Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy
Abstract
1. Introduction
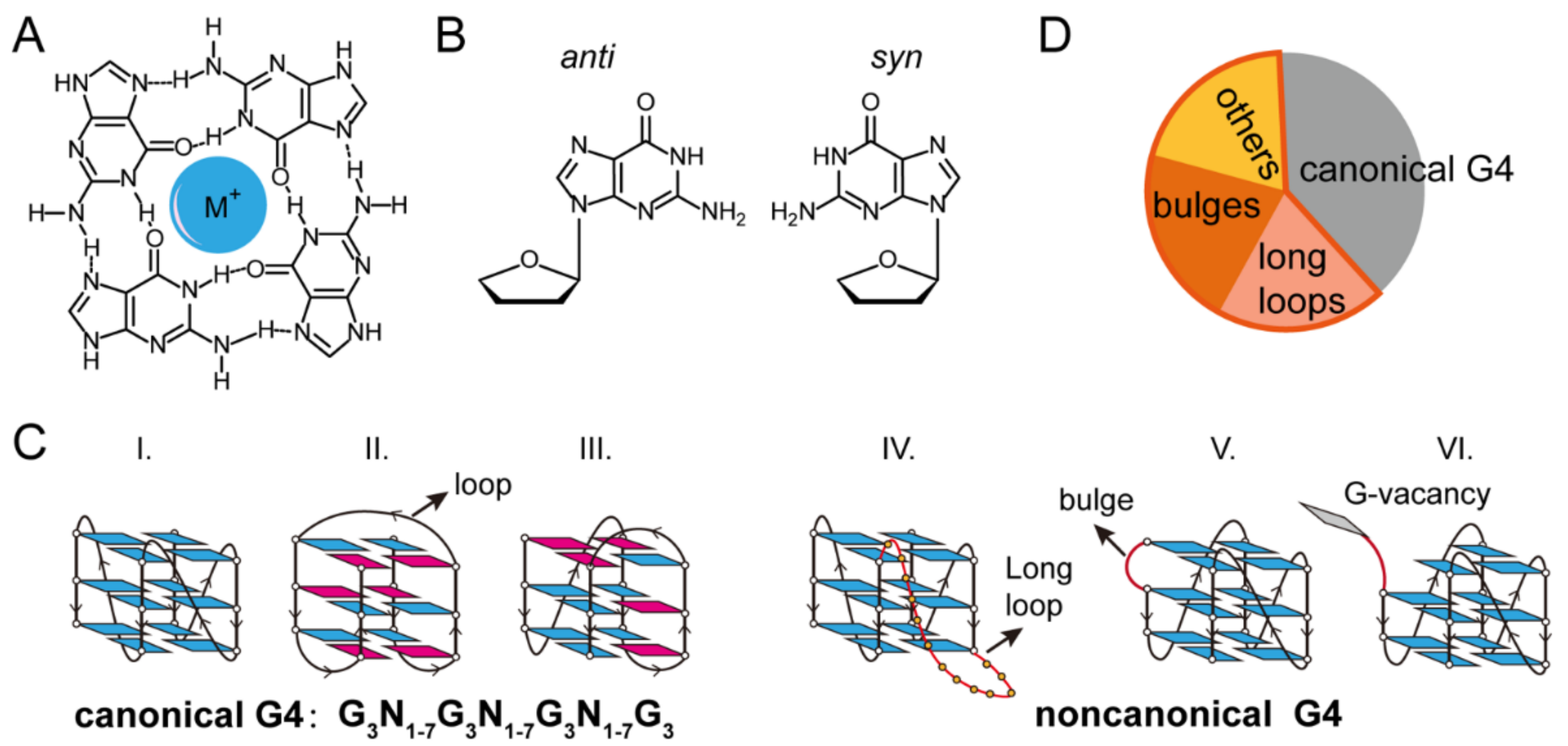
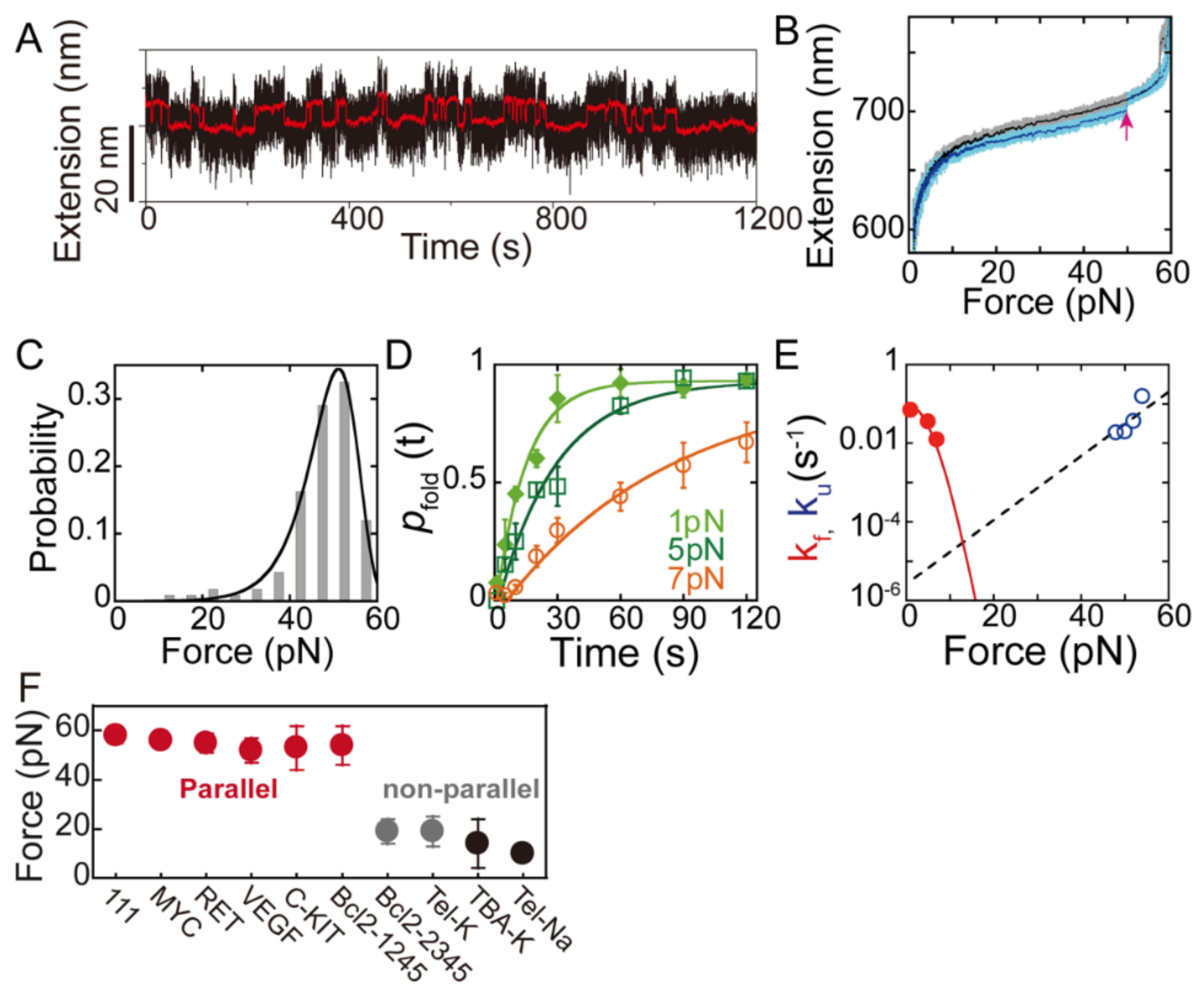
2. Single-Molecule Force Spectroscopy for the Investigation of G-Quadruplexes
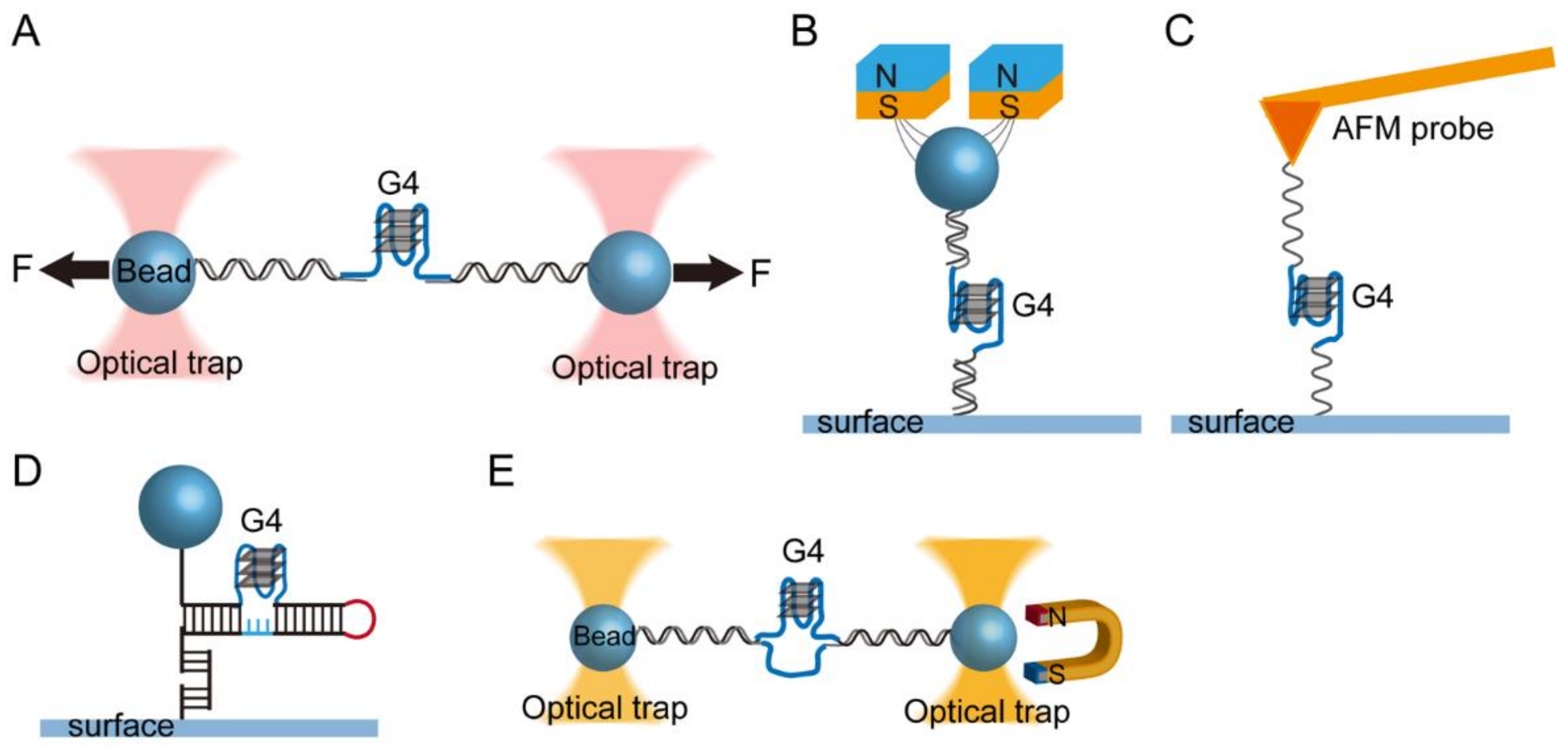
2.1. Single-Molecule Force Spectroscopy
2.2. Thermodynamics and Kinetics of G4s Obtained from Single-Molecule Force Spectroscopy
2.3. Folding/Unfolding Dynamics and Polymorphism of G4s
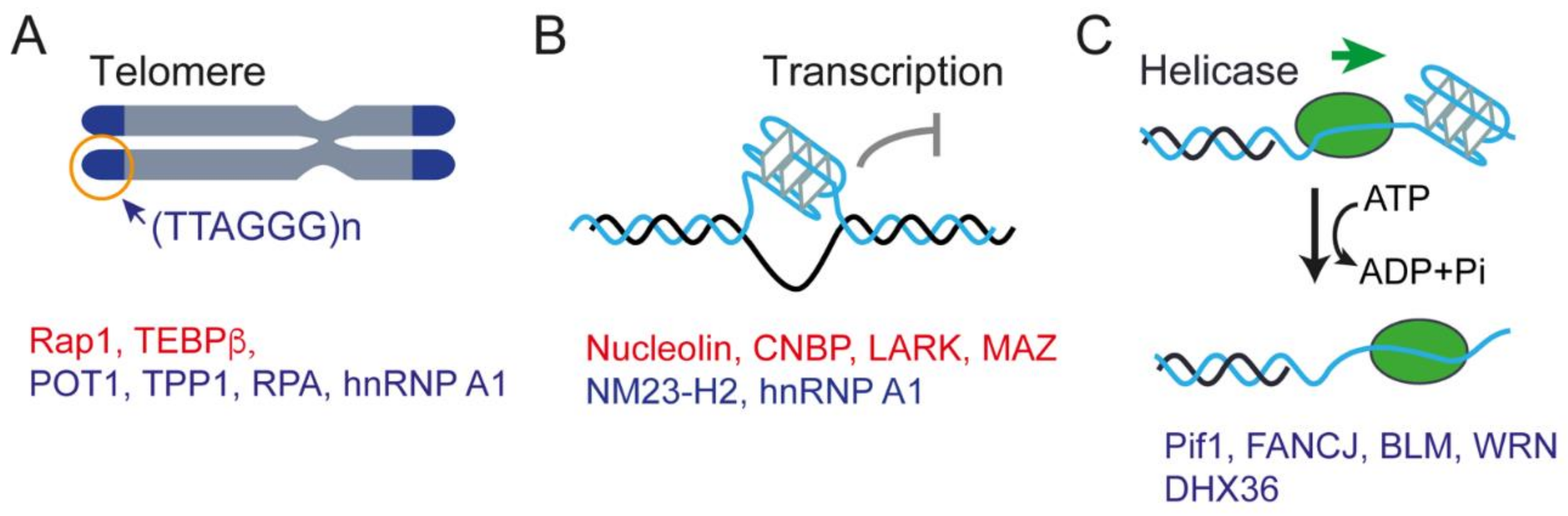
3. Single-Molecule Force Spectroscopy Reveals the Interactions between Proteins and G4s
| Function | Proteins 1 | Bind G4s | Promote G4s | Stabilize G4s | Unwind G4s | Structure 2 (PDB) | Reference |
|---|---|---|---|---|---|---|---|
| Telomere | Telomerase 3 | ✓ | ✓ | [73,130] | |||
| RAP1 | ✓ | ✓ | 6LDM | [131,132,133] | |||
| TEBP-β | ✓ | ✓ | 1JB7 | [128,134,135,136] | |||
| POT1-TPP1 | ✓ | ✓ | [69,137,138,139] | ||||
| RPA | ✓ | ✓ | [69,123,140] | ||||
| ATRX | ✓ | [141] | |||||
| hnRNP A1 | ✓ | ✓ | [142,143] | ||||
| hnRNP A2 | ✓ | ✓ | [144,145] | ||||
| Transcription | Nucleolin | ✓ | ✓ | ✓ | [146,147] | ||
| NM23-H2 | ✓ | ✓ | [148] | ||||
| CNBP | ✓ | ✓ | [149] | ||||
| LARK | ✓ | ✓ | [150] | ||||
| MAZ | ✓ | ✓ | [151] | ||||
| hnRNP A1 | ✓ | ✓ | [143] | ||||
| YY1 | ✓ | [152] | |||||
| DNA repair | PARP1 | ✓ | [153,154] | ||||
| Replication | RPA | ✓ | ✓ | [125] | |||
| Helicase | Pif1 | ✓ | ✓ | [155] | |||
| FANCJ | ✓ | ✓ | [72,125,156] | ||||
| BLM | ✓ | ✓ | [157] | ||||
| WRN | ✓ | ✓ | [158,159,160] | ||||
| DHX36 (RHAU) | ✓ | ✓ | ✓ | 5VHE | [63,161,162] | ||
| Viral protein | HIV-1 NCp | ✓ | ✓ | [163] | |||
| Nsp3 | ✓ | [126,127] | |||||
| Others | Topo I | ✓ | ✓ | [164] | |||
| BG4 antibody | ✓ | ✓ | ✓ | [64] | |||
| thrombin | ✓ | ✓ | 1HAO | [105,165,166] |
3.1. G4 Resolving/Unwinding Proteins
3.2. G4 Chaperone and G4 Stabilizer
3.3. The Effects of G4s on DNA and RNA Polymerases
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation byhh guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.B. Fundamentals of G-quadruplex biology. Annu. Rep. Med. Chem. 2020, 54, 3–44. [Google Scholar]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Di Antonio, M. DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Lam, E.Y.N.; Beraldi, D.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013, 4, 1796. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef]
