Observing Protein One-Dimensional Sliding: Methodology and Biological Significance
Abstract
1. Introduction
2. Modes of Protein 1D Sliding
2.1. Translocation
2.2. Facilitated Diffusion
2.3. Sliding as a Clamp
2.4. Hopping or Jumping
2.5. Intersegmental Transfer
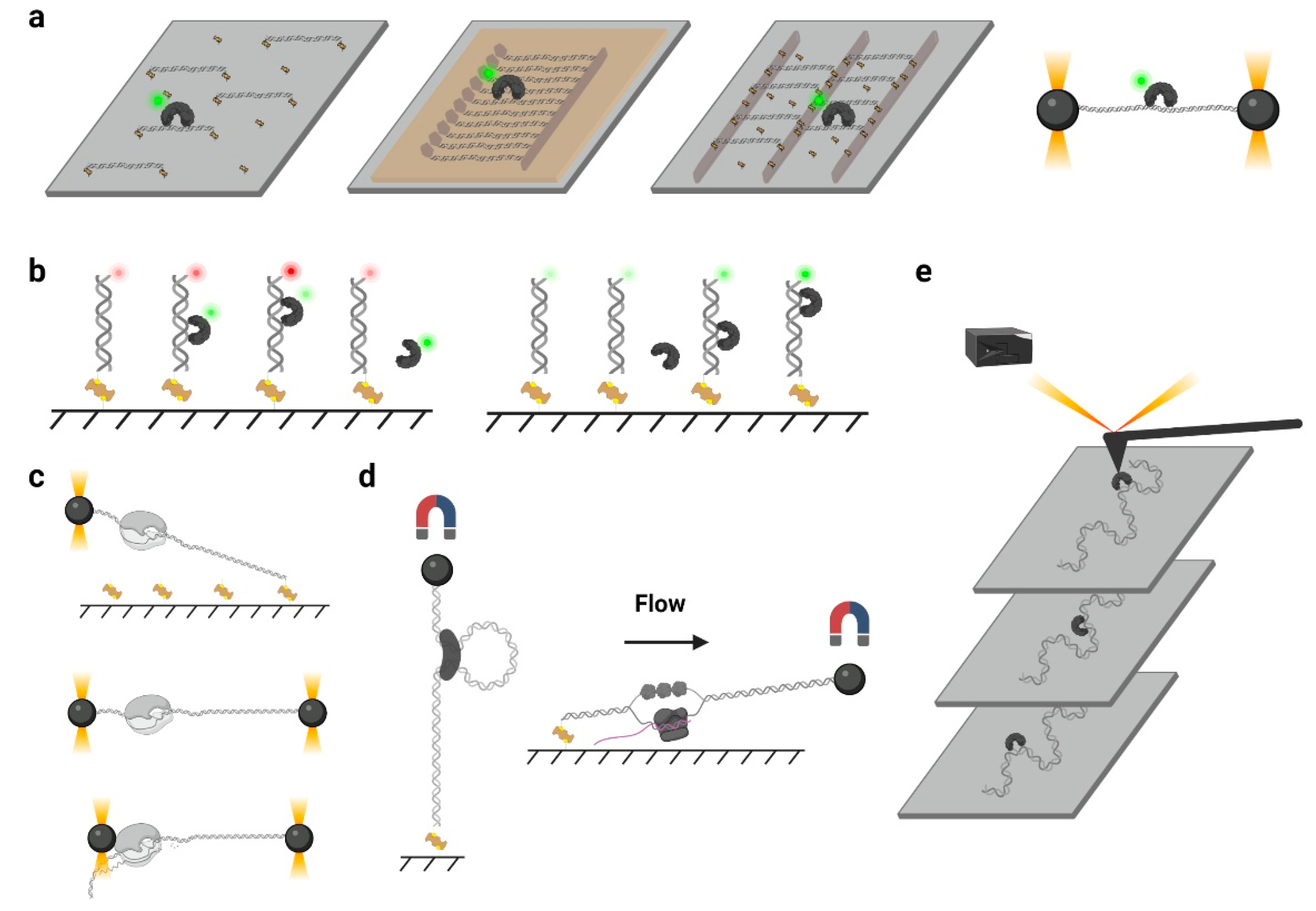
3. Approaches for Observing Protein 1D Sliding
3.1. Biochemical Assays
3.2. Single-Molecule Tracking
3.3. smFRET and PIFE
3.4. Single-Molecule Force Spectroscopy
4. Biological Significance of Protein 1D Sliding
4.1. Facilitation of Target Search
4.2. Processivity Regulation
4.3. DNA Duplex Interrogation
4.4. Distant Communications
4.5. Loop Extrusion
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustamante, C.; Marko, J.; Siggia, E.; Smith, S. Entropic elasticity of lambda-phage DNA. Science 1994, 265, 1599–1600. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.B.; Cui, Y.; Bustamante, C. Overstretching B-DNA: The Elastic Response of Individual Double-Stranded and Single-Stranded DNA Molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef]
- Berg, O.G.; Winter, R.B.; Von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 1981, 20, 6929–6948. [Google Scholar] [CrossRef] [PubMed]
- Khoury, A.M.; Lee, H.J.; Lillis, M.; Lu, P. Lac repressor-operator interaction: DNA length dependence. Biochim. Biophys. Acta Gene Struct. Expr. 1990, 1087, 55–60. [Google Scholar] [CrossRef]
- Park, C.S.; Wu, F.Y.; Wu, C.W. Molecular mechanism of promoter selection in gene transcription. II. Kinetic evidence for promoter search by a one-dimensional diffusion of RNA polymerase molecule along the DNA template. J. Biol. Chem. 1982, 257, 6950–6956. [Google Scholar] [CrossRef]
- Ricchetti, M.; Metzger, W.; Heumann, H. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: A mechanism to facilitate promoter location. Proc. Natl. Acad. Sci. USA 1988, 85, 4610–4614. [Google Scholar] [CrossRef]
- Kabata, H.; Kurosawa, O.; Arai, I.; Washizu, M.; Margarson, S.A.; Glass, R.E.; Shimamoto, N. Visualization of Single Molecules of RNA Polymerase Sliding Along DNA. Science 1993, 262, 1561–1563. [Google Scholar] [CrossRef]
- Eggleston, A.; Rahim, N.A.; Kowalczykowski, S.C. A Helicase Assay Based on the Displacement of Fluorescent, Nucleic Acid-Binding Ligands. Nucleic Acids Res. 1996, 24, 1179–1186. [Google Scholar] [CrossRef][Green Version]
- Bonnet, I.; Biebricher, A.; Porté, P.-L.; Loverdo, C.; Bénichou, O.; Voituriez, R.; Escudé, C.; Wende, W.; Pingoud, A.; Desbiolles, P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008, 36, 4118–4127. [Google Scholar] [CrossRef] [PubMed]
- Rau, D.C.; Sidorova, N.Y. Diffusion of the Restriction Nuclease EcoRI along DNA. J. Mol. Biol. 2010, 395, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Tafvizi, A.; Huang, F.; Leith, J.S.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. Tumor Suppressor p53 Slides on DNA with Low Friction and High Stability. Biophys. J. 2008, 95, L01–L03. [Google Scholar] [CrossRef] [PubMed]
