Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Primary Samples
2.2. Isolation, Cell Culture, and Characterization
2.3. Ribosome Profiling, Library Preparation, and Sequencing
2.4. Bioinformatics and Computational Analysis
2.5. Gene Ontology Analysis
2.6. RNA Minimum Free-Energy Calculation
2.7. Microprotein Features
3. Results
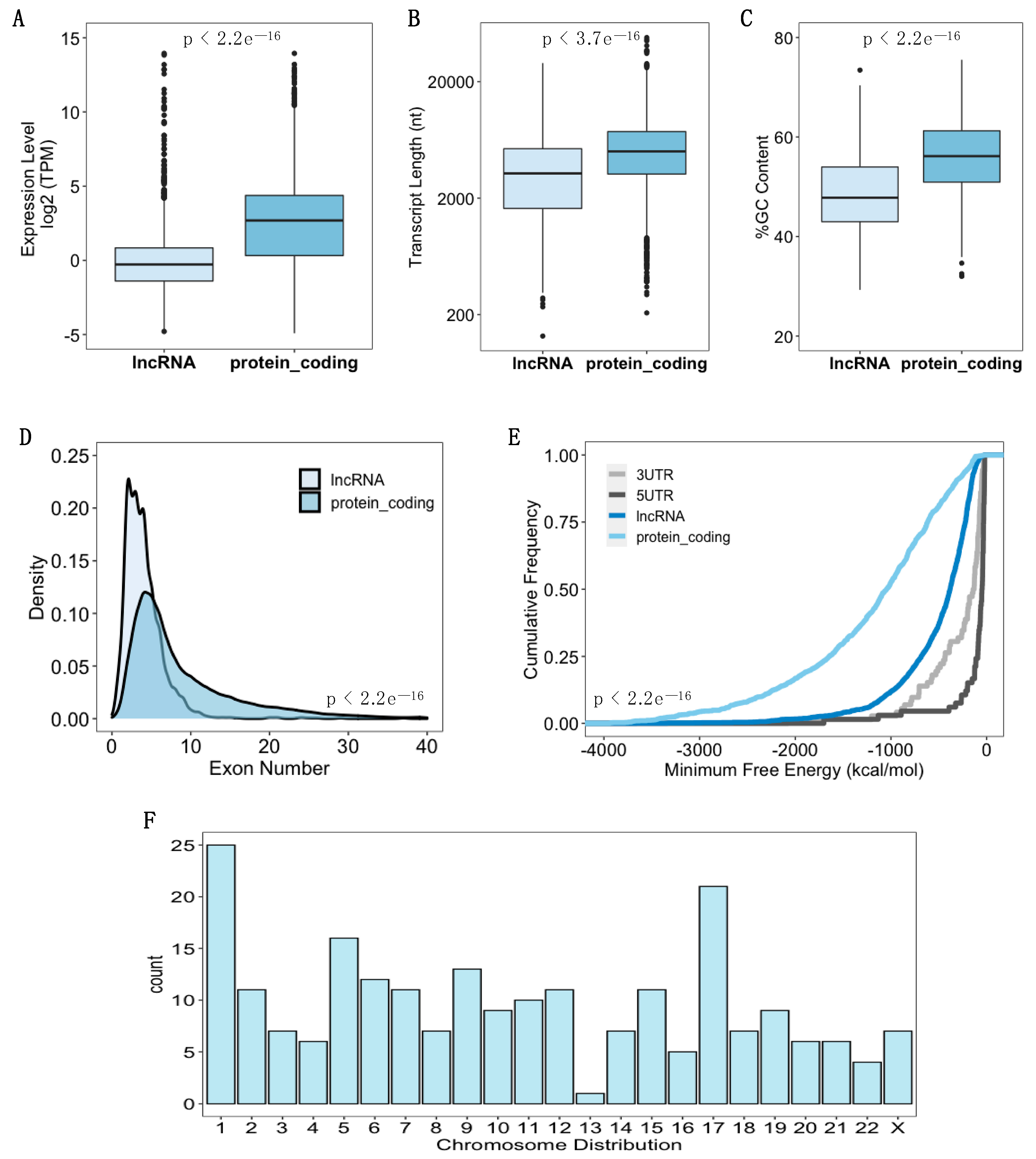
3.1. Study Overview: Searching for translated lncRNAs in hASC
3.2. Identification of Ribosome-Associated lncRNAs in hASC
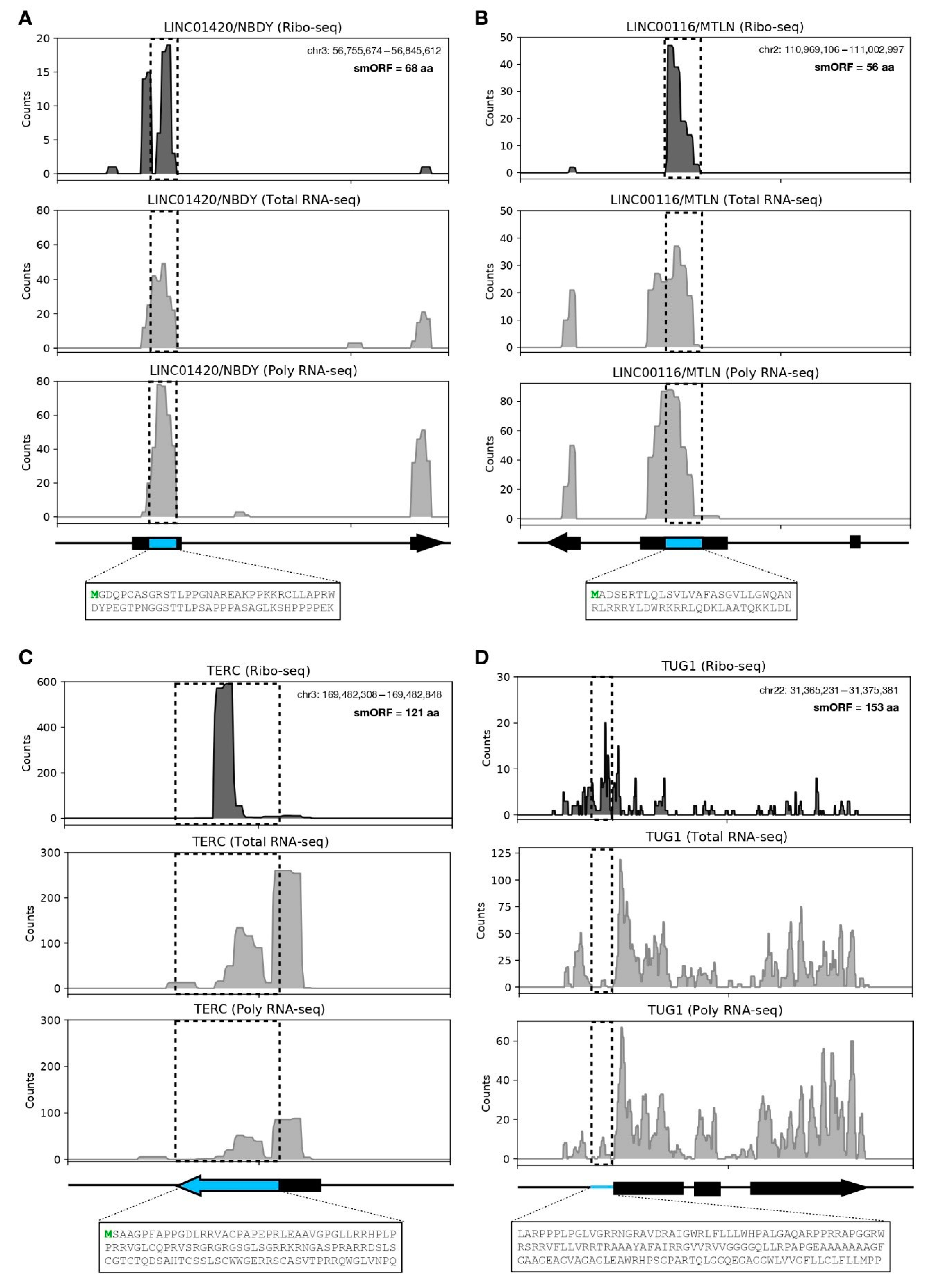
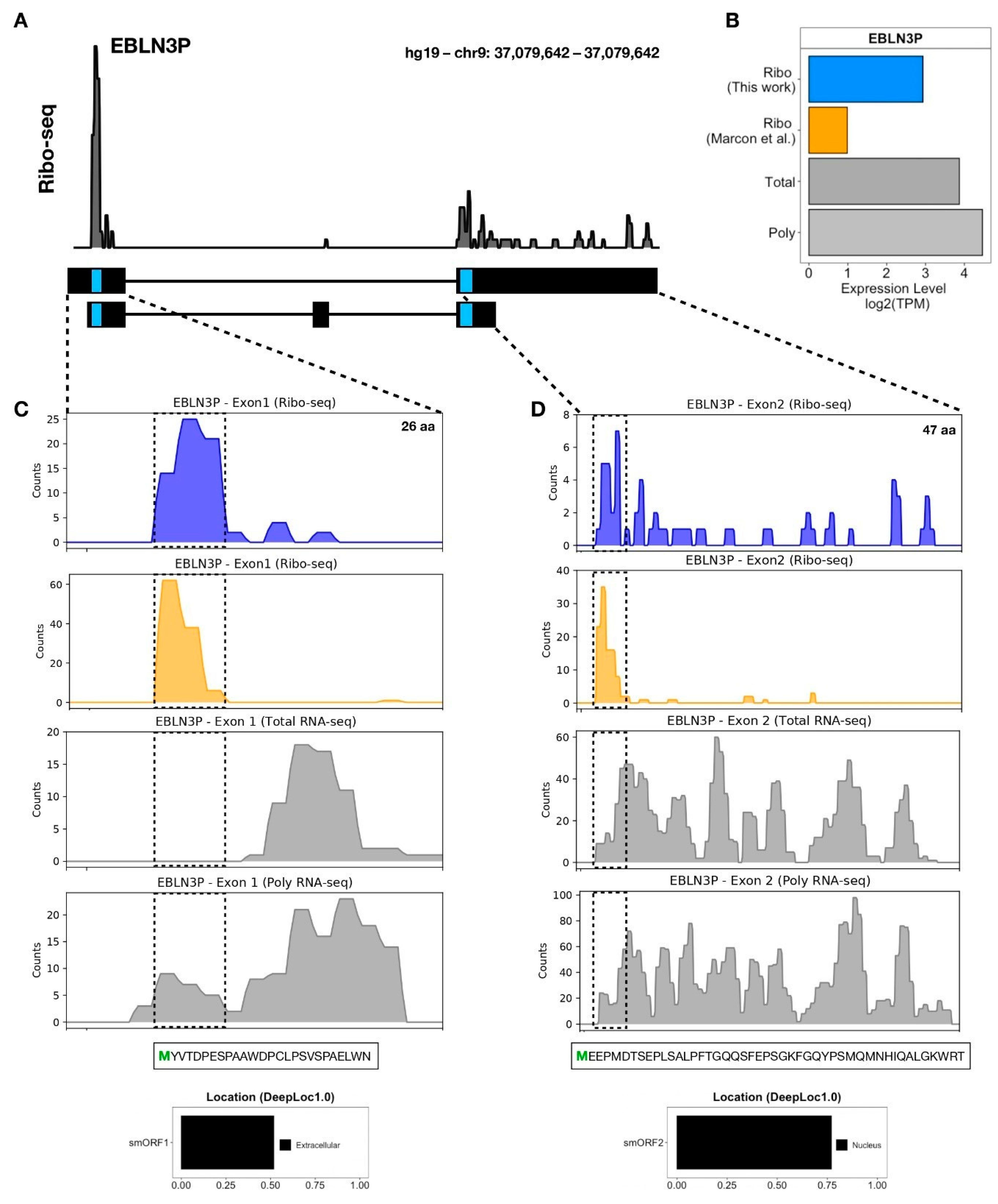
3.3. LncRNA-Encoded Microproteins in hASC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Merkel, A.; Gonzalez, D.; Lagarde, J.; et al. The GENCODE v7 Catalogue of Human Long Non-Coding RNAs: Analysis of Their Structure, Evolution and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Brown, C.; Hendrich, B.; Rupert, J.L.; Lafrenière, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nat. Cell Biol. 2011, 477, 295–300. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Khurana, S.; Kubben, N.; Abdelmohsen, K.; Oberdoerffer, P.; Gorospe, M.; Misteli, T. A BRCA 1-interacting lnc RNA regulates homologous recombination. EMBO Rep. 2015, 16, 1520–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 2017, 170, 72–85.e14. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Meng, H.; Bai, Y.; Wang, K. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2016, 23, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Bonilauri, B.; Dallagiovanna, B. Long Non-coding RNAs Are Differentially Expressed after Different Exercise Training Programs. Front. Physiol. 2020, 11, 567614. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef] [Green Version]
- Robert, A.; Angulski, A.B.B.; Spangenberg, L.; Shigunov, P.; Pereira, I.T.; Bettes, P.S.L.; Naya, H.; Correa, A.; Dallagiovanna, B.; Stimamiglio, M.A. Gene expression analysis of human adipose tissue-derived stem cells during the initial steps of in vitro osteogenesis. Sci. Rep. 2018, 8, 4739. [Google Scholar] [CrossRef]
- Robert, A.W.; Marcon, B.H.; Dallagiovanna, B.; Shigunov, P. Adipogenesis, Osteogenesis, and Chondrogenesis of Human Mesenchymal Stem/Stromal Cells: A Comparative Transcriptome Approach. Front. Cell Dev. Biol. 2020, 8, 561. [Google Scholar] [CrossRef]
- Si, Z.; Wang, X.; Sun, C.; Kang, Y.; Xu, J.; Wang, X.; Hui, Y. Adipose-derived stem cells: Sources, potency, and implications for regenerative therapies. Biomed. Pharmacother. 2019, 114, 108765. [Google Scholar] [CrossRef]