- Zheng, K.W.; Zhang, J.Y.; He, Y.D.; Gong, J.Y.; Wen, C.J.; Chen, J.N.; Hao, Y.H.; Zhao, Y.; Tan, Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020, 48, 11706–11720. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Miglietta, G.; Russo, M.; Capranico, G. G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acids Res. 2020, 48, 11942–11957. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid-Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef]
- Teng, F.Y.; Jiang, Z.Z.; Guo, M.; Tan, X.Z.; Chen, F.; Xi, X.G.; Xu, Y. G-quadruplex DNA: A novel target for drug design. Cell Mol. Life Sci. 2021, 2021, 682. [Google Scholar]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef]
- Yang, D. G-Quadruplex DNA and RNA. Methods Mol. Biol. 2019, 2035, 1–24. [Google Scholar]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef]
- Lombardi, E.P.; Londono-Vallejo, A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020, 48, 1–15. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Mergny, J.L.; Sen, D. DNA Quadruple Helices in Nanotechnology. Chem. Rev. 2019, 119, 6290–6325. [Google Scholar] [CrossRef]
- Grun, J.T.; Blumler, A.; Burkhart, I.; Wirmer-Bartoschek, J.; Heckel, A.; Schwalbe, H. Unraveling the Kinetics of Spare-Tire DNA G-Quadruplex Folding. J. Am. Chem. Soc. 2021, 143, 6185–6193. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Karsisiotis, A.I.; Hessari, N.M.; Novellino, E.; Spada, G.P.; Randazzo, A.; da Silva, M.W. Topological Characterization of Nucleic Acid G-Quadruplexes by UV Absorption and Circular Dichroism. Angew. Chem. Int. Ed. Engl. 2011, 50, 10645–10648. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, E.; Okamoto, K.; Yang, D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: Effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 2010, 49, 9152–9160. [Google Scholar]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef]
- Mendoza, O.; Elezgaray, J.; Mergny, J.L. Kinetics of quadruplex to duplex conversion. Biochimie 2015, 118, 225–233. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef]
- Chaires, J.B. Human telomeric G-quadruplex: Thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010, 277, 1098–1106. [Google Scholar] [CrossRef]
- Jana, J.; Weisz, K. Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment. Chembiochem 2021, 22, 2848–2856. [Google Scholar] [CrossRef]
