E3 Ubiquitin Ligase Regulators of Notch Receptor Endocytosis: From Flies to Humans
Abstract
1. Introduction
2. Lessons from the Fly: Deltex and Su(dx) Act as Both Positive and Negative Regulators of Notch
3. Vertebrate Deltex Proteins
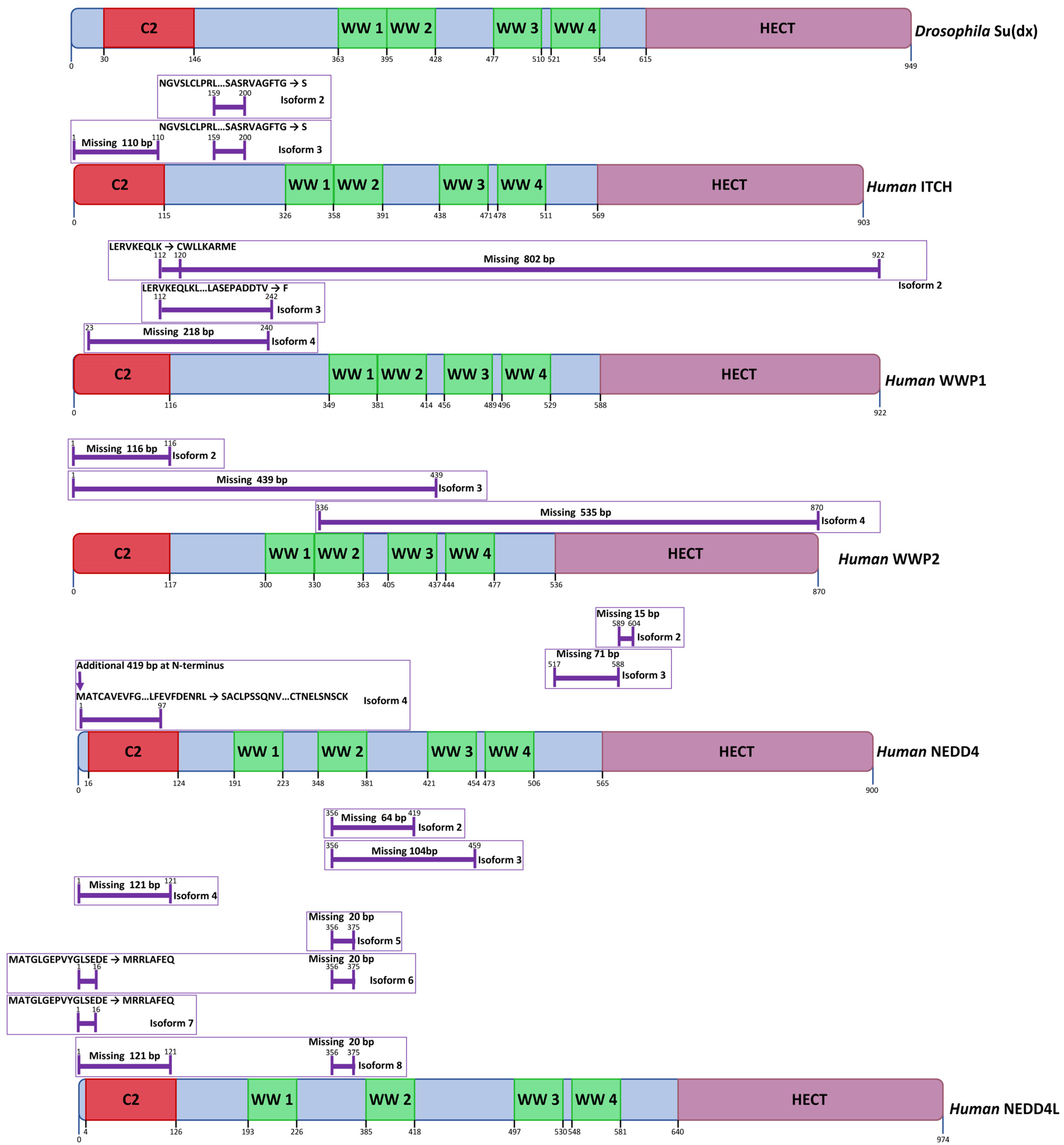
3.1. Domain Structure of Deltex Proteins
3.2. DTX1
3.3. DTX2
3.4. DTX3
3.5. DTX3L
3.6. DTX4
3.7. Regulation of DTX Proteins in Development and Disease
4. Vertebrate Su(dx)-Related Proteins
4.1. Domain Structure of Su(dx)-Related Proteins
4.2. ITCH/AIP4
4.3. NEDD4
4.4. NEDD4-like (NEDD4-L)
4.5. WWP1
4.6. WWP2
4.7. Therapeutic Targeting of Su(dx)-Related Proteins
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Go, M.J.; Eastman, D.S.; Artavanis-Tsakonas, S. Cell proliferation control by Notch signaling in Drosophila development. Development 1998, 125, 2031–2040. [Google Scholar] [CrossRef] [PubMed]
- Fre, S.; Huyghe, M.; Mourikis, P.; Robine, S.; Louvard, D.; Artavanis-Tsakonas, S. Notch signals control the fate of immature progenitor cells in the intestine. Nature 2005, 435, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.P.; Zheng, G.; Zou, L.; Liu, H.L.; Hou, L.H.; Zhou, P.; Yin, D.D.; Zheng, Q.J.; Liang, L.; Zhang, S.Z.; et al. Notch activation promotes cell proliferation and the formation of neural stem cell-like colonies in human glioma cells. Mol. Cell. Biochem. 2008, 307, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Ohlstein, B.; Spradling, A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science 2007, 315, 988–992. [Google Scholar] [CrossRef] [PubMed]
- Lowell, S.; Jones, P.; Le Roux, I.; Dunne, J.; Watt, F.M. Stimulation of human epidermal differentiation by Delta–Notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 2000, 10, 491–500. [Google Scholar] [CrossRef]
- Yang, X.; Klein, R.; Tian, X.; Cheng, H.T.; Kopan, R.; Shen, J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 2004, 269, 81–94. [Google Scholar] [CrossRef]
- Fortini, M.E.; Artavanis-Tsakonas, S. The suppressor of hairless protein participates in notch receptor signaling. Cell 1994, 79, 273–282. [Google Scholar] [CrossRef]
- Lubman, O.Y.; Ilagan, M.X.G.; Kopan, R.; Barrick, D. Quantitative dissection of the Notch: CSL interaction: Insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 2007, 365, 577–589. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Wolf, D.; Smylla, T.K.; Reichmuth, J.; Hoffmeister, P.; Kober, L.; Zimmermann, M.; Turkiewicz, A.; Borggrefe, T.; Nagel, A.C.; Oswald, F.; et al. Nucleo-cytoplasmic shuttling of Drosophila Hairless/Su(H) heterodimer as a means of regulating Notch dependent transcription. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1520–1532. [Google Scholar] [CrossRef]
- Wolf, D.B.; Maier, D.; Nagel, A.C. Nucleo-cytoplasmic shuttling of murine RBPJ by Hairless protein matches that of Su(H) protein in the model system Drosophila melanogaster. Hereditas 2021, 158, 11. [Google Scholar] [CrossRef] [PubMed]
- Baron, M. Endocytic routes to Notch activation. Semin. Cell Dev. Biol. 2012, 23, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Cornell, M.; Evans, D.A.; Mann, R.; Fostier, M.; Flasza, M.; Monthatong, M.; Artavanis-Tsakonas, S.; Baron, M. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 1999, 152, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K.; Diederich, R.J.; Go, M.J.; Blaumueller, C.M.; Artavanis-Tsakonas, S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 1995, 121, 2633–2644. [Google Scholar] [CrossRef]
- Wilkin, M.B.; Carbery, A.M.; Fostier, M.; Aslam, H.; Mazaleyrat, S.L.; Higgs, J.; Myat, A.; Evans, D.A.; Cornell, M.; Baron, M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 2004, 14, 2237–2244. [Google Scholar] [CrossRef]
- Wilkin, M.; Tongngok, P.; Gensch, N.; Clemence, S.; Motoki, M.; Yamada, K.; Hori, K.; Taniguchi-Kanai, M.; Franklin, E.; Matsuno, K.; et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell 2008, 15, 762–772. [Google Scholar] [CrossRef]