- Tye, C.; Gordon, J.A.; Martin-Buley, L.A.; Stein, J.L.; Lian, J.B.; Stein, G.S. Could lncRNAs be the Missing Links in Control of Mesenchymal Stem Cell Differentiation? J. Cell. Physiol. 2015, 230, 526–534. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.-Y.; Wang, P.; Wu, Y.-F.; Shen, H.-Y. Long non-coding RNA: The functional regulator of mesenchymal stem cells. World J. Stem Cells 2019, 11, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.; Weissman, J.S. Ribosome Profiling Reveals Pervasive Translation Outside of Annotated Protein-Coding Genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Orera, J.; Villanueva-Cañas, J.L.; Albà, M.M. Evolution of new proteins from translated sORFs in long non-coding RNAs. Exp. Cell Res. 2020, 391, 111940. [Google Scholar] [CrossRef]
- Van Heesch, S.; Van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; De Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef] [Green Version]
- Dallagiovanna, B.; Pereira, I.T.; Origa-Alves, A.C.; Shigunov, P.; Naya, H.; Spangenberg, L. lncRNAs are associated with polysomes during adipose-derived stem cell differentiation. Gene 2017, 610, 103–111. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Niazi, F.; Valadkhan, S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 2012, 18, 825–843. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Fukunaga, T.; Hamada, M. Identification and analysis of ribosome-associated lncRNAs using ribosome profiling data. BMC Genom. 2018, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Song, R.; Regev, A.; Struhl, K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 2015, 4, e08890. [Google Scholar] [CrossRef] [PubMed]
- Yeasmin, F.; Yada, T.; Akimitsu, N. Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Front. Genet. 2018, 9, 144. [Google Scholar] [CrossRef]
- Huang, J.; Chen, M.; Chen, D.; Gao, X.-C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.-R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Horinouchi, C.D.; Barisón, M.J.; Robert, A.W.; Kuligovski, C.; Aguiar, A.M.; Dallagiovanna, B. Influence of donor age on the differentiation and division capacity of human adipose-derived stem cells. World J. Stem Cells 2020, 12, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Marcon, B.H.; Spangenberg, L.; Bonilauri, B.; Robert, A.W.; Angulski, A.B.B.; Cabo, G.C.; Cofré, A.R.; Bettes, P.S.L.; Dallagiovanna, B.; Shigunov, P. Data describing the experimental design and quality control of RNA-Seq of human adipose-derived stem cells undergoing early adipogenesis and osteogenesis. Data Brief 2020, 28, 105053. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Marcon, B.H.; Holetz, F.B.; Eastman, G.; Origa-Alves, A.C.; Amorós, M.A.; de Aguiar, A.M.; Rebelatto, C.K.; Brofman, P.R.; Sotelo-Silveira, J.; Dallagiovanna, B. Downregulation of the protein synthesis machinery is a major regulatory event during early adipogenic differentiation of human adipose-derived stromal cells. Stem Cell Res. 2017, 25, 191–201. [Google Scholar] [CrossRef]
- Hsu, P.; Calviello, L.; Wu, H.-Y.L.; Li, F.-W.; Rothfels, C.J.; Ohler, U.; Benfey, P.N. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E7126–E7135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Clote, P.; Ferre’, F.; Kranakis, E.; Krizanc, D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA 2005, 11, 578–591. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Calviello, L.; Ohler, U. Beyond Read-Counts: Ribo-seq Data Analysis to Understand the Functions of the Transcriptome. Trends Genet. 2017, 33, 728–744. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Hussmann, J.; Weissman, J.S. Ribosome Profiling: Global Views of Translation. Cold Spring Harb. Perspect. Biol. 2019, 11, a032698. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Xie, S.; Liu, Y.; Xie, Z. Global and cell-type specific properties of lincRNAs with ribosome occupancy. Nucleic Acids Res. 2016, 45, 2786–2796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.-R.; Zhang, J. Human Long Noncoding RNAs Are Substantially Less Folded than Messenger RNAs. Mol. Biol. Evol. 2015, 32, 970–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Lima, N.G.; Ma, J.; Winkler, L.; Chu, Q.; Loh, K.H.; Corpuz, E.O.; Budnik, B.A.; Lykke-Andersen, J.; Saghatelian, J.M.Q.C.A.; A Slavoff, N.G.D.L.W.S. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017, 13, 174–180. [Google Scholar] [CrossRef]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.; Abouassaly, G.M.; Witmer, N.; Anderson, E.J.; Elrod, J.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e8. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.; Naraykina, Y.; Vasilkova, D.; Meerson, M.; Zvereva, M.; Prassolov, V.; Lazarev, V.; Manuvera, V.; Kovalchuk, S.; Anikanov, N.; et al. Protein encoded in human telomerase RNA is involved in cell protective pathways. Nucleic Acids Res. 2018, 46, 8966–8977. [Google Scholar] [CrossRef] [Green Version]
