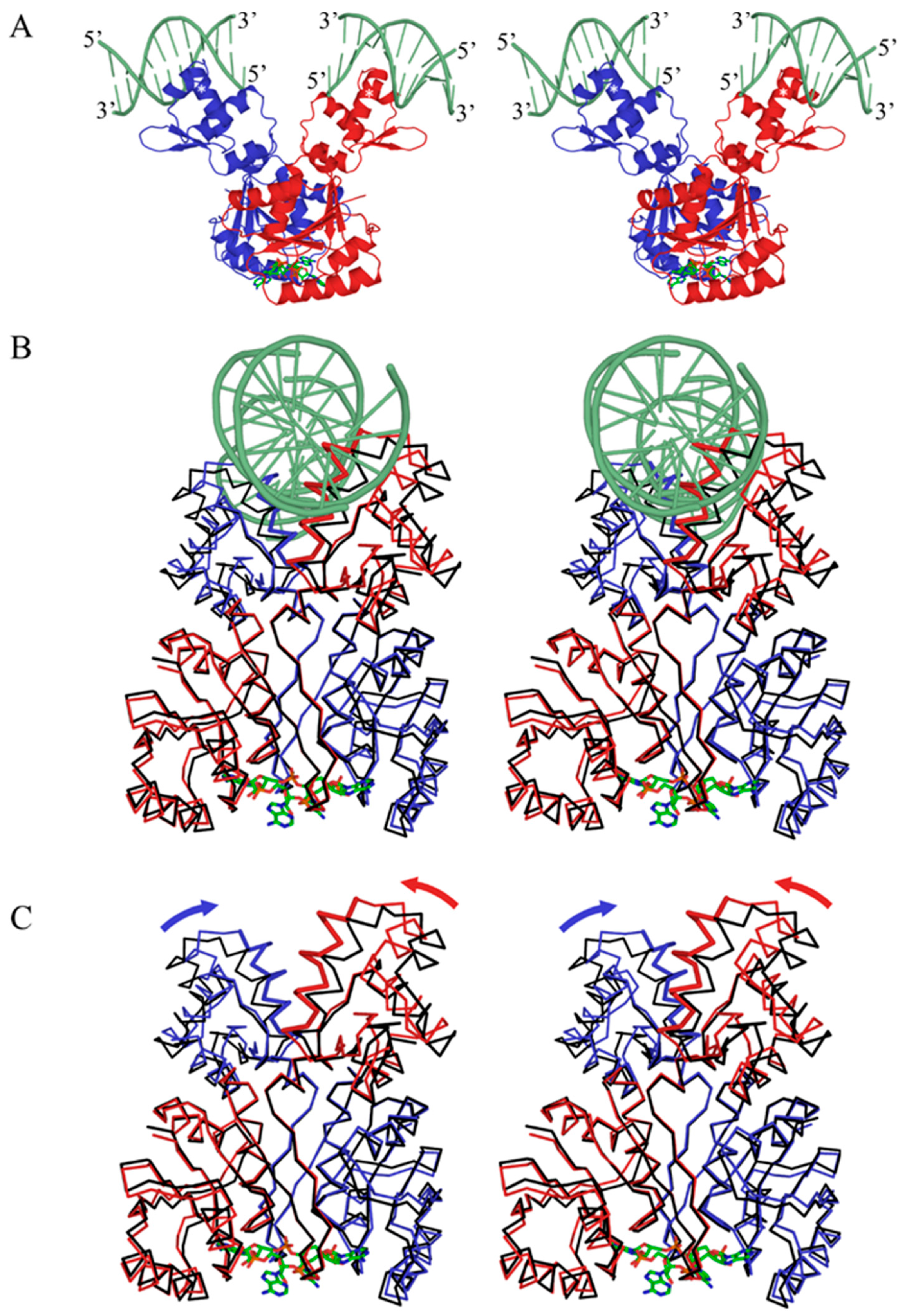
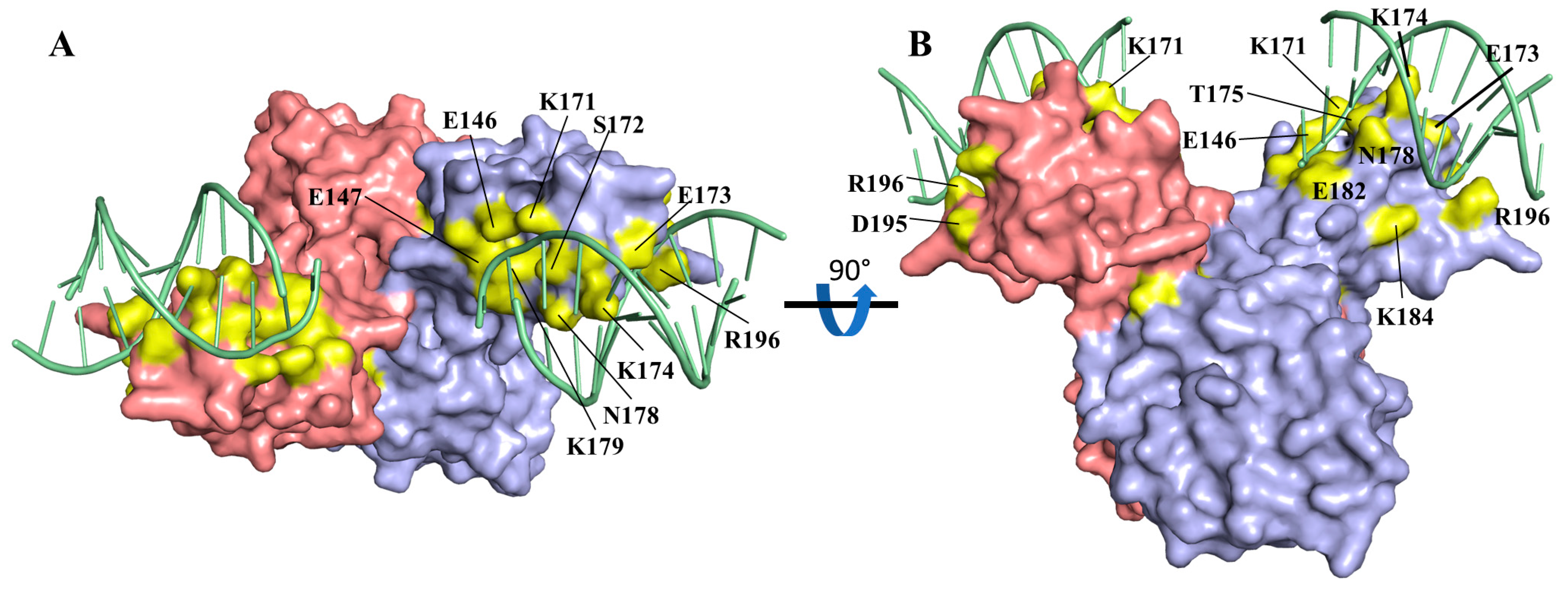
3.5. Csa3a Binds cA4 in the CARF-Binding Pocket
3.6. cA4-Induced Conformational Changes
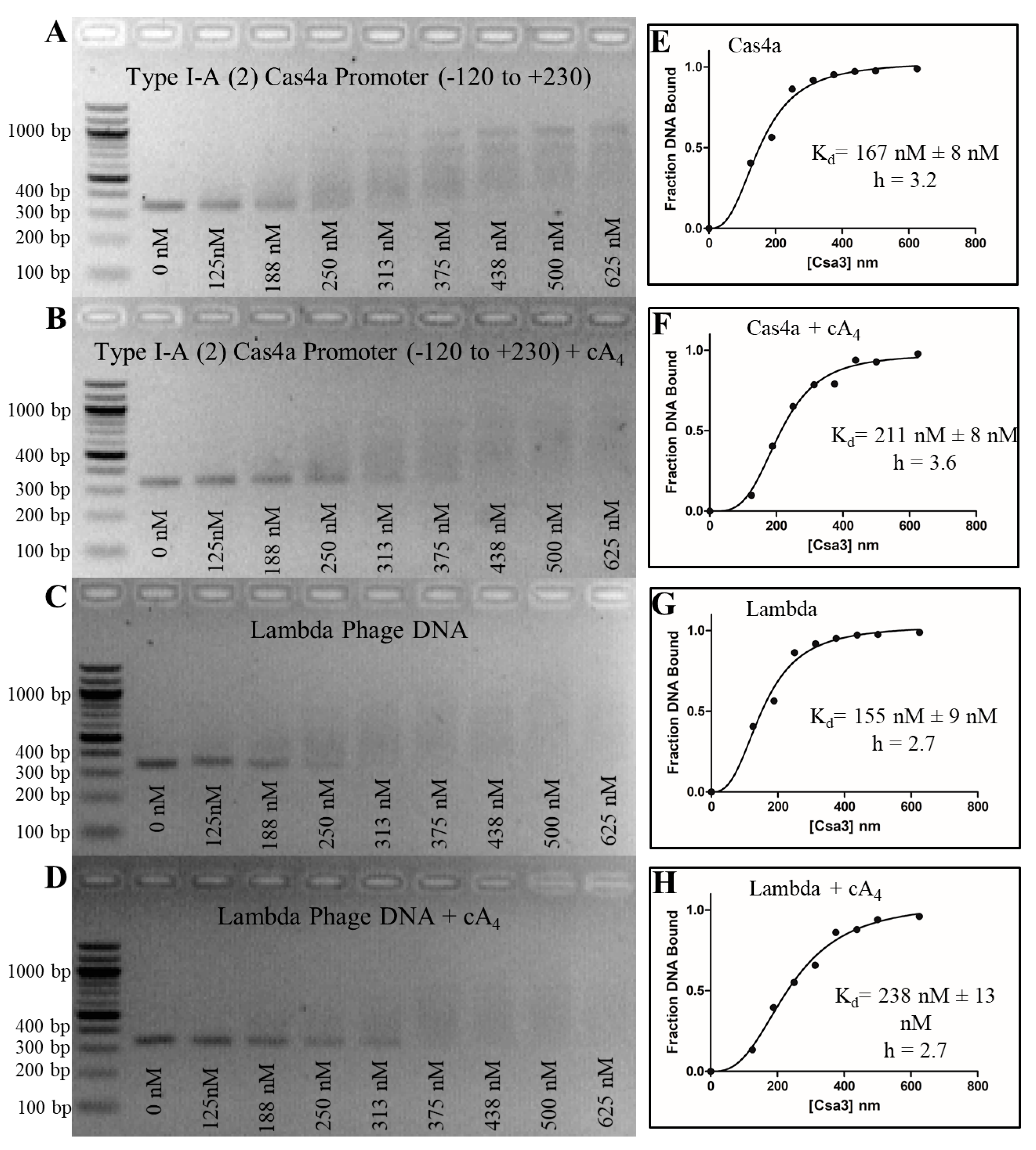
3.7. Csa3a Binds CAPPa Nonspecifically, in a Cooperative and cA4-Independent Manner
3.8. Csa3a Overexpresssion Does Not Stimulate Spacer Acquisition in S. solfataricus
4. Discussion
4.1. Physiological Implications of Low-Micromolar cA4 Affinity
4.2. cA4-Induced Conformational Changes
4.3. cA4 Recognition Is Entropy-Driven
4.4. Transcriptional Regulation of Spacer Acquisition and CRISPR Loci by Csa3
4.5. An Integrated, Synergistic Class 1 CRISPR-Cas Immune Response
4.6. Caveats and Additional Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, A.A.; Gauvin, C.C.; Lawrence, C.M. Encyclopedia of Biological, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 3, pp. 717–741. [Google Scholar]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Kurosawa, N. Saccharolobus caldissimus gen. nov., sp. nov., a facultatively anaerobic iron-reducing hyperthermophilic archaeon isolated from an acidic terrestrial hot spring, and reclassification of Sulfolobus solfataricus as Saccharolobus solfataricus comb. nov. and Sulfolobus shibatae as Saccharolobus shibatae comb. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; Frankel, K.A.; Tsutakawa, S.E.; Alsbury, D.L.; Copié, V.; Young, M.J.; Tainer, J.A.; Lawrence, C.M. The Structure of the CRISPR-Associated Protein Csa3 Provides Insight into the Regulation of the CRISPR/Cas System. J. Mol. Biol. 2011, 405, 939–955. [Google Scholar] [CrossRef] [PubMed]