- Shimizu, H.; Woodcock, S.A.; Wilkin, M.B.; Trubenová, B.; Monk, N.A.; Baron, M. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 2014, 157, 1160–1174. [Google Scholar] [CrossRef]
- Sakata, T.; Sakaguchi, H.; Tsuda, L.; Higashitani, A.; Aigaki, T.; Matsuno, K.; Hayashi, S. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr. Biol. 2004, 14, 2228–2236. [Google Scholar] [CrossRef]
- Xu, T.; Artavanis-Tsakonas, S. deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics 1990, 126, 665–677. [Google Scholar] [CrossRef]
- Diederich, R.J.; Matsuno, K.; Hing, H.; Artavanis-Tsakonas, S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development 1994, 120, 473–481. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Leahy, D.J.; Barrick, D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure 2005, 13, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.J.; Girton, J.R. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics 1992, 131, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Wilkin, M.B.; Woodcock, S.A.; Bonfini, A.; Hung, Y.; Mazaleyrat, S.; Baron, M. The Drosophila ZO-1 protein Polychaetoid suppresses Deltex-regulated Notch activity to modulate germline stem cell niche formation. Open Biol. 2017, 7, 160322. [Google Scholar] [CrossRef] [PubMed]
- Fuwa, T.J.; Hori, K.; Sasamura, T.; Higgs, J.; Baron, M.; Matsuno, K. The first deltex null mutant indicates tissue-specific deltex-dependent Notch signaling in Drosophila. Mol. Genet. Genom. 2006, 275, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Fostier, M.; Ito, M.; Fuwa, T.J.; Go, M.J.; Okano, H.; Baron, M.; Matsuno, K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 2004, 131, 5527–5537. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Veraksa, A.; Bauer, A.; Rosse, C.; Camonis, J.; Artavanis-Tsakonas, S. Regulation of Notch signalling by non-visual β-arrestin. Nat. Cell Biol. 2005, 7, 1191–1201. [Google Scholar] [CrossRef]
- Mazaleyrat, S.L.; Fostier, M.; Wilkin, M.B.; Aslam, H.; Evans, D.A.; Cornell, M.; Baron, M. Down-regulation of Notch target gene expression by Suppressor of deltex. Dev. Biol. 2003, 255, 363–372. [Google Scholar] [CrossRef]
- Busseau, I.; Diederich, R.J.; Xu, T.; Artavanis-Tsakonas, S. A member of the Notch group of interacting loci, deltex encodes a cytoplasmic basic protein. Genetics 1994, 136, 585–596. [Google Scholar] [CrossRef]
- Aravind, L. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001, 26, 273–275. [Google Scholar] [CrossRef]
- Matsuno, K.; Ito, M.; Hori, K.; Miyashita, F.; Suzuki, S.; Kishi, N.; Artavanis-Tsakonas, S.; Okano, H. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development 2002, 129, 1049–1059. [Google Scholar] [CrossRef]
- Obiero, J.; Walker, J.R.; Dhe-Paganon, S. Fold of the conserved DTC domain in Deltex proteins. Proteins 2012, 80, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Takeyama, K.; Aguiar, R.C.; Gu, L.; He, C.; Freeman, G.J.; Kutok, J.L.; Aster, J.C.; Shipp, M.A. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 2003, 278, 21930–21937. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K.; Eastman, D.; Mitsiades, T.; Quinn, A.M.; Carcanciu, M.L.; Ordentlich, P.; Kadesch, T.; Artavanis-Tsakonas, S. Human deltex is a conserved regulator of Notch signalling. Nat. Genet. 1998, 19, 74–78. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Buetow, L.; Gabrielsen, M.; Lilla, S.; Chatrin, C.; Sibbet, G.J.; Zanivan, S.; Huang, D.T. DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination. Sci. Adv. 2020, 6, eabc0629. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Huang, Y.C.; Yeh, T.H.; Shih, H.Y.; Lin, C.Y.; Lin, S.J.; Chiu, C.C.; Huang, C.W.; Jiang, Y.J. Deltex1 is inhibited by the Notch-Hairy/E(Spl) signaling pathway and induces neuronal and glial differentiation. Neural Dev. 2015, 10, 28. [Google Scholar] [CrossRef]