- Lewandowski, J.P.; Dumbović, G.; Watson, A.R.; Hwang, T.; Jacobs-Palmer, E.; Chang, N.; Much, C.; Turner, K.M.; Kirby, C.; Rubinstein, N.D.; et al. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020, 21, 1–35. [Google Scholar] [CrossRef]
- Na, Z.; Luo, Y.; Schofield, J.A.; Smelyansky, S.; Khitun, A.; Muthukumar, S.; Valkov, E.; Simon, M.D.; Slavoff, S.A. The NBDY Microprotein Regulates Cellular RNA Decapping. Biochemistry 2020, 59, 4131–4142. [Google Scholar] [CrossRef]
- Marcon, B.H.; Rebelatto, C.K.; Cofré, A.R.; Dallagiovanna, B.; Correa, A. DDX6 Helicase Behavior and Protein Partners in Human Adipose Tissue-Derived Stem Cells during Early Adipogenesis and Osteogenesis. Int. J. Mol. Sci. 2020, 21, 2607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarewich, C.A.; Baskin, K.K.; Munir, A.Z.; Bezprozvannaya, S.; Sharma, G.; Khemtong, C.; Shah, A.M.; McAnally, J.R.; Malloy, C.; Szweda, L.I.; et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018, 23, 3701–3709. [Google Scholar] [CrossRef]
- Friesen, M.; Warren, C.R.; Yu, H.; Toyohara, T.; Ding, Q.; Florido, M.H.; Sayre, C.; Pope, B.; Goff, L.A.; Rinn, J.L.; et al. Mitoregulin Controls β-Oxidation in Human and Mouse Adipocytes. Stem Cell Rep. 2020, 14, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Marcon, B.H.; Shigunov, P.; Spangenberg, L.; Pereira, I.; De Aguiar, A.M.; Amorín, R.; Rebelatto, C.K.; Correa, A.; Dallagiovanna, B. Cell cycle genes are downregulated after adipogenic triggering in human adipose tissue-derived stem cells by regulation of mRNA abundance. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Michel, A.M.; Fox, G.; Kiran, A.M.; De Bo, C.; O’Connor, P.B.F.; Heaphy, S.M.; Mullan, J.P.A.; Donohue, C.A.; Higgins, D.G.; Baranov, P.V. GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res. 2014, 42, D859–D864. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Xin, Y.; Zhang, Z.; Zou, X.; Xue, K.; Zhang, H.; Zhang, W.; Liu, K. Extracellular vesicles from adipose-derived stem cells ameliorate ultraviolet B-induced skin photoaging by attenuating reactive oxygen species production and inflammation. Stem Cell Res. Ther. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, T.; Sun, J.; Shen, M.; Xue, X.; Chen, Y.; Zhang, Z. Harnessing the secretome of adipose-derived stem cells in the treatment of ischemic heart diseases. Stem Cell Res. Ther. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Mitchell, R.; Mellows, B.; Sheard, J.; Antonioli, M.; Kretz, O.; Chambers, D.; Zeuner, M.-T.; Tomkins, J.; Denecke, B.; Musante, L.; et al. Secretome of adipose-derived mesenchymal stem cells promotes skeletal muscle regeneration through synergistic action of extracellular vesicle cargo and soluble proteins. Stem Cell Res. Ther. 2019, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Brunner, A.-D.; Cogan, J.Z.; Nuñez, J.K.; Fields, A.P.; Adamson, B.; Itzhak, D.N.; Li, J.Y.; Mann, M.; Leonetti, M.D.; et al. Pervasive functional translation of noncanonical human open reading frames. Science 2020, 367, 1140–1146. [Google Scholar] [CrossRef]
- Schlesinger, D.; Elsässer, S.J. Revisiting sORFs: Overcoming challenges to identify and characterize functional microproteins. FEBS J. 2021, 15769. [Google Scholar] [CrossRef]
- Martinez, T.F.; Chu, Q.; Donaldson, C.; Tan, D.; Shokhirev, M.N.; Saghatelian, A. Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 2020, 16, 458–468. [Google Scholar] [CrossRef]
- Prensner, J.R.; Enache, O.M.; Luria, V.; Krug, K.; Clauser, K.R.; Dempster, J.M.; Karger, A.; Wang, L.; Stumbraite, K.; Wang, V.M.; et al. Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat. Biotechnol. 2021, 39, 697–704. [Google Scholar] [CrossRef]
- Choi, S.-W.; Kim, H.-W.; Nam, J.-W. The small peptide world in long noncoding RNAs. Briefings Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef] [Green Version]
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell. Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef] [Green Version]
- Slavoff, S.; Mitchell, A.J.; Schwaid, A.G.; Cabili, M.N.; Ma, J.; Levin, J.; Karger, A.; Budnik, B.A.; Rinn, J.; Saghatelian, A. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat. Chem. Biol. 2012, 9, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.-L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, C.; Müller, M.; Pak, H.; Harnett, D.; Huber, F.; Grun, D.; Leleu, M.; Auger, A.; Arnaud, M.; Stevenson, B.J.; et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 2020, 11, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaertner, B.; van Heesch, S.; Schneider-Lunitz, V.; Schulz, J.F.; Witte, F.; Blachut, S.; Nguyen, S.; Wong, R.; Matta, I.; Hübner, N.; et al. A human ESC-based screen identifies a role for the translated lncRNA LINC00261 in pancreatic endocrine differentiation. eLife 2020, 9, 58659. [Google Scholar] [CrossRef]