- Lillestøl, R.K.; Shah, S.; Brügger, K.; Redder, P.; Phan, H.; Christiansen, J.; Garrett, R.A. CRISPR families of the crenarchaeal genus Sulfolobus: Bidirectional transcription and dynamic properties. Mol. Microbiol. 2009, 72, 259–272. [Google Scholar] [CrossRef]
- Zink, I.A.; Wimmer, E.; Schleper, C. Heavily Armed Ancestors: CRISPR Immunity and Applications in Archaea with a Comparative Analysis of CRISPR Types in Sulfolobales. Biomolecules 2020, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; Kerou, M.; Brumfield, S.K.; Graham, S.; Liu, H.; Naismith, J.; Sdano, M.; Peng, N.; She, Q.; Copié, V.; et al. Structural and Functional Characterization of an Archaeal Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Complex for Antiviral Defense (CASCADE). J. Biol. Chem. 2011, 286, 21643–21656. [Google Scholar] [CrossRef]
- Haft, D.; Selengut, J.; Mongodin, E.; Nelson, K. A Guild of 45 CRISPR-Associated (Cas) Protien Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef]
- Athukoralage, J.S.; McQuarrie, S.; Gruschow, S.; Graham, S.; Gloster, T.M.; White, M.F. Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage. Elife 2020, 9, e57627. [Google Scholar] [CrossRef]
- Makarova, K.S.; Anantharaman, V.; Grishin, N.V.; Koonin, E.V.; Aravind, L. CARF and WYL domains: Ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014, 5, 102. [Google Scholar] [CrossRef]
- Makarova, K.S.; Timinskas, A.; Wolf, Y.I.; Gussow, A.B.; Siksnys, V.; Venclovas, Č.; Koonin, E.V. Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res. 2020, 48, 8828–8847. [Google Scholar] [CrossRef]