- Stadlbauer, P.; Kuhrova, P.; Vicherek, L.; Banas, P.; Otyepka, M.; Trantirek, L.; Sponer, J. Parallel G-triplexes and G-hairpins as potential transitory ensembles in the folding of parallel-stranded DNA G-Quadruplexes. Nucleic Acids Res. 2019, 47, 7276–7293. [Google Scholar] [CrossRef]
- Sponer, J.; Islam, B.; Stadlbauer, P.; Haider, S. Molecular dynamics simulations of G-quadruplexes: The basic principles and their application to folding and ligand binding. Annu. Rep. Med. Chem. 2020, 54, 197–241. [Google Scholar]
- Ortiz de Luzuriaga, I.; Lopez, X.; Gil, A. Learning to Model G-Quadruplexes: Current Methods and Perspectives. Annu. Rev. Biophys. 2021, 50, 209–243. [Google Scholar] [CrossRef]
- Yu, Z.; Mao, H. Non-B DNA structures show diverse conformations and complex transition kinetics comparable to RNA or proteins--a perspective from mechanical unfolding and refolding experiments. Chem. Rec. 2013, 13, 102–116. [Google Scholar] [CrossRef]
- Mandal, S.; Hoque, M.E.; Mao, H. Single-Molecule Investigations of G-Quadruplex. Methods Mol. Biol. 2019, 2035, 275–298. [Google Scholar]
- Maleki, P.; Budhathoki, J.B.; Roy, W.A.; Balci, H. A practical guide to studying G-quadruplex structures using single-molecule FRET. Mol. Genet. Genomics 2017, 292, 483–498. [Google Scholar] [CrossRef]
- Lee, C.Y.; McNerney, C.; Myong, S. G-Quadruplex and Protein Binding by Single-Molecule FRET Microscopy. Methods Mol. Biol. 2019, 2035, 309–322. [Google Scholar]
- Parks, J.W.; Stone, M.D. Single-Molecule Studies of Telomeres and Telomerase. Annu. Rev. Biophys. 2017, 46, 357–377. [Google Scholar] [CrossRef][Green Version]
- You, H.; Zhou, Y.; Yan, J. Using Magnetic Tweezers to Unravel the Mechanism of the G-quadruplex Binding and Unwinding Activities of DHX36 Helicase. Methods Mol. Biol. 2021, 2209, 175–191. [Google Scholar]
- Bustamante, C.; Bryant, Z.; Smith, S.B. Ten years of tension: Single-molecule DNA mechanics. Nature 2003, 421, 423–427. [Google Scholar] [CrossRef]
- Woodside, M.T.; Block, S.M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 2014, 43, 19–39. [Google Scholar] [CrossRef]
- Bustamante, C.; Alexander, L.; Maciuba, K.; Kaiser, C.M. Single-Molecule Studies of Protein Folding with Optical Tweezers. Annu. Rev. Biochem. 2020, 89, 443–470. [Google Scholar] [CrossRef]
- Almaqwashi, A.A.; Paramanathan, T.; Rouzina, I.; Williams, M.C. Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy. Nucleic Acids Res. 2016, 44, 3971–3988. [Google Scholar] [CrossRef]