- Zheng, L.; Conner, S.D. PI5P4Kγ functions in DTX1-mediated Notch signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E1983–E1990. [Google Scholar] [CrossRef]
- Hu, Q.D.; Ang, B.T.; Karsak, M.; Hu, W.P.; Cui, X.Y.; Duka, T.; Takeda, Y.; Chia, W.; Sankar, N.; Ng, Y.K.; et al. F3/Contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 2003, 115, 163–175. [Google Scholar] [CrossRef]
- Bizzoca, A.; Corsi, P.; Polizzi, A.; Pinto, M.F.; Xenaki, D.; Furley, A.J.; Gennarini, G. F3/Contactin acts as a modulator of neurogenesis during cerebral cortex development. Dev. Biol. 2012, 365, 133–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steinbuck, M.P.; Arakcheeva, K.; Winandy, S. Novel TCR-Mediated Mechanisms of Notch Activation and Signaling. J. Immunol. 2018, 200, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Ordentlich, P.; Lin, A.; Shen, C.P.; Blaumueller, C.; Matsuno, K.; Artavanis-Tsakonas, S.; Kadesch, T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 1998, 18, 2230–2239. [Google Scholar] [CrossRef] [PubMed]
- Kishi, N.; Tang, Z.; Maeda, Y.; Hirai, A.; Mo, R.; Ito, M.; Suzuki, S.; Nakao, K.; Kinoshita, T.; Kadesch, T.; et al. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci. 2001, 19, 21–35. [Google Scholar] [CrossRef]
- Yamamoto, N.; Yamamoto, S.I.; Inagaki, F.; Kawaichi, M.; Fukamizu, A.; Kishi, N.; Matsuno, K.; Nakamura, K.; Weinmaster, G.; Okano, H.; et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 2001, 276, 45031–45040. [Google Scholar] [CrossRef] [PubMed]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef]
- Wilson, A.; MacDonald, H.R.; Radtke, F. Notch 1–deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001, 194, 1003–1012. [Google Scholar] [CrossRef]
- Amsen, D.; Helbig, C.; Backer, R.A. Notch in T cell differentiation: All things considered. Trends Immunol. 2015, 36, 802–814. [Google Scholar] [CrossRef]
- Izon, D.J.; Aster, J.C.; He, Y.; Weng, A.; Karnell, F.G.; Patriub, V.; Xu, L.; Bakkour, S.; Rodriguez, C.; Allman, D.; et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 2002, 16, 231–243. [Google Scholar] [CrossRef]
- Hsiao, H.W.; Hsu, T.S.; Liu, W.H.; Hsieh, W.C.; Chou, T.F.; Wu, Y.J.; Jiang, S.T.; Lai, M.Z. Deltex1 antagonizes HIF-1α and sustains the stability of regulatory T cells in vivo. Nat. Commun. 2015, 6, 6353. [Google Scholar] [CrossRef]
- Liu, W.H.; Lai, M.Z. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell. Biol. 2005, 25, 1367–1378. [Google Scholar] [CrossRef]
- Hsu, T.S.; Mo, S.T.; Hsu, P.N.; Lai, M.Z. c-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis. 2018, 9, 135. [Google Scholar] [CrossRef]
- Acar, A.; Hidalgo-Sastre, A.; Leverentz, M.K.; Mills, C.G.; Woodcock, S.; Baron, M.; Collu, G.M.; Brennan, K. Inhibition of Wnt signalling by Notch via two distinct mechanisms. Sci. Rep. 2021, 11, 9096. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Yi, Z.; Yi, T.; Yi, Z.; Yi, T.; Wu, Z. cDNA cloning, characterization and expression analysis of DTX2, a human WWE and RING-finger gene, in human embryos: Full length research paper. DNA Seq. 2006, 17, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Marracino, L.; Fortini, F.; Bouhamida, E.; Camponogara, F.; Severi, P.; Mazzoni, E.; Patergnani, S.; D’Aniello, E.; Campana, R.; Pinton, P.; et al. Adding a “Notch” to Cardiovascular Disease Therapeutics: A MicroRNA-Based Approach. Front. Cell Dev. Biol. 2021, 9, 695114. [Google Scholar] [CrossRef]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Luo, D.; de Morree, A.; Boutet, S.; Quach, N.; Natu, V.; Rustagi, A.; Rando, T.A. Deltex2 represses MyoD expression and inhibits myogenic differentiation by acting as a negative regulator of Jmjd1c. Proc. Natl. Acad. Sci. USA 2017, 114, E3071–E3080. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.Y.; Hu, H.Y.; Huang, K.N.; Wei, R.Q.; Min, J.; Qi, C.; Tang, H.; Qin, X. Ubiquitination of NOTCH2 by DTX3 suppresses the proliferation and migration of human esophageal carcinoma. Cancer Sci. 2020, 111, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, Y.; Liu, C.; Guo, M.; Ma, Z.; He, J.; Wang, A.; Sun, X.; Liu, Z. Deltex3 inhibits Epithelial Mesenchymal Transition in Papillary Thyroid Carcinoma via promoting ubiquitination of XRCC5 to regulate the AKT signal pathway. J. Cancer 2021, 12, 860–873. [Google Scholar] [CrossRef]