- Spencer, H.L.; Sanders, R.; Boulberdaa, M.; Meloni, M.; Cochrane, A.; Spiroski, A.-M.; Mountford, J.; Emanueli, C.; Caporali, A.; Brittan, M.; et al. The LINC00961 transcript and its encoded micropeptide, small regulatory polypeptide of amino acid response, regulate endothelial cell function. Cardiovasc. Res. 2020, 116, 1981–1994. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, M.; Zhou, H.; Lu, S.; Zhang, B. Long Noncoding RNA EBLN3P Promotes the Progression of Liver Cancer via Alteration of microRNA-144-3p/DOCK4 Signal. Cancer Manag. Res. 2020, 12, 9339–9349. [Google Scholar] [CrossRef]
- Jiang, W.; Peng, A.; Chen, Y.; Pang, B.; Zhang, Z. Long non-coding RNA EBLN3P promotes the recovery of the function of impaired spiral ganglion neurons by competitively binding to miR-204-5p and regulating TMPRSS3 expression. Int. J. Mol. Med. 2020, 45, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Mathias, C.; Muzzi, J.C.D.; Antunes, B.B.; Gradia, D.F.; Castro, M.A.A.; de Oliveira, J.C. Unraveling Immune-Related lncRNAs in Breast Cancer Molecular Subtypes. Front. Oncol. 2021, 11, 692170. [Google Scholar] [CrossRef]
- Dai, S.; Li, N.; Zhou, M.; Yuan, Y.; Yue, D.; Li, T.; Zhang, X. LncRNA EBLN3P promotes the progression of osteosarcoma through modifying the miR-224-5p/Rab10 signaling axis. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Razooky, B.S.; Obermayer, B.; O’May, J.B.; Tarakhovsky, A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes 2017, 8, 206. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Jia, L.; Tang, Z.; Zheng, Y. Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via PTEN/ AKT pathway. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Asila, A.; Yang, X.; Kaisaer, Y.; Ma, L. SNHG16/miR-485-5p/BMP7 axis modulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. J. Gene Med. 2021, 23, e3296. [Google Scholar] [CrossRef]
- Huang, N.; Li, F.; Zhang, M.; Zhou, H.; Chen, Z.; Ma, X.; Yang, L.; Wu, X.; Zhong, J.; Xiao, F.; et al. An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab. 2021, 33, 128–144.e9. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef]

| SUBJECT | DONOR1 | DONOR2 | DONOR3 | Mean ± SD |
|---|---|---|---|---|
| Age | 46 | 20 | 48 | 38 ± 12.75 |
| Gender | F | F | F | F |
| Weight (kg) | 74.5 | 75 | 90 | 79.8 ± 7.19 |
| Height (cm) | 166 | 174 | 175 | 171 ± 4.02 |
| BMI | 27.04 | 24.77 | 29.39 | 27.06 ± 1.88 |
| lncRNA | GeneID (Ensembl) | ORF Length (nt) | MP* Length (aa) | DeepLoc1.0 Pred. | DLoc† Score | Microprotein Sequence |