- Guajardoa, R.; Sousa, R. A model for the mechanism of polymerase translocation. J. Mol. Biol. 1997, 265, 8–19. [Google Scholar] [CrossRef]
- Gelles, J.; Landick, R. RNA Polymerase as a Molecular Motor. Cell 1998, 93, 13–16. [Google Scholar] [CrossRef]
- Moore, K.J.; Lohman, T.M. Helicase-catalyzed DNA unwinding: Energy coupling by DNA motor proteins. Biophys. J. 1995, 68, 180S–185S. [Google Scholar]
- Lohman, T.M.; Bjornson, K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996, 65, 169–214. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, T.; Bisht, S.; Eeftens, J.M.; Dekker, C.; Haering, C.H.; Greene, E.C. The condensin complex is a mechanochemical motor that translocates along DNA. Science 2017, 358, 672–676. [Google Scholar] [CrossRef]
- Bath, J.; Wu, L.J.; Errington, J.; Wang, J.C. Role of Bacillus subtilis SpoIIIE in DNA Transport Across the Mother Cell-Prespore Division Septum. Science 2000, 290, 995–997. [Google Scholar] [CrossRef]
- Aussel, L.; Barre, F.-X.; Aroyo, M.; Stasiak, A.; Stasiak, A.Z.; Sherratt, D. FtsK Is a DNA Motor Protein that Activates Chromosome Dimer Resolution by Switching the Catalytic State of the XerC and XerD Recombinases. Cell 2002, 108, 195–205. [Google Scholar] [CrossRef]
- Riggs, A.; Bourgeois, S.; Cohn, M. The lac represser-operator interaction: III. Kinetic studies. J. Mol. Biol. 1970, 53, 401–417. [Google Scholar] [CrossRef]
- Wang, Y.M.; Austin, R.H.; Cox, E.C. Single Molecule Measurements of Repressor Protein 1D Diffusion on DNA. Phys. Rev. Lett. 2006, 97, 048302. [Google Scholar] [CrossRef]
- Hammar, P.; Leroy, P.; Mahmutovic, A.; Marklund, E.G.; Berg, O.G.; Elf, J. The lac Repressor Displays Facilitated Diffusion in Living Cells. Science 2012, 336, 1595–1598. [Google Scholar] [CrossRef]
- Kong, X.-P.; Onrust, R.; O’Donnell, M.; Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 1992, 69, 425–437. [Google Scholar] [CrossRef]
- Hingorani, M.M.; O’Donnell, M. ATP Binding to the Escherichia coli Clamp Loader Powers Opening of the Ring-shaped Clamp of DNA Polymerase III Holoenzyme. J. Biol. Chem. 1998, 273, 24550–24563. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hanne, J.; Britton, B.M.; Bennett, J.; Kim, D.; Lee, J.-B.; Fishel, R. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature. 2016, 539, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Gradia, S.; Acharya, S.; Fishel, R. The Human Mismatch Recognition Complex hMSH2-hMSH6 Functions as a Novel Molecular Switch. Cell 1997, 91, 995–1005. [Google Scholar] [CrossRef]
- Gradia, S.; Subramanian, D.; Wilson, T.; Acharya, S.; Makhov, A.; Griffith, J.; Fishel, R. hMSH2–hMSH6 Forms a Hydrolysis-Independent Sliding Clamp on Mismatched DNA. Mol. Cell 1999, 3, 255–261. [Google Scholar] [CrossRef]
- London, J.; Martín-López, J.; Yang, I.; Liu, J.; Lee, J.-B.; Fishel, R. Linker domain function predicts pathogenic MLH1 missense variants. Proc. Natl. Acad. Sci. USA 2021, 118, e2019215118. [Google Scholar] [CrossRef]