- Yu, Z.; Schonhoft, J.D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 2009, 131, 1876–1882. [Google Scholar] [CrossRef]
- Schonhoft, J.D.; Bajracharya, R.; Dhakal, S.; Yu, Z.; Mao, H.; Basu, S. Direct experimental evidence for quadruplex-quadruplex interaction within the human ILPR. Nucleic Acids Res. 2009, 37, 3310–3320. [Google Scholar] [CrossRef] [PubMed]
- Koirala, D.; Dhakal, S.; Ashbridge, B.; Sannohe, Y.; Rodriguez, R.; Sugiyama, H.; Balasubramanian, S.; Mao, H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011, 3, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Gaerig, V.; Cui, Y.; Kang, H.; Gokhale, V.; Zhao, Y.; Hurley, L.H.; Mao, H. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J. Am. Chem. Soc. 2012, 134, 5157–5164. [Google Scholar] [CrossRef] [PubMed]
- Mitra, J.; Ha, T. Streamlining effects of extra telomeric repeat on telomeric DNA folding revealed by fluorescence-force spectroscopy. Nucleic Acids Res. 2019, 47, 11044–11056. [Google Scholar] [CrossRef]
- Lynch, S.; Baker, H.; Byker, S.G.; Zhou, D.; Sinniah, K. Single molecule force spectroscopy on G-quadruplex DNA. Chemistry 2009, 15, 8113–8116. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, W. Dynamic topology of double-stranded telomeric DNA studied by single-molecule manipulation in vitro. Nucleic Acids Res. 2020, 48, 6458–6470. [Google Scholar] [CrossRef]
- Funayama, R.; Nakahara, Y.; Kado, S.; Tanaka, M.; Kimura, K. A single-molecule force-spectroscopic study on stabilization of G-quadruplex DNA by a telomerase inhibitor. Analyst 2014, 139, 4037–4043. [Google Scholar] [CrossRef]
- Li, S.; Wang, X.; Li, Z.; Huang, Z.; Lin, S.; Hu, J.; Tu, Y. Research progress of single molecule force spectroscopy technology based on atomic force microscopy in polymer materials: Structure, design strategy and probe modification. Nano Select. 2021, 2, 909–931. [Google Scholar] [CrossRef]
- Li, W.; Hou, X.M.; Wang, P.Y.; Xi, X.G.; Li, M. Direct Measurement of Sequential Folding Pathway and Energy Landscape of Human Telomeric G-quadruplex Structures. J. Am. Chem. Soc. 2013, 135, 6423–6426. [Google Scholar] [CrossRef]
- Long, X.; Parks, J.W.; Bagshaw, C.R.; Stone, M.D. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013, 41, 2746–2755. [Google Scholar] [CrossRef]