- Choi, D.; Park, E.; Jung, E.; Seong, Y.J.; Yoo, J.; Lee, E.; Hong, M.; Lee, S.; Ishida, H.; Burford, J.; et al. Laminar flow downregulates Notch activity to promote lymphatic sprouting. J. Clin. Investig. 2017, 127, 1225–1240. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, D.; Roswit, W.T.; Jin, X.; Patel, A.C.; Patel, D.A.; Agapov, E.; Wang, Z.; Tidwell, R.M.; Atkinson, J.J.; et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 2015, 16, 1215–1227. [Google Scholar] [CrossRef]
- Yan, Q.; Dutt, S.; Xu, R.; Graves, K.; Juszczynski, P.; Manis, J.P.; Shipp, M.A. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell 2009, 36, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Jividen, K.; Spencer, A.; Dworak, N.; Ni, L.; Oostdyk, L.T.; Chatterjee, M.; Kuśmider, B.; Reon, B.; Parlak, M.; et al. Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516. [Google Scholar] [CrossRef]
- Chastagner, P.; Rubinstein, E.; Brou, C. Ligand-activated Notch undergoes DTX4-mediated ubiquitylation and bilateral endocytosis before ADAM10 processing. Sci. Signal. 2017, 10, eaag2989. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, N.L.; Avis, J.M. Mutational analysis of the Notch2 negative regulatory region identifies key structural elements for mechanical stability. FEBS Open Bio 2012, 5, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Gordon, W.R.; Zimmerman, B.; He, L.; Miles, L.J.; Huang, J.; Tiyanont, K.; McArthur, D.G.; Aster, J.C.; Perrimon, N.; Loparo, J.J.; et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell 2015, 33, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, Y.; Zhu, L.; Liu, D.; Songyang, Z.; Wang, H.Y.; Wang, R.F. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 2012, 13, 387–395. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, Z.; Pan, Y.; Wu, S.; Li, Z.; Zuo, C. E3 ubiquitin ligase DTX4 is required for adipogenic differentiation in 3T3-L1 preadipocytes cell line. Biochem. Biophys. Res. Commun. 2017, 492, 419–424. [Google Scholar] [CrossRef]
- Lehar, S.M.; Bevan, M.J. T cells develop normally in the absence of both Deltex1 and Deltex2. Mol. Cell. Biol. 2006, 26, 7358–7371. [Google Scholar] [CrossRef]
- Yun, T.J.; Bevan, M.J. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: Multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 2003, 170, 5834–5841. [Google Scholar] [CrossRef]
- Moran, S.T.; Cariappa, A.; Liu, H.; Muir, B.; Sgroi, D.; Boboila, C.; Pillai, S. Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J. Immunol. 2007, 179, 195–200. [Google Scholar] [CrossRef]
- Fujita, K.; Yasui, S.; Shinohara, T.; Ito, K. Interaction between NF-κB signaling and Notch signaling in gliogenesis of mouse mesencephalic neural crest cells. Mech. Dev. 2011, 128, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yang, Y.; Nolo, R.; Zweidler-McKay, P.A.; Hughes, D.P. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene 2010, 29, 2916–2926. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Perry, S.S.; Sun, X.H. Id1 attenuates Notch signaling and impairs T-cell commitment by elevating Deltex1 expression. Mol. Cell. Biol. 2009, 29, 4640–4652. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Ohtsuka, T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Pampeno, C.; Vukmanovic, S.; Meruelo, D. Characterization of the transcriptional expression of Notch-1 signaling pathway members, Deltex and HES-1, in developing mouse thymocytes. Dev. Comp. Immunol. 2002, 26, 575–588. [Google Scholar] [CrossRef]