- Halford, S.E.; Marko, J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004, 32, 3040–3052. [Google Scholar] [CrossRef]
- Doucleff, M.; Clore, G.M. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl. Acad. Sci. USA 2008, 105, 13871–13876. [Google Scholar] [CrossRef]
- Suzuki, Y.; Gilmore, J.L.; Yoshimura, S.H.; Henderson, R.M.; Lyubchenko, Y.L.; Takeyasu, K. Visual Analysis of Concerted Cleavage by Type IIF Restriction Enzyme SfiI in Subsecond Time Region. Biophys. J. 2011, 101, 2992–2998. [Google Scholar] [CrossRef]
- Sugaya, Y.; Ihara, K.; Masuda, Y.; Ohtsubo, E.; Maki, H. Hyper-processive and slower DNA chain elongation catalysed by DNA polymerase III holoenzyme purified from the dnaE173 mutator mutant of Escherichia coli. Genes Cells 2002, 7, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Harmon, F.G.; Kowalczykowski, S.C. Biochemical Characterization of the DNA Helicase Activity of the Escherichia coli RecQ Helicase. J. Biol. Chem. 2001, 276, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Joo, C.; Ha, T. Preparing Sample Chambers for Single-Molecule FRET. Cold Spring Harb. Protoc. 2012, 2012, 1104–1108. [Google Scholar] [CrossRef]
- Collins, B.E.; Ye, L.F.; Duzdevich, D.; Greene, E.C. DNA curtains: Novel tools for imaging protein-nucleic acid interactions at the single-molecule level. Methods Cell Biol. 2014, 123, 217–234. [Google Scholar] [CrossRef]
- Kim, D.; Rashid, F.; Cho, Y.; Zaher, M.S.; Cho, I.I.H.; Hamdan, S.M.; Jeong, C.; Lee, J.-B. DNA skybridge: 3D structure producing a light sheet for high-throughput single-molecule imaging. Nucleic Acids Res. 2019, 47, e107. [Google Scholar] [CrossRef]
- Qin, Z.; Bi, L.; Hou, X.-M.; Zhang, S.; Zhang, X.; Lu, Y.; Li, M.; Modesti, M.; Xi, X.-G.; Sun, B. Human RPA activates BLM’s bidirectional DNA unwinding from a nick. eLife 2020, 9, e54098. [Google Scholar] [CrossRef]
- Liu, J.; Hanne, J.; Britton, B.M.; Shoffner, M.A.; Albers, A.E.; Bennett, J.; Zatezalo, R.; Barfield, R.M.; Rabuka, D.; Lee, J.-B.; et al. An Efficient Site-Specific Method for Irreversible Covalent Labeling of Proteins with a Fluorophore. Sci. Rep. 2015, 5, 16883. [Google Scholar] [CrossRef]
- Theile, C.S.; Witte, M.D.; Blom, A.E.M.; Kundrat, L.; Ploegh, H.L.; Guimaraes, C.P. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc. 2013, 8, 1800–1807. [Google Scholar] [CrossRef]
- Seefeldt, B.; Kasper, R.; Seidel, T.; Tinnefeld, P.; Dietz, K.-J.; Heilemann, M.; Sauer, M. Fluorescent proteins for single-molecule fluorescence applications. J. Biophotonics 2008, 1, 74–82. [Google Scholar] [CrossRef]
- Jeong, C.; Cho, W.-K.; Song, K.-M.; Cook, C.; Yoon, T.-Y.; Ban, C.; Fishel, R.; Lee, J.-B. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat. Struct. Mol. Biol. 2011, 18, 379–385. [Google Scholar] [CrossRef]