|---|---|---|---|---|---|---|
| CYTOR | ENSG00000222041.11 | 156 | 52 | Nucleus | 0.37 | MTDTENHDSAPSSTSTCCPPITAGMQLKDSLGPGSNRPLWTLRPLHLRVVCL |
| EBLN3P | ENSG00000281649.2 | 141 | 47 | Nucleus | 0.77 | MEEPMDTSEPLSALPFTGQQSFEPSGKFGQYPSMQMNHIQALGKWRT |
| EBLN3P | ENSG00000281649.2 | 78 | 26 | Extracellular | 0.52 | MYVTDPESPAAWDPCLPSVSPAELWN |
| GAS5 | ENSG00000234741.8 | 150 | 50 | Extracellular | 0.43 | MVLGADAVWLWIAPYGQLCPQGRMRIATEVLKSKPNSSHWHTGIRQKAGS |
| GAS5 | ENSG00000234741.8 | 81 | 27 | Extracellular | 1 | MTCLGKDMKTVPVIPFKGTCFIDVNVN |
| LINC00968 | ENSG00000246430.7 | 132 | 44 | Extracellular | 0.53 | MFLQKLKSCLVKAFHKMVCVWDQEDRRLLKKRTGTLTHFRLLHV |
| LINC01116 | ENSG00000163364.10 | 249 | 83 | Nucleus | 0.71 | MGPRFLADARGRGRVPGSRFSQAPIPAHARGPRPTHEAPTPIVEAPPGKEVRLPLQAAPRGMGNRQEMTRTASLRLCSRPSLC |
| MEG3 | ENSG00000214548.18 | 168 | 56 | Nucleus | 0.50 | MPFERLEAKSIKHSWENTTGGTTRFSYTLGSHGEDRREKKEVEREERAGETGEENN |
| MEG3 | ENSG00000214548.18 | 444 | 148 | Nucleus/ Cytoplasm | 0.418/ 0.417 | MRRLSIVMKNPWHSPHPQTHGSHSHTGPKATVSAAVAPVDIGKPGEGVEEISWPPAGSLGFCAQGSWSPKNFQKLTPHVPILLGFLDFSEAPAEGSRCSLECRGSPLTWLLESLLFLLLLPSSSSSSLSISPSLCPSPVPDLAIPGCP |
| MEG3 | ENSG00000214548.18 | 252 | 84 | Cytoplasm | 0.37 | MEAAEEALMGPTIPDPSLLPGGPLVSFLVWAEAITWMPTWEGTSNVGPQPLSSSKSLHSHGDTLHLFPRDRLDPETLDPGPPLE |
| MIR22HG | ENSG00000186594.14 | 279 | 93 | Mitochondria | 0.60 | MGWEGPNSRVDDTFWASWRAFAQIGPARSGFRLETLAGLRSRRLKQPKRLQEAVSVRFGG |
| MIR22HG | ENSG00000186594.14 | 66 | 22 | Mitochondria | 0.50 | MIRFGQVGEPLPRLAQQGAVLD |
| MSC-AS1 | ENSG00000235531.10 | 192 | 64 | Nucleus | 0.42 | MSLETTGPQERQALSVLLLPWKKPAPTMPSATSKSSLRPPQKQMLSCFLYSCRTTSNHPNTREH |
| SNHG1 | ENSG00000255717.7 | 87 | 29 | Extracellular | 0.47 | MSYWAPVCRIYAHVGTEESSVVAPTRAYW |
| SNHG1 | ENSG00000255717.7 | 153 | 51 | Extracellular | 0.73 | MFSPQELTGEGMGQDPSLCKASVTVMFQVGVHGLCSYRGDLVDNHSMMNTK |
| SNHG16 | ENSG00000163597.15 | 99 | 33 | Nucleus | 0.78 | MATPVGVEHGEQSQAFSDDGAVSLSFQSRKRIL |
| SNHG16 | ENSG00000163597.15 | 108 | 36 | Nucleus | 0.58 | MATPVGVEHGEQSQAFSDDGWLGGLKVLDEKMLSKR |
| SNHG29 | ENSG00000175061.18 | 405 | 135 | Mitochondria | 0.69 | MFPGSLSRGRRAAVEMAWLPGSCARVAFAAGAAARYWTAWQGSAGPNPAAVAEAHGSLFCGRATSARAWSLRRPGPGSPAHSGGVQTRENWVSWGRLAVWGTPRAVYVGKIVTVLLEDLFDCPDDTCNRKCRQKR |
| SNHG29 | ENSG00000175061.18 | 285 | 95 | Mitochondria | 0.66 | MFPGSLSRGRRAAVEMAWLPGSCARVAFAAGAAARYWTAWQGSAGPNPAAVAEAHGSLFCGRATSARAWSLRRPGPGSPAHSGGVQTRENWVANS |
| SNHG29 | ENSG00000175061.18 | 111 | 37 | Extracellular | 0.68 | MDHSFVVGPHLPEPGVCEGRDPVPRPTVGVCKPERTG |