- You, H.; Zeng, X.; Xu, Y.; Lim, C.J.; Efremov, A.K.; Phan, A.T.; Yan, J. Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 2014, 42, 8789–8795. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Cheng, Y.; Chen, J.; Zheng, K.; You, H. Mechanical diversity and folding intermediates of parallel-stranded G-quadruplexes with a bulge. Nucleic Acids Res. 2021, 49, 7179–7188. [Google Scholar] [CrossRef]
- You, H.; Lattmann, S.; Rhodes, D.; Yan, J. RHAU helicase stabilizes G4 in its nucleotide-free state and destabilizes G4 upon ATP hydrolysis. Nucleic Acids Res. 2017, 45, 206–214. [Google Scholar] [CrossRef]
- Yangyuoru, P.M.; Di Antonio, M.; Ghimire, C.; Biffi, G.; Balasubramanian, S.; Mao, H. Dual Binding of an Antibody and a Small Molecule Increases the Stability of TERRA G-Quadruplex. Angew. Chem. Int. Ed. Engl. 2015, 127, 924–927. [Google Scholar] [CrossRef]
- Patrick, E.M.; Slivka, J.D.; Payne, B.; Comstock, M.J.; Schmidt, J.C. Observation of processive telomerase catalysis using high-resolution optical tweezers. Nat. Chem. Biol. 2020, 16, 801–809. [Google Scholar] [CrossRef]
- Lerner, E.; Barth, A.; Hendrix, J.; Ambrose, B.; Birkedal, V.; Blanchard, S.C.; Borner, R.; Sung Chung, H.; Cordes, T.; Craggs, T.D.; et al. FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices. Elife 2021, 10, e60416. [Google Scholar] [CrossRef]
- Lee, J.Y.; Okumus, B.; Kim, D.S.; Ha, T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 18938–18943. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef]
- Ray, S.; Bandaria, J.N.; Qureshi, M.H.; Yildiz, A.; Balci, H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. USA 2014, 111, 2990–2995. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, J.; Bochman, M.L.; Zakian, V.A.; Ha, T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife 2014, 3, e02190. [Google Scholar] [CrossRef]
- Hou, X.M.; Wu, W.Q.; Duan, X.L.; Liu, N.N.; Li, H.H.; Fu, J.; Dou, S.X.; Li, M.; Xi, X.G. Molecular mechanism of G-quadruplex unwinding helicase: Sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015, 466, 189–199. [Google Scholar] [CrossRef]
- Wu, C.G.; Spies, M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016, 44, 8742–8753. [Google Scholar] [CrossRef]
- Paudel, B.P.; Moye, A.L.; Abou Assi, H.; El-Khoury, R.; Cohen, S.B.; Holien, J.K.; Birrento, M.L.; Samosorn, S.; Intharapichai, K.; Tomlinson, C.G.; et al. A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. Elife 2020, 9, e56428. [Google Scholar] [CrossRef]