- Juszczynski, P.; Kutok, J.L.; Li, C.; Mitra, J.; Aguiar, R.C.; Shipp, M.A. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol. Cell. Biol. 2006, 26, 5348–5359. [Google Scholar] [CrossRef]
- Russo, L.C.; Tomasin, R.; Matos, I.A.; Manucci, A.C.; Sowa, S.T.; Dale, K.; Caldecott, K.W.; Lehtiö, L.; Schechtman, D.; Meotti, F.C.; et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021, 297, 101041. [Google Scholar] [CrossRef]
- Che, K.F.; Shankar, E.M.; Muthu, S.; Zandi, S.; Sigvardsson, M.; Hinkula, J.; Messmer, D.; Larsson, M. p38 Mitogen-activated protein kinase/signal transducer and activator of transcription-3 pathway signaling regulates expression of inhibitory molecules in T cells activated by HIV-1-exposed dendritic cells. Mol. Med. 2012, 18, 1169–1182. [Google Scholar] [CrossRef]
- Wu, C.C.; Hsu, S.C.; Shih, H.M.; Lai, M.Z. Nuclear factor of activated T cells c is a target of p38 mitogen-activated protein kinase in T cells. Mol. Cell. Biol. 2003, 23, 6442–6454. [Google Scholar] [CrossRef]
- Hsiao, H.W.; Liu, W.H.; Wang, C.J.; Lo, Y.H.; Wu, Y.H.; Jiang, S.T.; Lai, M.Z. Deltex1 is a target of the transcription factor NFAT that promotes T cell anergy. Immunity 2009, 31, 72–83. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Mao, A.; Lu, J.; Liu, W.; Qie, J.; Pan, G. DCAF13 promotes triple-negative breast cancer metastasis by mediating DTX3 mRNA degradation. Cell Cycle 2020, 19, 3622–3631. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, T.; Casemayou, A.; Neau, E.; Breuil, B.; Caubet, C.; Calise, D.; Thornhill, B.A.; Bachvarova, M.; Belliere, J.; Chevalier, R.L.; et al. Systems biology combining human- and animal-data miRNA and mRNA data identifies new targets in ureteropelvic junction obstruction. BMC Syst. Biol. 2017, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Xue, Y.; Huo, R.; Yan, Z.; Xu, H.; Li, H.; Wang, J.; Zhang, Q.; Cao, Y.; Zhao, J.Z. N6-methyladenosine methyltransferase METTL3 affects the phenotype of cerebral arteriovenous malformation via modulating Notch signaling pathway. J. Biomed. Sci. 2020, 27, 62. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Welsh, E.A.; Oguz, U.; Fang, B.; Bai, Y.; Kinose, F.; Bronk, C.; Remsing Rix, L.L.; Beg, A.A.; Rix, U.; et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12414–12419. [Google Scholar] [CrossRef]
- Chien, Y.; Kim, S.; Bumeister, R.; Loo, Y.M.; Kwon, S.W.; Johnson, C.L.; Balakireva, M.G.; Romeo, Y.; Kopelovich, L.; Gale, M., Jr.; et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006, 127, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Chastagner, P.; Israël, A.; Brou, C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006, 7, 1147–1153. [Google Scholar] [CrossRef]
- Ingham, R.J.; Colwill, K.; Howard, C.; Dettwiler, S.; Lim, C.S.; Yu, J.; Hersi, K.; Raaijmakers, J.; Gish, G.; Mbamalu, G.; et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell Biol. 2005, 25, 7092–7106. [Google Scholar] [CrossRef][Green Version]
- Nalefski, E.A.; Falke, J.J. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996, 5, 2375–2390. [Google Scholar] [CrossRef]
- Staub, O.; Rotin, D. WW domains. Structure 1996, 15, 495–499. [Google Scholar] [CrossRef]
- Lorenz, S. Structural mechanisms of HECT-type ubiquitin ligases. Biol. Chem. 2018, 399, 127–145. [Google Scholar] [CrossRef]