- Anantharaman, V.; Makarova, K.S.; Burroughs, A.M.; Koonin, E.V.; Aravind, L. Comprehensive analysis of the HEPN superfamily: Identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 2013, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, Y.G.; Oh, B.H. Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus. Proteins 2013, 81, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, N.F.; Glover, C.V., 3rd; Terns, R.M.; Terns, M.P. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 2016, 22, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Guo, W.; Yuan, Y.A. Crystal Structures of CRISPR-Associated Csx3 Reveal a Manganese-Dependent Deadenylation Exoribonuclease. RNA Biol. 2015, 12, 749–760. [Google Scholar] [CrossRef][Green Version]
- Topuzlu, E.; Lawrence, C.M. Recognition of a pseudo-symmetric RNA tetranucleotide by Csx3, a new member of the CRISPR associated Rossmann fold superfamily. RNA Biol. 2016, 13, 254–257. [Google Scholar] [CrossRef]
- Zhu, X.; Ye, K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012, 586, 939–945. [Google Scholar] [CrossRef]
- Kazlauskiene, M.; Kostiuk, G.; Venclovas, Č.; Tamulaitis, G.; Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 2017, 357, 605–609. [Google Scholar] [CrossRef]
- Niewoehner, O.; Doval, C.G.; Rostøl, J.T.; Berk, C.; Schwede, F.; Bigler, L.; Hall, J.; Marraffini, L.A.; Jinek, M. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 2017, 548, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Wang, X.; Ye, Q.; Li, H.; Liang, Y.; She, Q.; Peng, N. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015, 43, 1044–1055. [Google Scholar] [CrossRef]