- Chatterjee, S.; Zagelbaum, J.; Savitsky, P.; Sturzenegger, A.; Huttner, D.; Janscak, P.; Hickson, I.D.; Gileadi, O.; Rothenberg, E. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 2014, 5, 5556. [Google Scholar] [CrossRef]
- Budhathoki, J.B.; Stafford, E.J.; Yodh, J.G.; Balci, H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015, 43, 5961–5970. [Google Scholar] [CrossRef]
- Tippana, R.; Hwang, H.; Opresko, P.L.; Bohr, V.A.; Myong, S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. USA 2016, 113, 8448–8453. [Google Scholar] [CrossRef]
- Tippana, R.; Chen, M.C.; Demeshkina, N.A.; Ferre-D’Amare, A.R.; Myong, S. RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nat. Commun. 2019, 10, 1855. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevicius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.Y.; Shen, J.Z.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Mitra, J.; Makurath, M.A.; Ngo, T.T.M.; Troitskaia, A.; Chemla, Y.R.; Ha, T. Extreme mechanical diversity of human telomeric DNA revealed by fluorescence-force spectroscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 8350–8359. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Gosse, C.; Croquette, V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys. J. 2002, 82, 3314–3329. [Google Scholar] [CrossRef]
- Garavis, M.; Bocanegra, R.; Herrero-Galan, E.; Gonzalez, C.; Villasante, A.; Arias-Gonzalez, J.R. Mechanical unfolding of long human telomeric RNA (TERRA). Chem. Commun. 2013, 49, 6397–6399. [Google Scholar] [CrossRef] [PubMed]
- Yangyuoru, P.M.; Zhang, A.Y.; Shi, Z.; Koirala, D.; Balasubramanian, S.; Mao, H. Mechanochemical properties of individual human telomeric RNA (TERRA) G-quadruplexes. Chembiochem 2013, 14, 1931–1935. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Koirala, D.; Cui, Y.; Easterling, L.F.; Zhao, Y.; Mao, H. Click chemistry assisted single-molecule fingerprinting reveals a 3D biomolecular folding funnel. J. Am. Chem. Soc. 2012, 134, 12338–12341. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, C.; Park, S.; Iida, K.; Yangyuoru, P.; Otomo, H.; Yu, Z.; Nagasawa, K.; Sugiyama, H.; Mao, H. Direct quantification of loop interaction and pi-pi stacking for G-quadruplex stability at the submolecular level. J. Am. Chem. Soc. 2014, 136, 15537–15544. [Google Scholar] [CrossRef] [PubMed]