- Cho, W.-K.; Jeong, C.; Kim, D.; Chang, M.; Song, K.-M.; Hanne, J.; Ban, C.; Fishel, R.; Lee, J.-B. ATP Alters the Diffusion Mechanics of MutS on Mismatched DNA. Structure 2012, 20, 1264–1274. [Google Scholar] [CrossRef]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef]
- Roy, R.; Kozlov, A.G.; Lohman, T.M.; Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009, 461, 1092–1097. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Myong, S. Protein induced fluorescence enhancement (PIFE) for probing protein–nucleic acid interactions. Chem. Soc. Rev. 2014, 43, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Myong, S.; Cui, S.; Cornish, P.V.; Kirchhofer, A.; Gack, M.U.; Jung, J.U.; Hopfner, K.-P.; Ha, T. Cytosolic Viral Sensor RIG-I Is a 5′-Triphosphate-Dependent Translocase on Double-Stranded RNA. Science 2009, 323, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, J.R.; Chemla, Y.; Izhaky, D.; Bustamante, C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA 2006, 103, 9006–9011. [Google Scholar] [CrossRef]
- Lee, K.S.; Marciel, A.; Kozlov, A.G.; Schroeder, C.M.; Lohman, T.M.; Ha, T. Ultrafast Redistribution of E. coli SSB along Long Single-Stranded DNA via Intersegment Transfer. J. Mol. Biol. 2014, 426, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Landry, M.P.; Zou, X.; Wang, L.; Huang, W.M.; Schulten, K.; Chemla, Y.R. DNA target sequence identification mechanism for dimer-active protein complexes. Nucleic Acids Res. 2012, 41, 2416–2427. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, K.S.; Balci, H.; Jia, H.; Lohman, T.M.; Ha, T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat. Commun. 2013, 4, 1878. [Google Scholar] [CrossRef]
- Comstock, M.J.; Whitley, K.; Jia, H.; Sokoloski, J.; Lohman, T.M.; Ha, T.; Chemla, Y.R. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science 2015, 348, 352–354. [Google Scholar] [CrossRef]
- Shaevitz, J.W.; Abbondanzieri, E.A.; Landick, R.; Block, S.M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 2003, 426, 684–687. [Google Scholar] [CrossRef]
- Saleh, O.A.; Pérals, C.; Barre, F.-X.; Allemand, J.-F. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004, 23, 2430–2439. [Google Scholar] [CrossRef]
- Strick, T.; Croquette, V.; Bensimon, D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 2000, 404, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Ohara, O.; Takatsuki, A.; Itoh, H.; Shimamoto, N.; Kinosita, K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature 2001, 409, 113–115. [Google Scholar] [CrossRef]
- Dessinges, M.-N.; Lionnet, T.; Xi, X.G.; Bensimon, D.; Croquette, V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl. Acad. Sci. USA 2004, 101, 6439–6444. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-B.; Hite, R.K.; Hamdan, S.; Xie, X.S.; Richardson, C.C.; van Oijen, A. DNA primase acts as a molecular brake in DNA replication. Nature 2006, 439, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Kim, D.; Martín-López, J.V.; Lee, R.; Oh, J.; Hanne, J.; Fishel, R.; Lee, J.