- Mari, S.; Ruetalo, N.; Maspero, E.; Stoffregen, M.C.; Pasqualato, S.; Polo, S.; Wiesner, S. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 2014, 22, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Soond, S.M.; Chantry, A. Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene 2011, 30, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Perry, W.L.; Hustad, C.M.; Swing, D.A.; O’Sullivan, T.N.; Jenkins, N.A.; Copeland, N.G. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998, 18, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Elly, C.; Gao, B.; Fang, N.; Altman, Y.; Joazeiro, C.; Hunter, T.; Copeland, N.; Jenkins, N.; Liu, Y.C. Dysregulation of T lymphocyte function in itchy mice: A role for Itch in TH2 differentiation. Nat. Immunol. 2002, 3, 281–287. [Google Scholar] [CrossRef]
- Moser, E.K.; Oliver, P.M. Regulation of autoimmune disease by the E3 ubiquitin ligase Itch. Cell. Immunol. 2019, 340, 103916. [Google Scholar] [CrossRef]
- Marchese, A.; Raiborg, C.; Santini, F.; Keen, J.H.; Stenmark, H.; Benovic, J.L. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell 2003, 5, 709–722. [Google Scholar] [CrossRef]
- Rossi, M.; De Simone, M.; Pollice, A.; Santoro, R.; La Mantia, G.; Guerrini, L.; Calabrò, V. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle 2006, 5, 1816–1822. [Google Scholar] [CrossRef]
- Qiu, L.; Joazeiro, C.; Fang, N.; Wang, H.Y.; Elly, C.; Altman, Y.; Fang, D.; Hunter, T.; Liu, Y.C. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 2000, 275, 35734–35737. [Google Scholar] [CrossRef]
- Chastagner, P.; Israël, A.; Brou, C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 2008, 3, e2735. [Google Scholar] [CrossRef]
- McGill, M.A.; McGlade, C.J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 2003, 278, 23196–23203. [Google Scholar] [CrossRef]
- McGill, M.A.; Dho, S.E.; Weinmaster, G.; McGlade, C.J. Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 2009, 284, 26427–26438. [Google Scholar] [CrossRef] [PubMed]
- Sapir, T.; Levy, T.; Kozer, N.; Shin, I.; Zamor, V.; Haffner-Krausz, R.; McGlade, J.C.; Reiner, O. Notch Activation by Shootin1 Opposing Activities on 2 Ubiquitin Ligases. Cereb. Cortex 2018, 28, 3115–3128. [Google Scholar] [CrossRef] [PubMed]
- Beres, B.J.; George, R.; Lougher, E.J.; Barton, M.; Verrelli, B.C.; McGlade, C.J.; Rawls, J.A.; Wilson-Rawls, J. Numb regulates Notch1, but not Notch3, during myogenesis. Mech. Dev. 2011, 128, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Puca, L.; Chastagner, P.; Meas-Yedid, V.; Israël, A.; Brou, C. α-arrestin 1 (ARRDC1) and β-arrestins cooperate to mediate Notch degradation in mammals. J. Cell Sci. 2013, 126, 4457–4468. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liu, A.D.; Hou, G.Q.; Zhang, X.; Ren, K.; Chen, X.Z.; Li, S.S.C.; Wu, Y.S.; Cao, X. N-acetylcysteine decreases malignant characteristics of glioblastoma cells by inhibiting Notch2 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 2. [Google Scholar] [CrossRef]
- Koncarevic, A.; Jackman, R.W.; Kandarian, S.C. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J. 2007, 21, 427–437. [Google Scholar] [CrossRef][Green Version]
- Shukla, V.; Shukla, A.; Joshi, S.S.; Lu, R. Interferon regulatory factor 4 attenuates Notch signaling to suppress the development of chronic lymphocytic leukemia. Oncotarget 2016, 7, 41081–41094. [Google Scholar] [CrossRef][Green Version]
- Zhang, P.; He, Q.; Chen, D.; Liu, W.; Wang, L.; Zhang, C.; Ma, D.; Li, W.; Liu, B.; Liu, F.G. Protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res. 2015, 25, 1093–1107. [Google Scholar] [CrossRef]
- Persaud, A.; Alberts, P.; Amsen, E.M.; Xiong, X.; Wasmuth, J.; Saadon, Z.; Fladd, C.; Parkinson, J.; Rotin, D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 2009, 5, 333. [Google Scholar] [CrossRef]