- He, F.; Vestergaard, G.; Peng, W.; She, Q.; Peng, X. CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 2016, 45, 1902–1913. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Ye, Q.; Pan, S.; Wang, X.; Li, Y.; Peng, W.; Liang, Y.; She, Q.; Peng, N. Coupling transcriptional activation of CRISPR–Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 2017, 45, 8978–8992. [Google Scholar] [CrossRef]
- Dlakić, M. HHsvm: Fast and accurate classification of profile–profile matches identified by HHsearch. Bioinformatics 2009, 25, 3071–3076. [Google Scholar] [CrossRef]
- Menon, S.K.; Eilers, B.J.; Young, M.J.; Lawrence, C.M. The Crystal Structure of D212 from Sulfolobus Spindle-Shaped Virus Ragged Hills Reveals a New Member of the PD-(D/E)XK Nuclease Superfamily. J. Virol. 2010, 84, 5890–5897. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brown, S.; Gauvin, C.C.; Charbonneau, A.A.; Burman, N.; Lawrence, C.M. Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR–Cas that degrades the second messenger cA4. J. Biol. Chem. 2020, 295, 14963–14972. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Haurwitz, R.E.; Doudna, J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 2012, 18, 661–672. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Mccoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.; Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.I.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The Pymol Molecular Graphics System; Version 1.7.4; Schrödinger, LLC.: New York, NY, USA, 2015; Available online: http://www.pymol.org (accessed on 30 November 2021).