| SNHG29 | ENSG00000175061.18 | 237 | 79 | Nucleus/ Extracellular | 0.243/ 0.230 | MDHSFVVGPHLPEPGVCEGRDPVPRPTVGVCKPERTGLQIREESASCLAAEYWSQEPAMRLYSQRMSVPRTSSCHQFGF |
| SNHG29 | ENSG00000175061.18 | 210 | 70 | Endoplasmic Reticulum | 0.49 | MLALCIRGHAQQIQEIYLATFSRKGTLGIIHYILEFFWVFFFFFETVLLYCPGWSVVAQSQLIASSITQA |
| SNHG29 | ENSG00000175061.18 | 54 | 18 | - | - | MYQRTCSADPRDIFGNFF |
| SNHG29 | ENSG00000175061.18 | 237 | 79 | Golgi Apparatus | 0.42 | MLSRSKRYIWQLFLEKAHWVSFITFLSFFGFFFFFLRQSCCIAQAGVWWHNHSSLHPQSPRPKQSSHLVAGTTAHSTPG |
| SNHG29 | ENSG00000175061.18 | 78 | 26 | Mitochondria | 0.97 | MLPRLVSGSWAQMVLLPQLPKAQAKL |
| SNHG5 | ENSG00000203875.12 | 105 | 35 | Mitochondria | 0.26 | MALSSVAQWSSSEDAKIHEKTSRTSGRIFNGKSLG |
| SNHG5 | ENSG00000203875.12 | 72 | 24 | Mitochondria | 0.57 | MQRYTKKLPEHLGEYLMENRLVKT |
| SNHG6 | ENSG00000245910.8 | 75 | 25 | Mitochondria | 0.86 | MPVWWRRRRLRARSWALRGARKPLR |
| SNHG8 | ENSG00000269893.8 | 156 | 52 | Mitochondria | 0.68 | MIIGPKLTALPKRQRSQDIGRSGAALETLKFTSMRGLECSLGRRASTCSPGP |
| SNHG8 | ENSG00000269893.8 | 108 | 36 | Mitochondria/ Nucleus | 0.309/ 0.303 | MDDGNIRLSRNPSGNGRSLFSIRQWTYRSWGNGCSE |
| ZFAS1 | ENSG00000177410.13 | 75 | 25 | Mitochondria | 0.34 | MDFGRGSHHWTSKEATCRHLQPSIS |
| ZFAS1 | ENSG00000177410.13 | 60 | 20 | - | - | MRVLEVEYIYTYKIETGDGI |
| ZFAS1 | ENSG00000177410.13 | 99 | 33 | Extracellular | 0.44 | MRVLEVEYIYTYKIGWEPRVPVCVDLGLIQSAL |
| ZFAS1 | ENSG00000177410.13 | 153 | 51 | Nucleus | 0.57 | MEYERSPLERKGQTLCFHESEDLAEPVPQGYCIHSLSLKGCAHFKNVIVRL |
| ZFAS1 | ENSG00000177410.13 | 99 | 33 | Extracellular | 0.51 | MRGALWKEKDRPCAFMKVKIWLNQFHKVTVYIA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonilauri, B.; Holetz, F.B.; Dallagiovanna, B. Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins. Biomolecules 2021, 11, 1673. https://doi.org/10.3390/biom11111673
Bonilauri B, Holetz FB, Dallagiovanna B. Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins. Biomolecules. 2021; 11(11):1673. https://doi.org/10.3390/biom11111673
Chicago/Turabian StyleBonilauri, Bernardo, Fabiola Barbieri Holetz, and Bruno Dallagiovanna. 2021. "Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins" Biomolecules 11, no. 11: 1673. https://doi.org/10.3390/biom11111673
APA StyleBonilauri, B., Holetz, F. B., & Dallagiovanna, B. (2021). Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins. Biomolecules, 11(11), 1673. https://doi.org/10.3390/biom11111673