-B. Dynamic control of strand excision during human DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2016, 113, 3281–3286. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, R.; Britton, B.M.; London, J.A.; Yang, K.; Hanne, J.; Lee, J.-B.; Fishel, R. MutL sliding clamps coordinate exonuclease-independent Escherichia coli mismatch repair. Nat. Commun. 2019, 10, 5294. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Woodside, M.T.; Block, S.M. High-Resolution, Single-Molecule Measurements of Biomolecular Motion. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 171–190. [Google Scholar] [CrossRef]
- Shibata, M.; Nishimasu, H.; Kodera, N.; Hirano, S.; Ando, T.; Uchihashi, T.; Nureki, O. Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun. 2017, 8, 1430. [Google Scholar] [CrossRef]
- Tafvizi, A.; Huang, F.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Biebricher, A.; Wende, W.; Escudé, C.; Pingoud, A.; Desbiolles, P. Tracking of Single Quantum Dot Labeled EcoRV Sliding along DNA Manipulated by Double Optical Tweezers. Biophys. J. 2009, 96, L50–L52. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef]
- Kostiuk, G.; Dikić, J.; Schwarz, F.W.; Sasnauskas, G.; Seidel, R.; Siksnys, V. The dynamics of the monomeric restriction endonuclease BcnI during its interaction with DNA. Nucleic Acids Res. 2017, 45, 5968–5979. [Google Scholar] [CrossRef]
- Ahmadi, A.; Rosnes, I.; Blicher, P.; Diekmann, R.; Schüttpelz, M.; Glette, K.; Tørresen, J.; Bjørås, M.; Dalhus, B.; Rowe, A.D. Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V. Nat. Commun. 2018, 9, 5381. [Google Scholar] [CrossRef]
- Terakawa, T.; Kenzaki, H.; Takada, S. p53 Searches on DNA by Rotation-Uncoupled Sliding at C-Terminal Tails and Restricted Hopping of Core Domains. J. Am. Chem. Soc. 2012, 134, 14555–14562. [Google Scholar] [CrossRef] [PubMed]
- Marklund, E.; Van Oosten, B.; Mao, G.; Amselem, E.; Kipper, K.; Sabantsev, A.; Emmerich, A.; Globisch, D.; Zheng, X.; Lehmann, L.C.; et al. DNA surface exploration and operator bypassing during target search. Nature 2020, 583, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Subekti, D.R.G.; Murata, A.; Itoh, Y.; Takahashi, S.; Kamagata, K. Transient binding and jumping dynamics of p53 along DNA revealed by sub-millisecond resolved single-molecule fluorescence tracking. Sci. Rep. 2020, 10, 13697. [Google Scholar] [CrossRef]
- Cuculis, L.; Abil, Z.; Zhao, H.; Schroeder, C.M. TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 2016, 12, 831–837. [Google Scholar] [CrossRef]
- Jo, M.H.; Shin, S.; Jung, S.-R.; Kim, E.; Song, J.-J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 2015, 59, 117–124. [Google Scholar] [CrossRef]