- Peng, N.; Deng, L.; Mei, Y.; Jiang, D.; Hu, Y.; Awayez, M.; Liang, Y.; She, Q. A Synthetic Arabinose-Inducible Promoter Confers High Levels of Recombinant Protein Expression in Hyperthermophilic Archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012, 78, 5630–5637. [Google Scholar] [CrossRef] [PubMed]
- Athukoralage, J.; Rouillon, C.; Graham, S.; Grüschow, S.; White, M.F. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 2018, 562, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Athukoralage, J.S.; McMahon, S.A.; Zhang, C.; Grüschow, S.; Graham, S.; Krupovic, M.; Whitaker, R.J.; Gloster, T.M.; White, M.F. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nat. Cell Biol. 2020, 577, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Athukoralage, J.S.; Graham, S.; Rouillon, C.; Grüschow, S.; Czekster, C.M.; White, M.F. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. eLife 2020, 9, 15. [Google Scholar] [CrossRef]
- Athukoralage, J.S.; Graham, S.; Grüschow, S.; Rouillon, C.; White, M.F. A Type III CRISPR Ancillary Ribonuclease Degrades Its Cyclic Oligoadenylate Activator. J. Mol. Biol. 2019, 431, 2894–2899. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Doval, C.; Schwede, F.; Berk, C.; Rostøl, J.T.; Niewoehner, O.; Tejero, O.; Hall, J.; Marraffini, L.A.; Jinek, M. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 2020, 11, 1596. [Google Scholar] [CrossRef]
- Jia, N.; Jones, R.; Yang, G.; Ouerfelli, O.; Patel, D.J. CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4 Cleavage with ApA>p Formation Terminating RNase Activity. Mol. Cell 2019, 75, 944–956. [Google Scholar] [CrossRef]
- Richardson, J.S.; Schneider, B.; Murray, L.W.; Kapral, G.J.; Immormino, R.M.; Headd, J.J.; Richardson, D.C.; Ham, D.; Hershkovits, E.; Williams, L.D.; et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA 2008, 14, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Fuangthong, M.; Helmann, J.D.; Brennan, R.G. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 2005, 20, 131–141. [Google Scholar] [CrossRef]
- Peng, W.; Feng, M.; Feng, X.; Liang, Y.X.; She, Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2014, 43, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.; Schäfer, G. Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1988, 232, 359–363. [Google Scholar] [CrossRef]
- Rostøl, J.T.; Xie, W.; Kuryavyi, V.; Maguin, P.; Kao, K.; Froom, R.; Patel, D.J.; Marraffini, L.A. The Card1 nuclease provides defence during type III CRISPR immunity. Nature 2021, 590, 624–629. [Google Scholar] [CrossRef]
- Ye, Q.; Zhao, X.; Liu, J.; Zeng, Z.; Zhang, Z.; Liu, T.; Li, Y.; Han, W.; Peng, N. CRISPR-Associated Factor Csa3b Regulates CRISPR Adaptation and Cmr-Mediated RNA Interference in Sulfolobus islandicus. Front. Microbiol. 2020, 11, 2038. [Google Scholar] [CrossRef]
- Rouillon, C.; Athukoralage, J.S.; Graham, S.; Grüschow, S.; White, M.F. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 2018, 7, e36734. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, M.; Liu, J.; Liu, T.; Ye, Q.; Li, Y.; Peng, N. A CRISPR-associated factor Csa3a regulates DNA damage repair in Crenarchaeon Sulfolobus islandicus. Nucleic Acids Res. 2020, 48, 9681–9693. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Lowey, B.; Whiteley, A.T.; Keszei, A.F.A.; Morehouse, B.R.; Mathews, I.T.; Antine, S.P.; Cabrera, V.J.; Kashin, D.; Niemann, P.; Jain, M.; et al. CBASS Immunity Uses CARF-Related Effectors to Sense 3′-5′- and 2′-5′-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 2020, 182, 38–49. [Google Scholar] [CrossRef]