- Selvam, S.; Yu, Z.; Mao, H. Exploded view of higher order G-quadruplex structures through click-chemistry assisted single-molecule mechanical unfolding. Nucleic Acids Res. 2016, 44, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q.; Wu, J.; Liang, J.H.; Yan, J.W.; Zhu, Z.; Yang, C.J.; Mao, B.W. Single-molecule force spectroscopic studies on intra- and intermolecular interactions of G-quadruplex aptamer with target Shp2 protein. J. Phys. Chem. B 2012, 116, 11397–11404. [Google Scholar] [CrossRef]
- Selvam, S.; Koirala, D.; Yu, Z.; Mao, H. Quantification of topological coupling between DNA superhelicity and G-quadruplex formation. J. Am. Chem. Soc. 2014, 136, 13967–13970. [Google Scholar] [CrossRef]
- Buglione, E.; Salerno, D.; Marrano, C.A.; Cassina, V.; Vesco, G.; Nardo, L.; Dacasto, M.; Rigo, R.; Sissi, C.; Mantegazza, F. Nanomechanics of G-quadruplexes within the promoter of the KIT oncogene. Nucleic Acids Res. 2021, 49, 4564–4573. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Rieu, M.; Hodeib, S.; Joubert, A.; Ouellet, J.; Alberti, P.; Bugaut, A.; Allemand, J.F.; Boule, J.B.; Croquette, V. Folding and persistence times of intramolecular G-quadruplexes transiently embedded in a DNA duplex. Nucleic Acids Res. 2021, 49, 5189–5201. [Google Scholar] [CrossRef]
- Shrestha, P.; Jonchhe, S.; Emura, T.; Hidaka, K.; Endo, M.; Sugiyama, H.; Mao, H. Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 2017, 12, 582–588. [Google Scholar] [CrossRef]
- Jonchhe, S.; Pandey, S.; Emura, T.; Hidaka, K.; Hossain, M.A.; Shrestha, P.; Sugiyama, H.; Endo, M.; Mao, H. Decreased water activity in nanoconfinement contributes to the folding of G-quadruplex and i-motif structures. Proc. Natl. Acad. Sci. USA 2018, 115, 9539–9544. [Google Scholar] [CrossRef]
- Jonchhe, S.; Pandey, S.; Karna, D.; Pokhrel, P.; Cui, Y.; Mishra, S.; Sugiyama, H.; Endo, M.; Mao, H. Duplex DNA Is Weakened in Nanoconfinement. J. Am. Chem. Soc. 2020, 142, 10042–10049. [Google Scholar] [CrossRef]
- You, H.; Wu, J.; Shao, F.; Yan, J. Stability and kinetics of c-MYC promoter G-quadruplexes studied by single-molecule manipulation. J. Am. Chem. Soc. 2015, 137, 2424–2427. [Google Scholar] [CrossRef]
- Marko, J.F.; Siggia, E.D. Stretching DNA. Macromolecules 1995, 28, 8759–8770. [Google Scholar] [CrossRef]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. [Google Scholar] [CrossRef]