- Song, G.; Chen, H.; Sheng, G.; Wang, Y.; Lou, J. Argonaute Facilitates the Lateral Diffusion of the Guide along Its Target and Prevents the Guide from Being Pushed Away by the Ribosome. Biochemistry 2018, 57, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.J.; Klein, M.; Hegge, J.W.; Chandradoss, S.D.; van der Oost, J.; Depken, M.; Joo, C. Argonaute bypasses cellular obstacles without hindrance during target search. Nat. Commun. 2019, 10, 4390. [Google Scholar] [CrossRef]
- Jeon, Y.; Choi, Y.H.; Jang, Y.; Yu, J.; Goo, J.; Lee, G.; Jeong, Y.K.; Lee, S.H.; Kim, I.-S.; Kim, J.-S.; et al. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018, 9, 2777. [Google Scholar] [CrossRef]
- Globyte, V.; Lee, S.H.; Bae, T.; Kim, J.; Joo, C. CRISPR /Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 2019, 38, e99466. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Treco, D.; Szostak, J.W. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 1991, 64, 1155–1161. [Google Scholar] [CrossRef]
- Radding, C.M. Helical RecA nucleoprotein filaments mediate homologous pairing and strand exchange. Biochim. Biophys. Acta Gene Struct. Expr. 1989, 1008, 131–145. [Google Scholar] [CrossRef]
- Forget, A.L.; Kowalczykowski, S.C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 2012, 482, 423–427. [Google Scholar] [CrossRef]
- Ragunathan, K.; Liu, C.; Ha, T. RecA filament sliding on DNA facilitates homology search. eLife 2012, 1, e00067. [Google Scholar] [CrossRef]
- Crickard, J.B.; Moevus, C.J.; Kwon, Y.; Sung, P.; Greene, E.C. Rad54 Drives ATP Hydrolysis-Dependent DNA Sequence Alignment during Homologous Recombination. Cell 2020, 181, 1380–1394. [Google Scholar] [CrossRef]
- Lionnet, T.; Spiering, M.M.; Benkovic, S.J.; Bensimon, D.; Croquette, V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 19790–19795. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Leonhardt, H.; Pichler, G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus 2011, 2, 392–402. [Google Scholar] [CrossRef]
- Turner, J.; Hingorani, M.M.; Kelman, Z.; O’Donnell, M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999, 18, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Ellison, V.; Stillman, B. Opening of the Clamp: An intimate view of an ATP-driven biological machine. Cell 2001, 106, 655–660. [Google Scholar] [CrossRef]
- Hedglin, M.; Kumar, R.; Benkovic, S.J. Replication Clamps and Clamp Loaders. Cold Spring Harb. Perspect. Biol. 2013, 5, a010165. [Google Scholar] [CrossRef]
- Sakato, M.; O’Donnell, M.; Hingorani, M.M. A Central Swivel Point in the RFC Clamp Loader Controls PCNA Opening and Loading on DNA. J. Mol. Biol. 2012, 416, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Kelch, B.A.; Makino, D.L.; O’Donnell, M.; Kuriyan, J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012, 10, 34. [Google Scholar] [CrossRef]
- Kochaniak, A.B.; Habuchi, S.; Loparo, J.J.; Chang, D.J.; Cimprich, K.A.; Walter, J.; van Oijen, A.M. Proliferating Cell Nuclear Antigen Uses Two Distinct Modes to Move along DNA. J. Biol. Chem. 2009, 284, 17700–17710. [Google Scholar] [CrossRef]
- De March, M.; Merino, N.; Barrera-Vilarmau, S.; Crehuet, R.; Onesti, S.; Blanco, F.J.; De Biasio, A. Structural basis of human PCNA sliding on DNA. Nat. Commun. 2017, 8, 13935. [Google Scholar] [CrossRef]
- Mizrahi, V.; Henrie, R.N.; Marlier, J.F.; Johnson, K.A.; Benkovic, S.J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry 1985, 24, 4010–4018. [Google Scholar] [CrossRef]
- Leu, F.P.; O’Donnell, M. Interplay of Clamp Loader Subunits in Opening the β Sliding Clamp of Escherichia coli DNA Polymerase III Holoenzyme. J. Biol. Chem. 2001, 276, 47185–47194. [Google Scholar] [CrossRef]
- Lammens, K.; Bemeleit, D.J.; Möckel, C.; Clausing, E.; Schele, A.; Hartung, S.; Schiller, C.B.; Lucas, M.; Angermüller, C.; Söding, J.; et al. The Mre11:Rad50 Structure Shows an ATP-Dependent Molecular Clamp in DNA Double-Strand Break Repair. Cell 2011, 145, 54–66. [Google Scholar] [CrossRef]
- Myler, L.; Gallardo, I.F.; Soniat, M.; Deshpande, R.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.R.; Dunn, A.R.; Kathe, S.D.; Warshaw, D.M.; Wallace, S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA 2014, 111, E2091–E2099. [Google Scholar] [CrossRef] [PubMed]
- Rowland, M.M.; Schonhoft, J.D.; McKibbin, P.L.; David, S.S.; Stivers, J.T. Microscopic mechanism of DNA damage searching by hOGG1. Nucleic Acids Res. 2014, 42, 9295–9303. [Google Scholar] [CrossRef]
- Peng, S.; Wang, X.; Zhang, L.; He, S.; Zhao, X.S.; Huang, X.; Chen, C. Target search and recognition mechanisms of glycosylase AlkD revealed by scanning FRET-FCS and Markov state models. Proc. Natl. Acad. Sci. USA 2020, 117, 21889–21895. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.; van Oijen, A.; Banerjee, A.; Verdine, G.L.; Xie, X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5752–5757. [Google Scholar] [CrossRef] [PubMed]
- Gorman, J.; Chowdhury, A.; Surtees, J.A.; Shimada, J.; Reichman, D.R.; Alani, E.; Greene, E.C. Dynamic Basis for One-Dimensional DNA Scanning by the Mismatch Repair Complex Msh2-Msh6. Mol. Cell 2007, 28, 359–370. [Google Scholar] [CrossRef]
- Springall, L.; Hughes, C.D.; Simons, M.; Azinas, S.; Van Houten, B.; Kad, N.M. Recruitment of UvrBC complexes to UV-induced damage in the absence of UvrA increases cell survival. Nucleic Acids Res. 2018, 46, 1256–1265. [Google Scholar] [CrossRef] [PubMed]
- Beckwitt, E.C.; Jang, S.; Detweiler, I.C.; Kuper, J.; Sauer, F.; Simon, N.; Bretzler, J.; Watkins, S.C.; Carell, T.; Kisker, C.; et al. Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search. Nat. Commun. 2020, 11, 1356. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.; Luo, G.; Kou, S.C.; Mangel, W.F.; Verdine, G.L.; Bagchi, B.; Xie, X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009, 16, 1224–1229. [Google Scholar] [CrossRef]
- Fishel, R. Mismatch Repair. J. Biol. Chem. 2015, 290, 26395–26403. [Google Scholar] [CrossRef]
- Modrich, P. Mechanisms in E. coli and Human Mismatch Repair (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 8490–8501. [Google Scholar] [CrossRef] [PubMed]