- Guarnieri, A.L.; Towers, C.G.; Drasin, D.J.; Oliphant, M.U.J.; Andrysik, Z.; Hotz, T.J.; Vartuli, R.L.; Linklater, E.S.; Pandey, A.; Khanal, S.; et al. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene 2018, 37, 3879–3893. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Heidersbach, A.; Kathiriya, I.S.; Garay, B.I.; Ivey, K.N.; Srivastava, D.; Han, Z.; King, I.N. The E3 ubiquitin ligase Nedd4/Nedd4L is directly regulated by microRNA 1. Development 2017, 144, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, J.; Lin, Z.; Feng, R.; Wang, Z.W.; Chen, G. The emerging role of WWP1 in cancer development and progression. Cell Death Discov. 2021, 7, 163. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Huu, N.S.; Ryder, W.D.; Zeps, N.; Flasza, M.; Chiu, M.; Hanby, A.M.; Poulsom, R.; Clarke, R.B.; Baron, M. Tumour-promoting activity of altered WWP1 expression in breast cancer and its utility as a prognostic indicator. J. Pathol. 2008, 216, 93–102. [Google Scholar] [CrossRef]
- Flasza, M.; Nguyen Huu, N.S.; Mazaleyrat, S.; Clémence, S.; Villemant, C.; Clarke, R.; Baron, M. Regulation of the nuclear localization of the human Nedd4-related WWP1 protein by Notch. Mol. Membr. Biol. 2006, 23, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.G.; Stoeck, A.; Guan, B.; Wu, R.C.; Zhu, H.; Blackshaw, S.; Shih, I.M.; Wang, T.L. Notch3 interactome analysis identified WWP2 as a negative regulator of Notch3 signaling in ovarian cancer. PLoS Genet. 2014, 10, e1004751. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.X.; Liu, H.T.; Zhang, N.; Niu, W.X.; Lu, Z.N.; Bao, Y.Y.; Huang, R.; Yu, D.K.; Shao, R.G.; He, H.W. Costunolide represses hepatic fibrosis through WW domain-containing protein 2-mediated Notch3 degradation. Br. J. Pharmacol. 2020, 177, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Mund, T.; Graeb, M.; Mieszczanek, J.; Gammons, M.; Pelham, H.R.; Bienz, M. Disinhibition of the HECT E3 ubiquitin ligase WWP2 by polymerized Dishevelled. Open Biol. 2015, 5, 150185. [Google Scholar] [CrossRef]
- Mund, T.; Pelham, H.R. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 2009, 10, 501–507. [Google Scholar] [CrossRef]
- Dalton, H.E.; Denton, D.; Foot, N.J.; Ho, K.; Mills, K.; Brou, C.; Kumar, S. Drosophila Ndfip is a novel regulator of Notch signaling. Cell Death Differ. 2011, 18, 1150–1160. [Google Scholar] [CrossRef]
- Persaud, A.; Alberts, P.; Mari, S.; Tong, J.; Murchie, R.; Maspero, E.; Safi, F.; Moran, M.F.; Polo, S.; Rotin, D. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci. Signal. 2014, 7, ra95. [Google Scholar] [CrossRef]
- Su, J.; Zhou, X.; Yin, X.; Wang, L.; Zhao, Z.; Hou, Y.; Zheng, N.; Xia, J.; Wang, Z. The effects of curcumin on proliferation, apoptosis, invasion, and NEDD4 expression in pancreatic cancer. Biochem. Pharm. 2017, 140, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, J.J.; Zhou, S.J.; Chen, J.; Hu, Q.; Pu, J.X.; Lu, J.L. Diosgenin inhibits the expression of NEDD4 in prostate cancer cells. Am. J. Transl. Res. 2019, 11, 3461–3471. [Google Scholar]
- Watt, J.E.; Hughes, G.R.; Walpole, S.; Monaco, S.; Stephenson, G.R.; Bulman Page, P.C.; Hemmings, A.M.; Angulo, J.; Chantry, A. Discovery of Small Molecule WWP2 Ubiquitin Ligase Inhibitors. Chemistry 2018, 24, 17677–17680. [Google Scholar] [CrossRef] [PubMed]
- Mund, T.; Lewis, M.J.; Maslen, S.; Pelham, H.R. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2014, 111, 16736–16741. [Google Scholar] [CrossRef] [PubMed]
- Bjij, I.; Ramharack, P.; Khan, S.; Cherqaoui, D.; Soliman, M.E.S. Tracing Potential Covalent Inhibitors of an E3 Ubiquitin Ligase through Target-Focused Modelling. Molecules 2019, 24, 3125. [Google Scholar] [CrossRef] [PubMed]
- Kathman, S.G.; Span, I.; Smith, A.T.; Xu, Z.; Zhan, J.; Rosenzweig, A.C.; Statsyuk, A.V. A Small Molecule That Switches a Ubiquitin Ligase from a Processive to a Distributive Enzymatic Mechanism. J. Am. Chem. Soc. 2015, 137, 12442–12445. [Google Scholar] [CrossRef]
- Quirit, J.G.; Lavrenov, S.N.; Poindexter, K.; Xu, J.; Kyauk, C.; Durkin, K.A.; Aronchik, I.; Tomasiak, T.; Solomatin, Y.A.; Preobrazhenskaya, M.N.; et al. Indole-3-carbinol (I3C) analogues are potent small molecule inhibitors of NEDD4-1 ubiquitin ligase activity that disrupt proliferation of human melanoma cells. Biochem. Pharm. 2017, 127, 13–27. [Google Scholar] [CrossRef]
- Lee, Y.R.; Chen, M.; Lee, J.D.; Zhang, J.; Lin, S.Y.; Fu, T.M.; Chen, H.; Ishikawa, T.; Chiang, S.Y.; Katon, J.; et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 2019, 364, eaau0159. [Google Scholar] [CrossRef]
- Novelli, G.; Liu, J.; Biancolella, M.; Alonzi, T.; Novelli, A.; Patten, J.J.; Cocciadiferro, D.; Agolini, E.; Colona, V.L.; Rizzacasa, B.; et al. Inhibition of HECT E3 ligases as potential therapy for COVID-19. Cell Death Dis. 2021, 12, 310. [Google Scholar] [CrossRef]
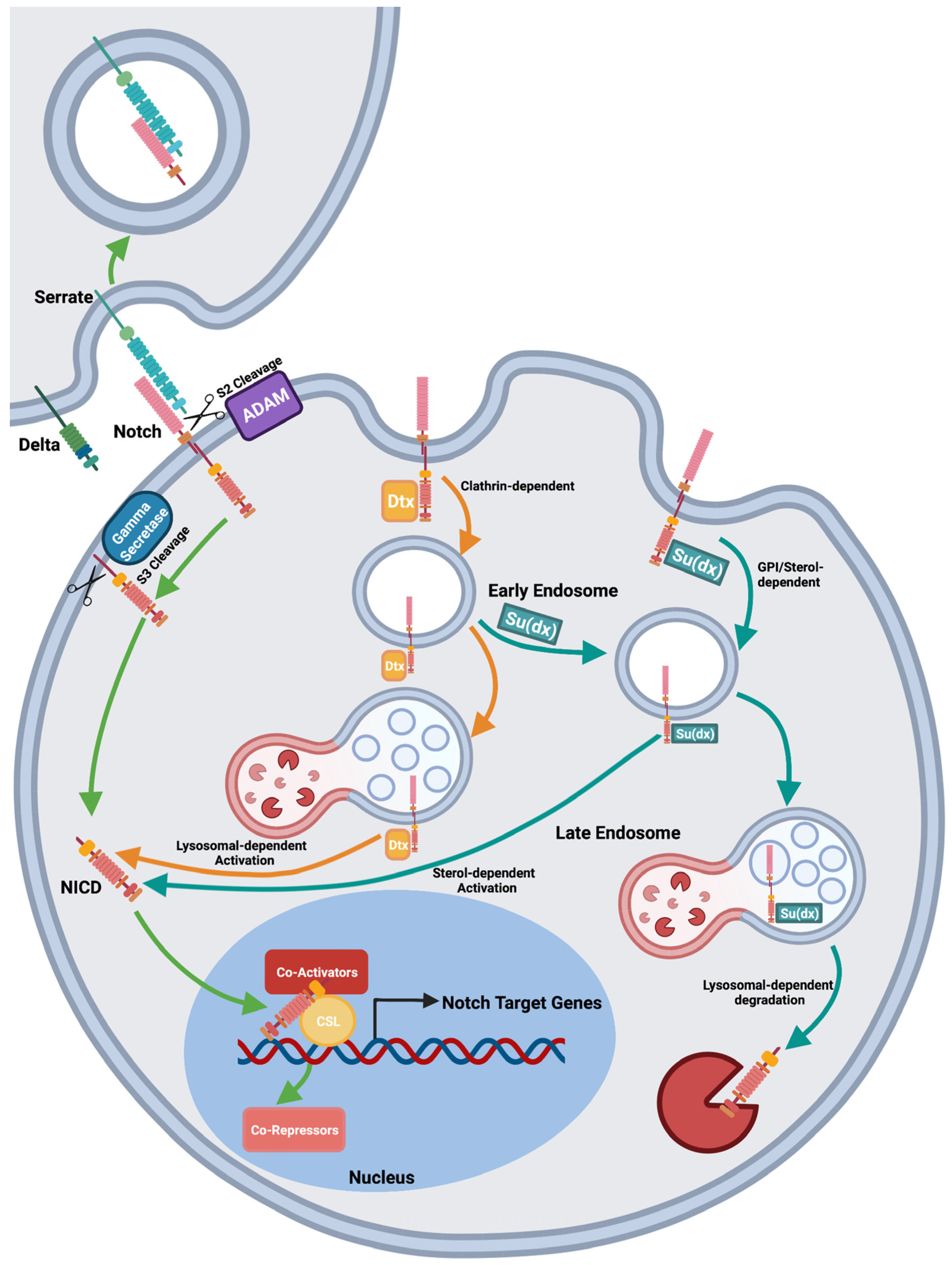

| Species | Homologuegue | Effect onEffect on Notch Signalling | Function | References |
|---|---|---|---|---|
| Drosophila melanogaster | Dx | Upregulates | Binds Notch ICD, promotes clathrin-dependent endocytosis of the receptor from the plasma membrane Stabilizes the receptor on endosome membrane to facilitate ligand-independent activation by ADAM 10-independent mechanism. | [16,17,20,25] |
| Downregulates | Promotes Notch removal from plasma membrane and downregulates ligand-dependent Notch activation. | [17] | ||
| Vertebrates | DTX1 | Upregulates | Enhances transcription of Notch target genes through interaction with NICD | [34] |
| Downregulates | Inhibits recycling of the receptor back to the plasma membrane to down regulate ligand-dependent signalling. | [38] | ||
| Inhibits Notch coactivator recruitment. | [48] | |||
| DTX2 | Not determined | Required for endocytosis-dependent downregulation of β-catenin by NOTCH1 | [52] | |
| DTX3 | Downregulates | Enhances NOTCH2 receptor degradation | [57] | |
| DTX3L | Downregulates | Heterodimerizes with DTX1 to downregulate NOTCH1. | [59] | |
| DTX4 | Upregulates | Triggers endocytosis of the NOTCH1 receptor and activates ECD shedding. | [63] | |
| Drosophila melanogaster | Su(dx) | Upregulates | Promotes Notch clathrin-independent endocytosis. When HECT domain is inactive, Su(dx) promotes ligand-independent signalling by ADAM10-dependent mechanism. | [17] |
| Downregulates | Promotes Notch clathrin-independent endocytosis, and, when HECT domain is active, Su(dx) promotes receptor-ubiquitination and degradation, to downregulate ligand-dependent and ligand-independent signalling. | [13,15,17] | ||
| Nedd4 | Downregulates | Promotes Notch endocytosis, Ubiquitinates and destabilizes Notch. | [15,18] | |
| Vertebrates | ITCH/AIP4 | Downregulates | Promotes endocytosis and directs the receptor towards lysosome-mediated degradation. | [101,105] |
| NEDD4 | Downregulates | NEDD4-mediated ubiquitination is necessary and sufficient for Notch1 down-regulation. | [106] | |
| NEDD4-L | Downregulates | NEDD4-L promotes NOTCH1 ubiquitination and degradation. | [109,110] | |
| WWP1 | Not determined | Colocalises with Notch1, which inhibits WWP1 localisation to nucleus. | [113,114] | |
| WWP2 | Downregulates | Promotes Notch3 degradation. | [115,116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Revici, R.; Hosseini-Alghaderi, S.; Haslam, F.; Whiteford, R.; Baron, M. E3 Ubiquitin Ligase Regulators of Notch Receptor Endocytosis: From Flies to Humans. Biomolecules 2022, 12, 224. https://doi.org/10.3390/biom12020224
Revici R, Hosseini-Alghaderi S, Haslam F, Whiteford R, Baron M. E3 Ubiquitin Ligase Regulators of Notch Receptor Endocytosis: From Flies to Humans. Biomolecules. 2022; 12(2):224. https://doi.org/10.3390/biom12020224
Chicago/Turabian StyleRevici, Raluca, Samira Hosseini-Alghaderi, Fabienne Haslam, Rory Whiteford, and Martin Baron. 2022. "E3 Ubiquitin Ligase Regulators of Notch Receptor Endocytosis: From Flies to Humans" Biomolecules 12, no. 2: 224. https://doi.org/10.3390/biom12020224
APA StyleRevici, R., Hosseini-Alghaderi, S., Haslam, F., Whiteford, R., & Baron, M. (2022). E3 Ubiquitin Ligase Regulators of Notch Receptor Endocytosis: From Flies to Humans. Biomolecules, 12(2), 224. https://doi.org/10.3390/biom12020224