- Lau, R.K.; Ye, Q.; Birkholz, E.A.; Berg, K.R.; Patel, L.; Mathews, I.T.; Watrous, J.D.; Ego, K.; Whiteley, A.T.; Lowey, B.; et al. Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity. Mol. Cell 2020, 77, 723–733.e6. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Zhang, D.; Schäffer, D.; Iyer, L.M.; Aravind, L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015, 43, 10633–10654. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, A.T.; Eaglesham, J.B.; de Oliveira Mann, C.C.; Morehouse, B.R.; Lowey, B.; Nieminen, E.A.; Danilchanka, O.; King, D.S.; Lee, A.S.Y.; Mekalanos, J.J.; et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 2019, 567, 194–199. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Makarova, K.S.; Wolf, Y.I.; Severinov, K.V.; Koonin, E.V. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. USA 2018, 115, E5307–E5316. [Google Scholar] [CrossRef] [PubMed]
- White, M.F. Bacteria SAVED from Viruses. Cell 2020, 182, 5–6. [Google Scholar] [CrossRef] [PubMed]




| Dataset | SsCsa3-cA4 |
|---|---|
| Wavelength (Å) | 1.54 |
| Space Group | P212121 |
| Cell Constants (a,b,c; Å) (α, β, γ; o) | 46.1, 93.4, 105.2 90, 90, 90 |
| Resolution Range a (Å) | 27.91–2.45 (2.49–2.45) |
| Unique Reflections a | 16,551 |
| Average Redundancy a | 10.4 (10.4) |
| I/σ a | 36.4 (2.5) |
| Completeness (%) | 96.8(100) |
| Rsym a,b (%) | 7.9 (82.0) |
| Rpim a (%) | 2.6 (26.5) |
| CC1/2 a | 0.99 (0.737) |
| Model Refinement | |
|---|---|
| Rwork c (%) | 20.3 (28.8) |
| Rfree c (%) | 24.3 (34.6) |
| Real Space CC d (%) | 84.8 |
| Mean B Value (overall; Å2) | 85.0 |
| Coordinate Error (based on maximum likelihood, Å) | 0.37 |
| RMSD from Ideality: | |
| Bonds (Å) | 0.005 |
| Angles (°) | 0.687 |
| Ramachandran Plot e | |
| Most Favored (%) | 96.0 |
| Additional Allowed (%) | 4.0 |
| PDB Accession Code | 6W11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charbonneau, A.A.; Eckert, D.M.; Gauvin, C.C.; Lintner, N.G.; Lawrence, C.M. Cyclic Tetra-Adenylate (cA4) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus. Biomolecules 2021, 11, 1852. https://doi.org/10.3390/biom11121852
Charbonneau AA, Eckert DM, Gauvin CC, Lintner NG, Lawrence CM. Cyclic Tetra-Adenylate (cA4) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus. Biomolecules. 2021; 11(12):1852. https://doi.org/10.3390/biom11121852
Chicago/Turabian StyleCharbonneau, Alexander A., Debra M. Eckert, Colin C. Gauvin, Nathanael G. Lintner, and C. Martin Lawrence. 2021. "Cyclic Tetra-Adenylate (cA4) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus" Biomolecules 11, no. 12: 1852. https://doi.org/10.3390/biom11121852
APA StyleCharbonneau, A. A., Eckert, D. M., Gauvin, C. C., Lintner, N. G., & Lawrence, C. M. (2021). Cyclic Tetra-Adenylate (cA4) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus. Biomolecules, 11(12), 1852. https://doi.org/10.3390/biom11121852