- Guarné, A. The Functions of MutL in Mismatch Repair: The power of multitasking. Progress in molecular biology and transla-tional science. Prog. Mol. Biol. Transl. Sci. 2012, 110, 41–70. [Google Scholar] [CrossRef]
- Friedhoff, P.; Li, P.; Gotthardt, J. Protein-protein interactions in DNA mismatch repair. DNA Repair 2016, 38, 50–57. [Google Scholar] [CrossRef]
- Groothuizen, F.S.; Winkler, I.; Cristóvão, M.; Fish, A.; Winterwerp, H.H.K.; Reumer, A.; Marx, A.D.; Hermans, N.; Nicholls, R.; Murshudov, G.N.; et al. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 2015, 4, e06744. [Google Scholar] [CrossRef] [PubMed]
- Graham, W.J.; Putnam, C.D.; Kolodner, R.D. The properties of Msh2–Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair. J. Biol. Chem. 2018, 293, 18055–18070. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Kong, M.; Gassman, N.; Freudenthal, B.D.; Prasad, R.; Zhen, S.; Watkins, S.; Wilson, S.; Van Houten, B. PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res. 2017, 45, 12834–12847. [Google Scholar] [CrossRef]
- Hassler, M.; Shaltiel, I.; Haering, C.H. Towards a Unified Model of SMC Complex Function. Curr. Biol. 2018, 28, R1266–R1281. [Google Scholar] [CrossRef] [PubMed]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging of DNA loop extrusion by condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Kong, M.; Cutts, E.E.; Pan, D.; Beuron, F.; Kaliyappan, T.; Xue, C.; Morris, E.P.; Musacchio, A.; Vannini, A.; Greene, E.C. Human Condensin I and II Drive Extensive ATP-Dependent Compaction of Nucleosome-Bound DNA. Mol. Cell 2020, 79, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human cohesin compacts DNA by loop extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef] [PubMed]
- Golfier, S.; Quail, T.; Kimura, H.; Brugués, J. Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. eLife 2020, 9, e53885. [Google Scholar] [CrossRef]
- Crocker, K.; London, J.; Medina, A.; Fishel, R.; Bundschuh, R. Evolutionary advantage of a dissociative search mechanism in DNA mismatch repair. Phys. Rev. E 2021, 103, 052404. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.-W.; Liu, J. Observing Protein One-Dimensional Sliding: Methodology and Biological Significance. Biomolecules 2021, 11, 1618. https://doi.org/10.3390/biom11111618
Yang X-W, Liu J. Observing Protein One-Dimensional Sliding: Methodology and Biological Significance. Biomolecules. 2021; 11(11):1618. https://doi.org/10.3390/biom11111618
Chicago/Turabian StyleYang, Xiao-Wen, and Jiaquan Liu. 2021. "Observing Protein One-Dimensional Sliding: Methodology and Biological Significance" Biomolecules 11, no. 11: 1618. https://doi.org/10.3390/biom11111618
APA StyleYang, X.-W., & Liu, J. (2021). Observing Protein One-Dimensional Sliding: Methodology and Biological Significance. Biomolecules, 11(11), 1618. https://doi.org/10.3390/biom11111618