- De Messieres, M.; Chang, J.C.; Brawn-Cinani, B.; La Porta, A. Single-Molecule Study of G-Quadruplex Disruption Using Dynamic Force Spectroscopy. Phys. Rev. Lett. 2012, 109, 058101. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, Y.; Gong, Z.; Zhang, X.; Li, Y.; Shi, X.; Pei, Y.; You, H. High Mechanical Stability and Slow Unfolding Rates Are Prevalent in Parallel-Stranded DNA G-Quadruplexes. J. Phys. Chem. Lett. 2020, 11, 7966–7971. [Google Scholar] [CrossRef]
- Jarzynski, C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997, 78, 2690–2693. [Google Scholar] [CrossRef]
- You, H.; Guo, S.; Le, S.; Tang, Q.; Yao, M.; Zhao, X.; Yan, J. Two-State Folding Energy Determination Based on Transition Points in Nonequilibrium Single-Molecule Experiments. J. Phys. Chem. Lett. 2018, 9, 811–816. [Google Scholar] [CrossRef]
- Koirala, D.; Mashimo, T.; Sannohe, Y.; Yu, Z.; Mao, H.; Sugiyama, H. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. 2012, 48, 2006–2008. [Google Scholar] [CrossRef]
- Jiang, H.X.; Cui, Y.; Zhao, T.; Fu, H.W.; Koirala, D.; Punnoose, J.A.; Kong, D.M.; Mao, H. Divalent cations and molecular crowding buffers stabilize G-triplex at physiologically relevant temperatures. Sci. Rep. 2015, 5, 9255. [Google Scholar] [CrossRef]
- Koirala, D.; Ghimire, C.; Bohrer, C.; Sannohe, Y.; Sugiyama, H.; Mao, H. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013, 135, 2235–2241. [Google Scholar] [CrossRef]
- Cheng, Y.; Tang, Q.; Li, Y.; Zhang, Y.; Zhao, C.; Yan, J.; You, H. Folding/unfolding kinetics of G-quadruplexes upstream of the P1 promoter of the human BCL-2 oncogene. J. Biol. Chem. 2019, 294, 5890–5895. [Google Scholar] [CrossRef]
- Liu, J.; Feng, W.; Zhang, W. A single-molecule study reveals novel rod-like structures formed by a thrombin aptamer repeat sequence. Nanoscale 2020, 12, 4159–4166. [Google Scholar] [CrossRef]
- Gutierrez, I.; Garavis, M.; de Lorenzo, S.; Villasante, A.; Gonzalez, C.; Arias-Gonzalez, J.R. Single-Stranded Condensation Stochastically Blocks G-Quadruplex Assembly in Human Telomeric RNA. J. Phys. Chem. Lett. 2018, 9, 2498–2503. [Google Scholar] [CrossRef]
- Ye, S.; Chen, Z.; Zhang, X.; Li, F.; Guo, L.; Hou, X.M.; Wu, W.Q.; Wang, J.; Liu, C.; Zheng, K.; et al. Proximal Single-Stranded RNA Destabilizes Human Telomerase RNA G-Quadruplex and Induces Its Distinct Conformers. J. Phys. Chem. Lett. 2021, 12, 3361–3366. [Google Scholar] [CrossRef]
- Abraham Punnoose, J.; Cui, Y.; Koirala, D.; Yangyuoru, P.M.; Ghimire, C.; Shrestha, P.; Mao, H. Interaction of G-quadruplexes in the full-length 3′ human telomeric overhang. J. Am. Chem. