The Membrane-Bound Notch Regulator Mnr Supports Notch Cleavage and Signaling Activity in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Work and Microscopy
2.2. Analysis of CG32521 Expression
2.3. Protein Fractionation
2.4. dsRNA Production against RA and RC
2.5. Aggregation Assay of S2 cells
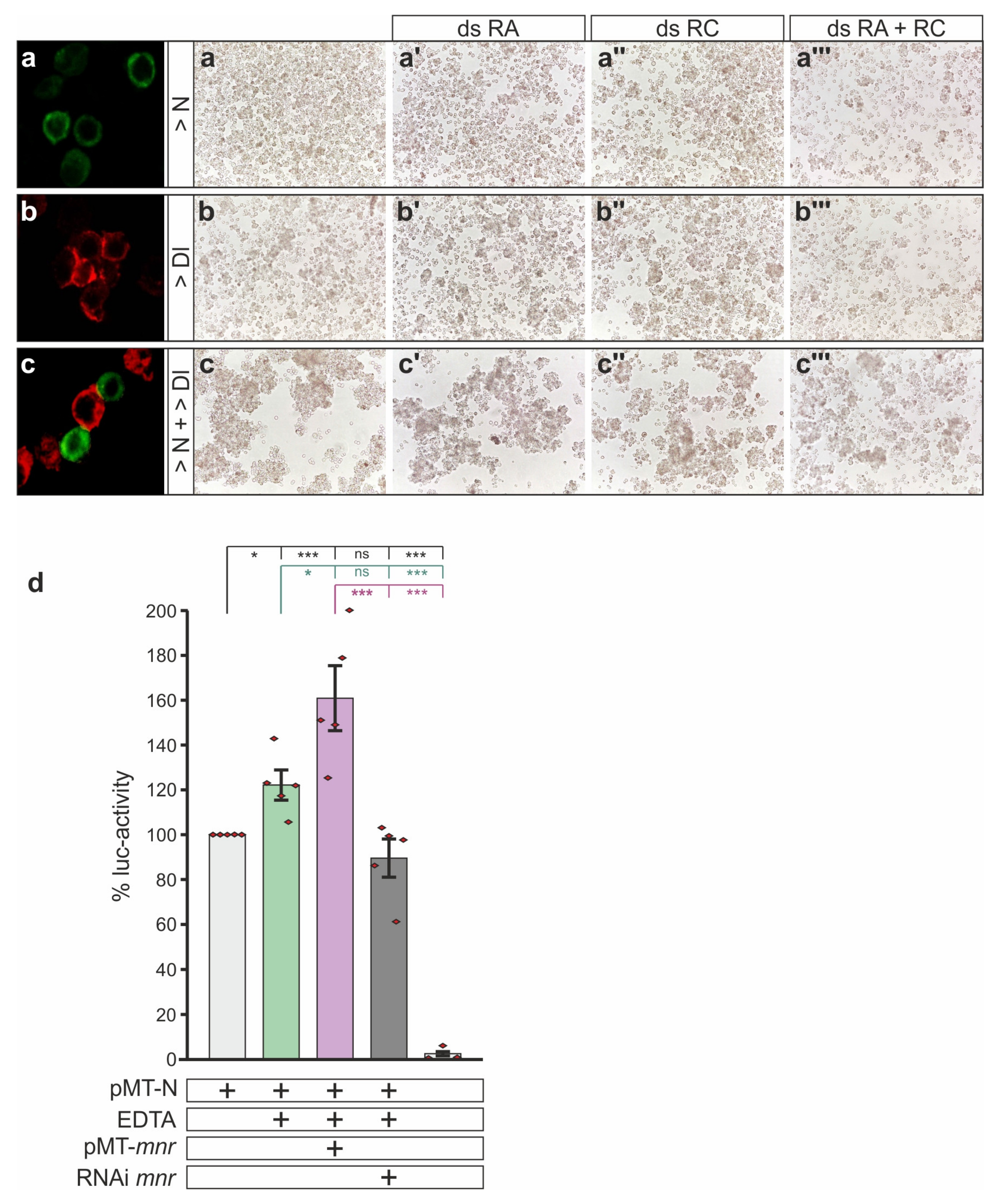
2.6. Notch Activity Assay in S2 Cells
2.7. Bioinformatics and Statistics
3. Results and Discussion
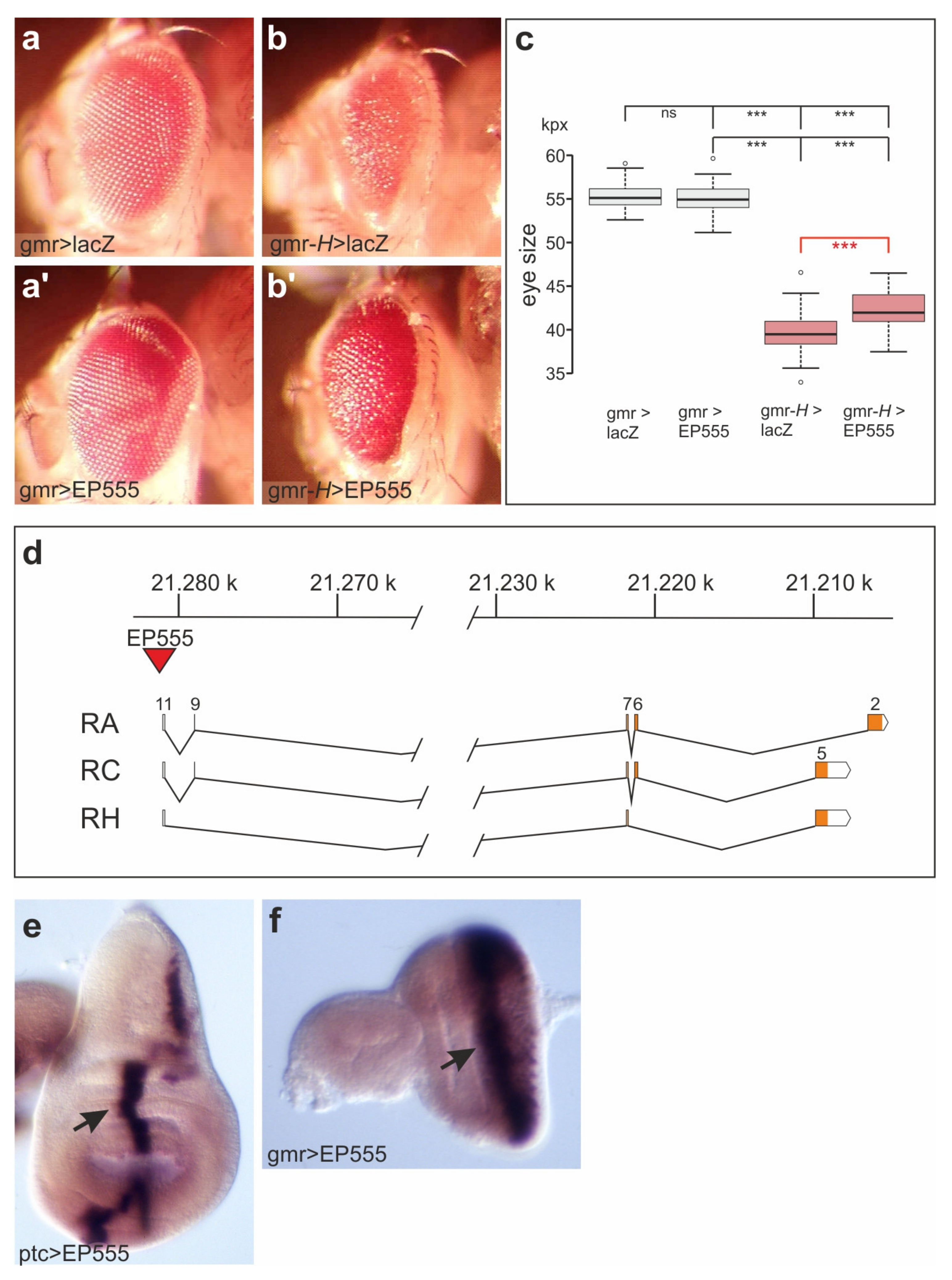
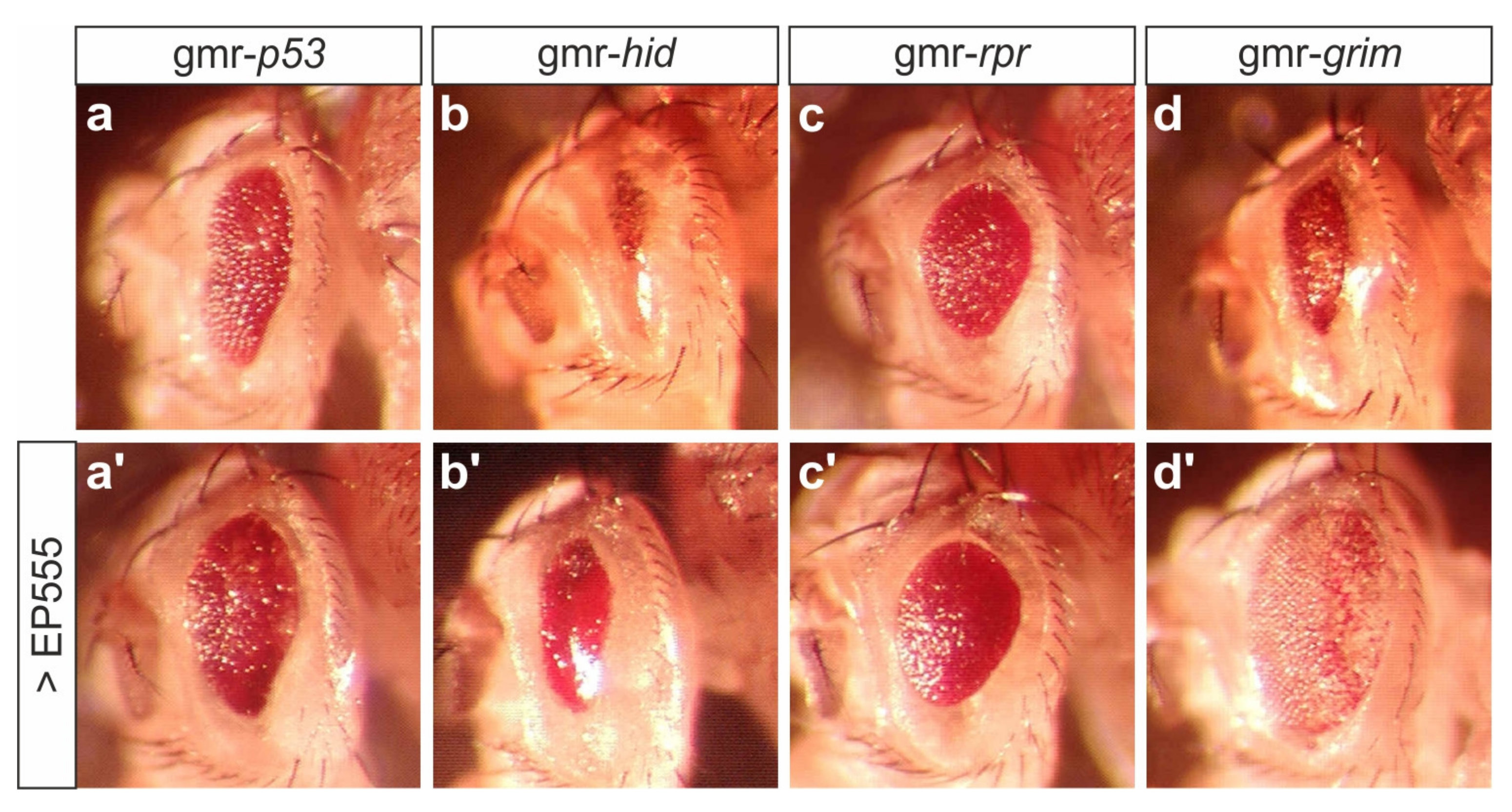
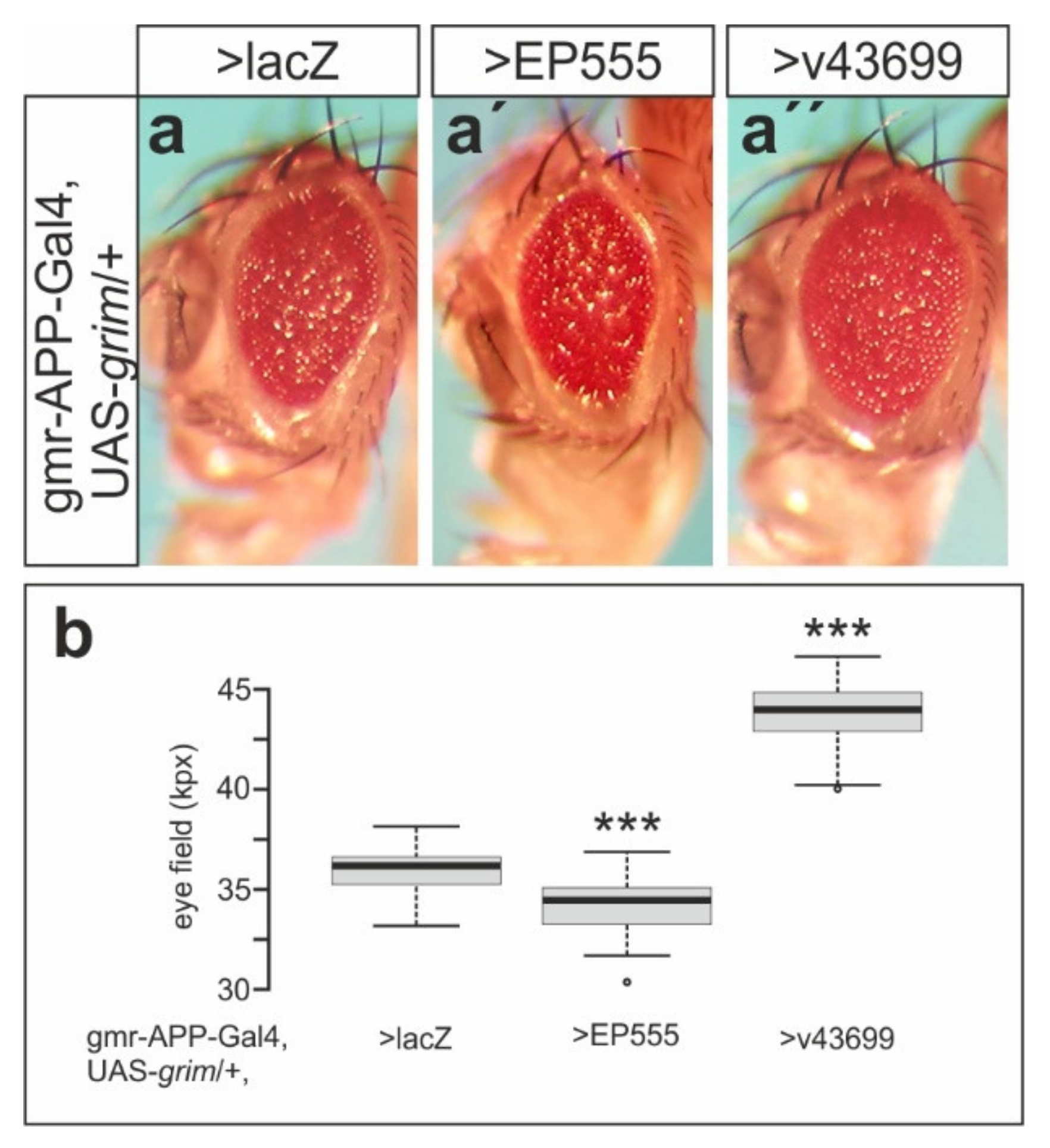
3.1. EP555 Drives Expression of CG32521 Supporting Notch Signaling Activity with an Apparent Anti-Apoptotic Role in Eye Development
3.2. CG32521 Provides Several Transcript Classes with a Broad Expression during Embryogenesis
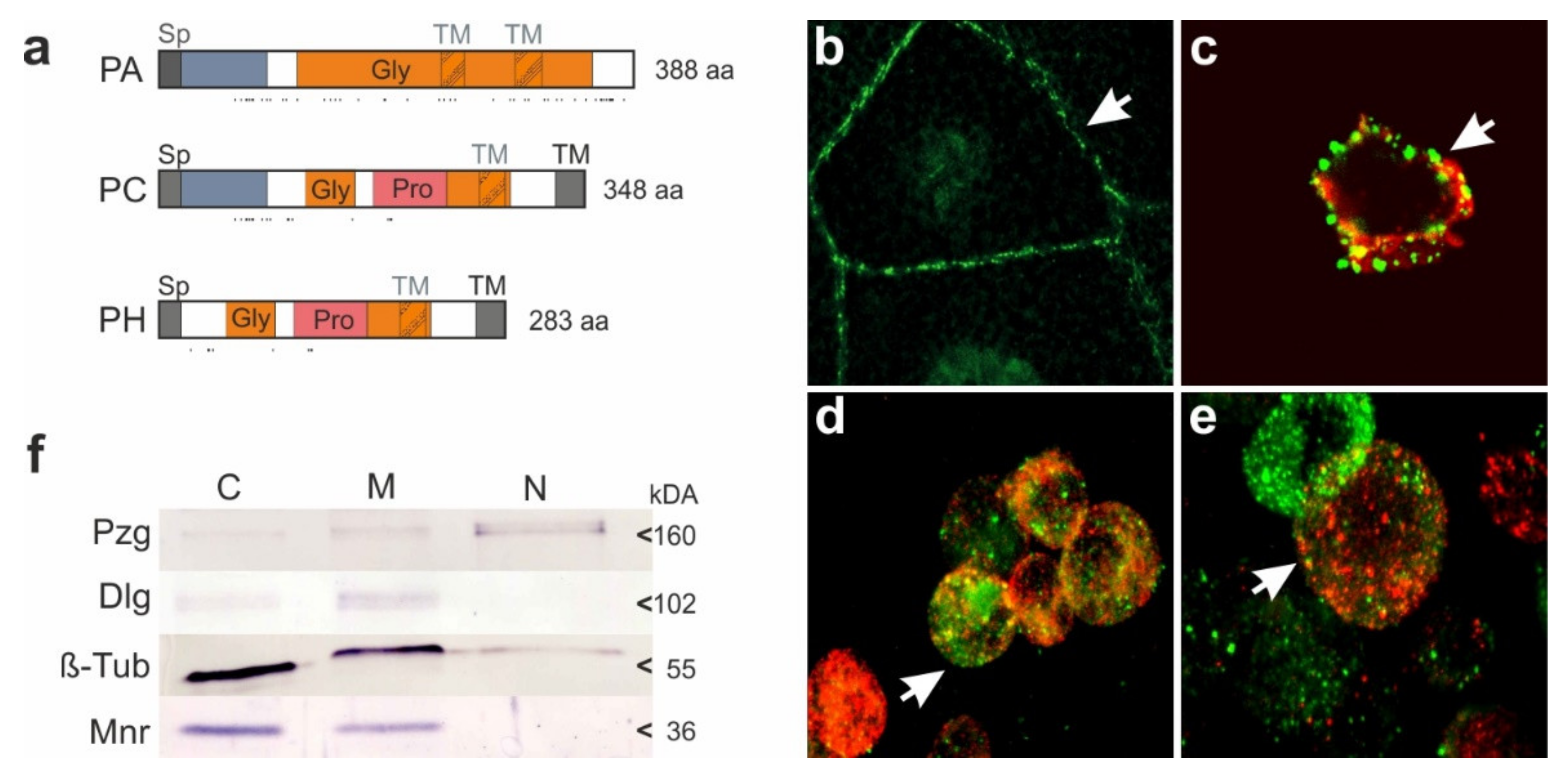
3.3. CG32521 Encodes Membrane-Bound Proteins
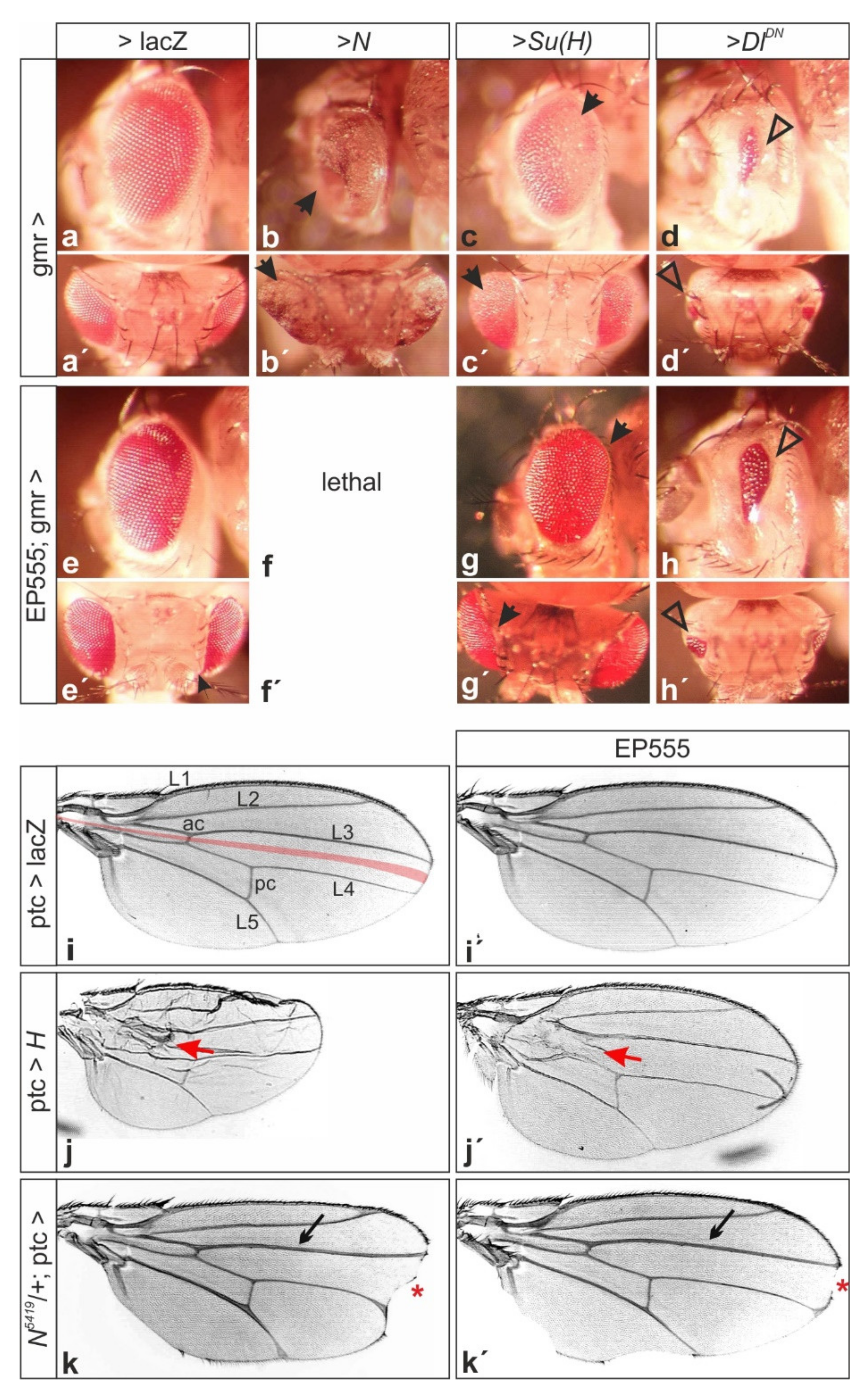
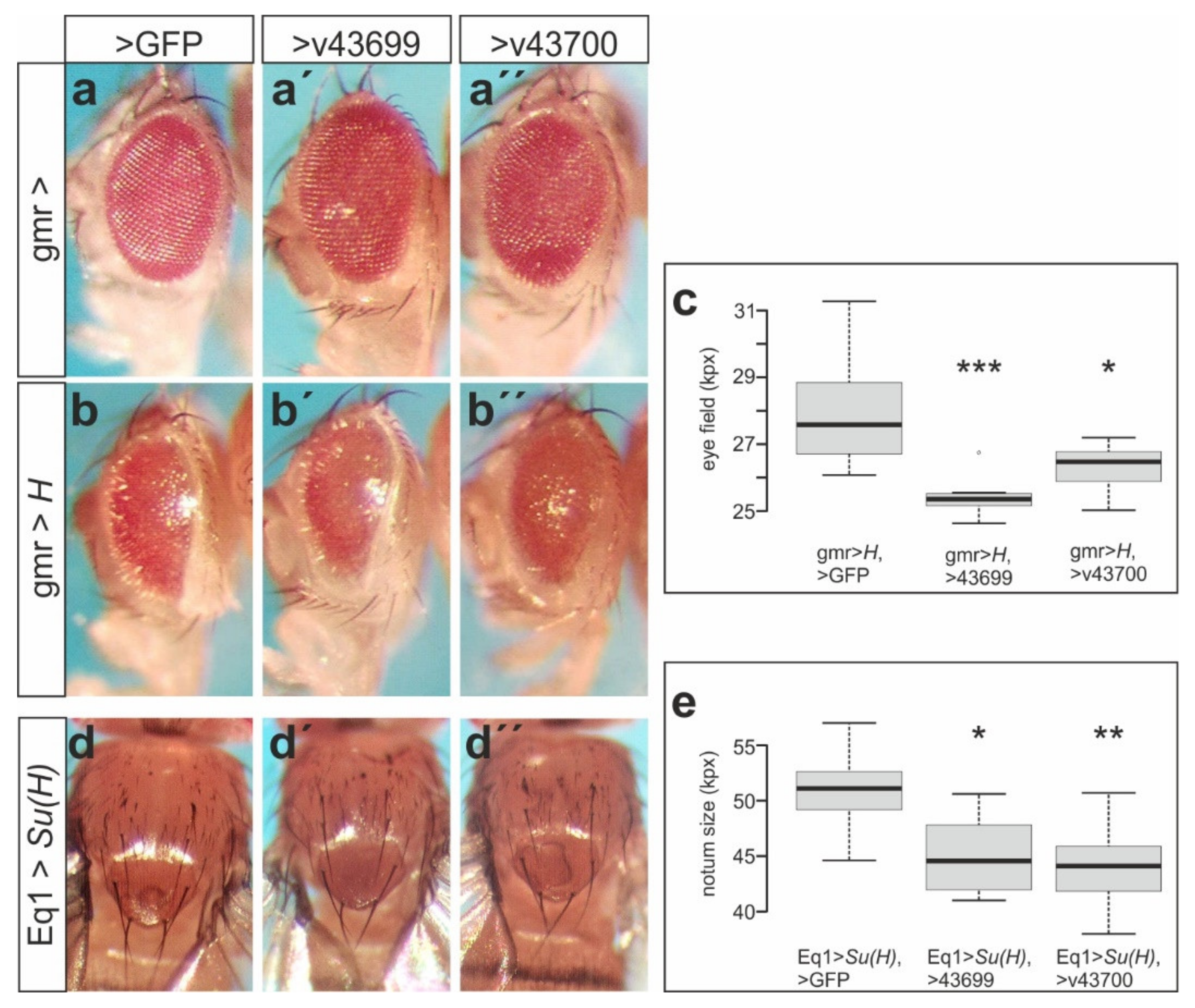
3.4. The Membrane-Bound Notch Regulator Mnr Encoded by CG32521 Boosts Notch Signaling Activity
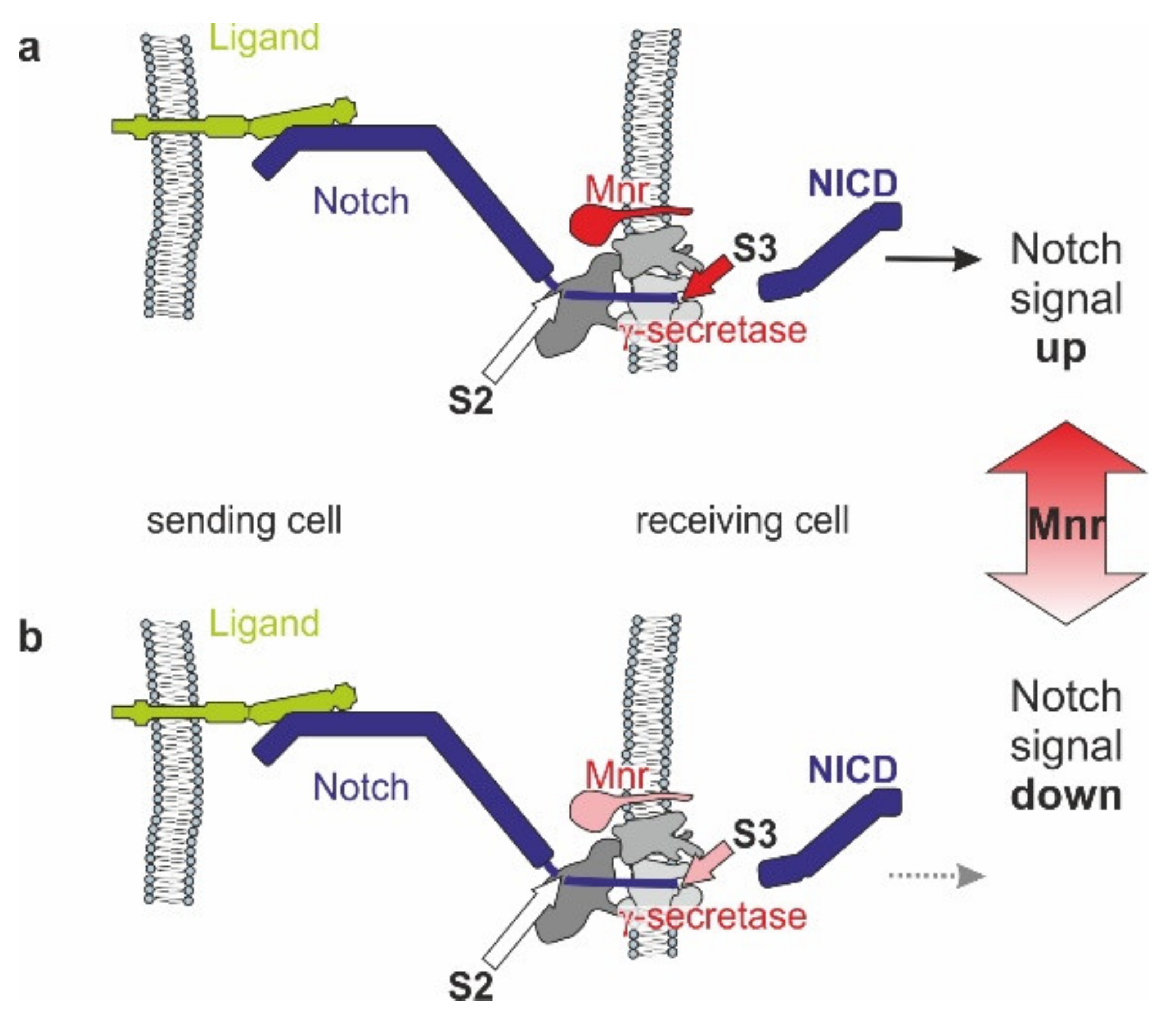
3.5. Mnr Promotes Notch Receptor Cleavage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louvi, A.; Artavanis-Tsakonas, S. Notch and disease: A growing field. Semin. Cell. Dev. Biol. 2012, 23, 473–480. [Google Scholar]
- Siebel, C.; Lendahl, U. Notch Signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [PubMed] [Green Version]
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012, 13, 654–666. [Google Scholar] [PubMed] [Green Version]
- Morgan, T.H.; Bridges, C.B. Sex-linked inheritance in Drosophila. Publs. Carnegie Instn. 1916, 237, 1–88. [Google Scholar]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell. Biol. 2016, 17, 722–735. [Google Scholar] [PubMed]
- Bray, S.J.; Gomez-Lamarca, M. Notch after cleavage. Curr. Opin. Cell. Biol. 2018, 51, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Baron, M.; Aslam, H.; Flasza, M.; Fostier, M.; Higgs, J.E.; Mazaleyrat, S.L.; Wilkin, M.B. Multiple levels of Notch signal regulation (review). Mol. Membr. Biol. 2002, 19, 27–38. [Google Scholar]
- Harvey, B.M.; Haltiwanger, R.S. Regulation of Notch Function by O-Glycosylation. Adv. Exp. Med. Biol. 2018, 1066, 59–78. [Google Scholar] [PubMed]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The canonical Notch signaling pathway: Structural and biochemical insights into shape, sugar, and force. Dev. Cell. 2017, 41, 28–241. [Google Scholar] [CrossRef] [Green Version]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar]
- Gordon, W.R.; Arnett, K.L.; Blacklow, S.C. The molecular logic of Notch signaling—A structural and biochemical perspective. J. Cell. Sci. 2008, 121 Pt 19, 3109–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beel, A.J.; Sanders, C.R. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 2008, 65, 1311–1334. [Google Scholar]
- Vetrivel, K.S.; Thinakaran, G. Membrane rafts in Alzheimer’s disease beta-amyloid production. Biochim. Biophys. Acta. 2010, 1801, 860–867. [Google Scholar] [CrossRef] [Green Version]
- Jorissen, E.; De Strooper, B. Gamma-secretase and the intramembrane proteolysis of Notch. Curr. Top. Dev. Biol. 2010, 92, 201–230. [Google Scholar]
- Haining, E.J.; Yang, J.; Bailey, R.L.; Khan, K.; Collier, R.; Tsai, S.; Watson, S.P.; Frampton, J.; Garcia, P.; Tomlinson, M.G. The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J. Biol. Chem. 2012, 287, 39753–39765. [Google Scholar] [CrossRef] [Green Version]
- Dornier, E.; Coumailleau, F.; Ottavi, J.F.; Moretti, J.; Boucheix, C.; Mauduit, P.; Schweisguth, F.; Rubinstein, E. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell. Biol. 2012, 199, 481–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seipold, L.; Saftig, P. The emerging role of tetraspanins in the proteolytic processing of the amyloid precursor protein. Front. Mol. Neurosci. 2016, 9, 149. [Google Scholar] [CrossRef] [Green Version]
- Herranz, H.; Stamataki, E.; Feiguin, F.; Milán, M. Self-refinement of Notch activity through the transmembrane protein Crumbs: Modulation of gamma-secretase activity. EMBO Rep. 2006, 7, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor-Giles, K.M.; Skeath, J.B. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 2003, 5, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, A.; Kandachar, V.; Zitserman, D.; Tong, X.; Roegiers, F. Sanpodo controls sensory organ precursor fate by directing Notch trafficking and binding γ-secretase. J. Cell. Biol. 2013, 201, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Roegiers, F.; Jan, Y.N. Asymmetric cell division. Curr. Opin. Cell. Biol. 2004, 16, 195–205. [Google Scholar] [CrossRef]
- Lindsley, D.; Zimm, G.G. The Genome of Drosophila melanogaster; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- Zacharioudaki, E.; Bray, S.J. Tools and methods for studying Notch signaling in Drosophila melanogaster. Methods 2014, 68, 173–182. [Google Scholar] [CrossRef]
- Müller, D.; Kugler, S.J.; Preiss, A.; Maier, D.; Nagel, A.C. Genetic modifier screens on Hairless gain-of-function phenotypes reveal genes involved in cell differentiation, cell growth and apoptosis in Drosophila melanogaster. Genetics 2005, 171, 1137–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, D. Hairless: The ignored antagonist of the Notch signalling pathway. Hereditas 2006, 143, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Oswald, F. Setting the stage for Notch: The Drosophila Su(H)-Hairless repressor complex. PLoS Biol. 2016, 14, e1002524. [Google Scholar]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.A.; Maile, R.; Rubin, G.M. P-element insertion dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 1997, 94, 5195–5200. [Google Scholar] [CrossRef] [Green Version]
- Speicher, S.A.; Thomas, U.; Hinz, U.; Knust, E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: Control of cell proliferation. Development 1994, 120, 535–544. [Google Scholar] [CrossRef]
- Tang, Y.C.; Sun, Y.H. Use of mini-white as a reporter gene to screen for GAL4 insertions with spatially restricted expression pattern in the developing eye in Drosophila. Genesis 2002, 34, 39–45. [Google Scholar] [CrossRef]
- Maier, D.; Nagel, A.C.; Johannes, B.B.; Preiss, A. Subcellular localization of Hairless protein shows a major focus of activity within the nucleus. Mech. Dev. 1999, 89, 195–199. [Google Scholar] [CrossRef]
- Fuerstenberg, S.; Giniger, E. Multiple roles for Notch in Drosophila myogenesis. Dev. Biol. 1998, 201, 66–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, A.C.; Krejci, A.; Tenin, G.; Bravo-Patiño, A.; Bray, S.; Maier, D.; Preiss, A. Hairless-mediated repression of Notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 2005, 25, 10433–10441. [Google Scholar] [CrossRef] [Green Version]
- Huppert, S.S.; Jacobsen, T.L.; Muskavitch, M.A. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development 1997, 124, 3283–3291. [Google Scholar] [CrossRef] [PubMed]
- Rørth, P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 1996, 93, 12418–12422. [Google Scholar] [CrossRef] [Green Version]
- White, K.; Tagaoglu, E.; Steller, H. Cell killing by the Drosophila gene reaper. Science 1996, 271, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Nordstrom, W.; Gish, B.; Abrams, J.M. grim, a novel cell death gene in Drosophila. Genes Dev. 1996, 10, 1773–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grether, M.E.; Abrams, J.M.; Agapite, J.; White, K.; Steller, H. The head involution defective gene of Drosophila melanogaster. Genes Dev. 1995, 9, 1694–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ollmann, M.; Young, L.M.; Di Como, C.J.; Karim, F.; Belvin, M.; Robertson, S.; Whittaker, K.; Demsky, M.; Fisher, W.W.; Buchman, A.; et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 2000, 101, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Hong., E.J.; Fernandes, J.; Zipursky, S.L.; Hay, B.A. A reporter for amyloid precursor protein gamma-secretase activity in Drosophila. Hum Mol Genet. 2003, 12, 2669–2678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef]
- Tautz, D.; Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 1989, 98, 81–85. [Google Scholar] [CrossRef]
- Cubas, P.; de Celis, J.F.; Campuzano, S.; Modolell, J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 1991, 5, 996–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggs, P. Expression and purification of Maltose-binding protein fusions. Expression and purification of maltose-binding protein fusions. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Fehon, R.G.; Kooh, P.J.; Rebay, I.; Regan, C.L.; Xu, T.; Muskavitch, M.A.; Artavanis-Tsakonas, S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 1990, 61, 523–534. [Google Scholar] [CrossRef]
- Krämer, H.; Phistry, M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell. Biol. 1996, 133, 1205–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, P.; La Rosa, M.K.; Schulz, A.; Preiss, A.; Nagel, A.C. Cyclin G functions as a positive regulator of growth and metabolism in Drosophila. PLoS Genet. 2015, 11, e1005440. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, H.; Krejcí, A.; Moshkin, Y.; Verrijzer, C.P.; Karch, F.; Bray, S.J. Gene-specific targeting of the histone chaperone Asf1 to mediate silencing. Dev. Cell. 2007, 13, 593–600. [Google Scholar] [CrossRef] [Green Version]
- Kugler, S.J.; Nagel, A.C. putzig is required for cell proliferation and regulates Notch activity in Drosophila. Mol. Biol. Cell. 2007, 18, 3733–3740. [Google Scholar] [CrossRef] [Green Version]
- Hortsch, M. Preparation and analysis of membranes and membrane proteins from Dosophila. In Drosophila melanogaster: Practical Uses in Cell and Molecular Biology; Goldstein, L.S.B., Fyrberg, E.A., Eds.; Academic Press: New York, NY, USA, 1994. [Google Scholar]
- Klueg, K.M.; Parody, T.R.; Muskavitch, M.A. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol. Biol. Cell 1998, 9, 1709–1723. [Google Scholar] [CrossRef] [Green Version]
- Clemens, J.C.; Worby, C.A.; Simonson-Leff, N.; Muda, M.; Maehama, T.; Hemmings, B.A.; Dixon, J.E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 2000, 97, 6499–6503. [Google Scholar] [CrossRef] [Green Version]
- Bray, S.; Musisi, H.; Bienz, M. Bre1 is required for Notch signaling and histone modification. Dev. Cell. 2005, 8, 279–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunch, T.A.; Grinblat, Y.; Goldstein, L.S. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988, 16, 1043–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kober, L.; Zimmermann, M.; Kurz, M.; Bayer, M.; Nagel, A.C. Loss of putzig in the germline impedes germ cell development by inducing cell death and new niche like microenvironments. Sci. Rep. 2019, 9, 9108. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for a group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl. Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Hu, Y.; Sopko, R.; Foos, F.; Kelley, C.; Flockhart, I.; Ammeux, N.; Wang, X.; Perkins, L.; Perrimon, N.; Mohr, S.E. FlyPrimerBank: An online database for Drosophila melanogaster gene expression analysis and knock-down evaluation of RNAi reagents. G3 (Bethesda) 2013, 3, 1607–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [Green Version]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Protzer, C.E.; Wech, I.; Nagel, A.C. Hairless induces cell death by downregulation of EGFR signalling activity. J. Cell Sci. 2008, 121 Pt 19, 3167–3176. [Google Scholar] [CrossRef] [Green Version]
- Arya, R.; White, K. Cell death in development: Signaling pathways and core mechanisms. Semin. Cell Dev. Biol. 2015, 39, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Ho, D.M.; Artavanis-Tsakonas, S. The Notch-mediated proliferation circuitry. Curr. Top. Dev. Biol. 2016, 116, 17–33. [Google Scholar] [PubMed]
- Housden, B.E.; Fu, A.Q.; Krejci, A.; Bernard, F.; Fischer, B.; Tavaré, S.; Russell, S.; Bray, S.J. Transcriptional dynamics elicited by a short pulse of Notch activation involves feed-forward regulation by E(spl)/Hes genes. PLoS Genet. 2013, 9, e1003162. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, M.; Takemoto, D.; Igaki, T. Interaction between Ras and Src clones causes interdependent tumor malignancy via Notch signaling in Drosophila. Dev. Cell. 2021, 56, 2223–2236.e5. [Google Scholar] [CrossRef]
- Aradska, J.; Bulat, T.; Sialana, F.J.; Birner-Gruenberger, R.; Erich, B.; Lubec, G. Gel-free mass spectrometry analysis of Drosophila melanogaster heads. Proteomics 2015, 15, 3356–3360. [Google Scholar] [CrossRef] [PubMed]
- Auer, J.S.; Nagel, A.C.; Schulz, A.; Wahl, V.; Preiss, A. MAPK-dependent phosphorylation modulates the activity of Suppressor of Hairless in Drosophila. Cell Signal. 2015, 27, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Cherbas, L.; Gong, L. Cell lines. Methods 2014, 68, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Bernard, F.; Krejci, A.; Housden, B.; Adryan, B.; Bray, S.J. Specificity of Notch pathway activation: Twist controls the transcriptional output in adult muscle progenitors. Development 2010, 137, 2633–2642. [Google Scholar] [CrossRef] [Green Version]
- Krejcí, A.; Bray, S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007, 21, 1322–1327. [Google Scholar] [CrossRef] [Green Version]
- Tagami, S.; Okochi, M.; Yanagida, K.; Ikuta, A.; Fukumori, A.; Matsumoto, N.; Ishizuka-Katsura, Y.; Nakayama, T.; Itoh, N.; Jiang, J.; et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 2008, 28, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.C.; Yan, C.; Yang, G.; Lu, P.; Ma, D.; Sun, L.; Zhou, R.; Scheres, S.; Shi, Y. An atomic structure of human γ-secretase. Nature 2015, 525, 212–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagel, A.C.; Müller, D.; Zimmermann, M.; Preiss, A. The Membrane-Bound Notch Regulator Mnr Supports Notch Cleavage and Signaling Activity in Drosophila melanogaster. Biomolecules 2021, 11, 1672. https://doi.org/10.3390/biom11111672
Nagel AC, Müller D, Zimmermann M, Preiss A. The Membrane-Bound Notch Regulator Mnr Supports Notch Cleavage and Signaling Activity in Drosophila melanogaster. Biomolecules. 2021; 11(11):1672. https://doi.org/10.3390/biom11111672
Chicago/Turabian StyleNagel, Anja C., Dominik Müller, Mirjam Zimmermann, and Anette Preiss. 2021. "The Membrane-Bound Notch Regulator Mnr Supports Notch Cleavage and Signaling Activity in Drosophila melanogaster" Biomolecules 11, no. 11: 1672. https://doi.org/10.3390/biom11111672
APA StyleNagel, A. C., Müller, D., Zimmermann, M., & Preiss, A. (2021). The Membrane-Bound Notch Regulator Mnr Supports Notch Cleavage and Signaling Activity in Drosophila melanogaster. Biomolecules, 11(11), 1672. https://doi.org/10.3390/biom11111672





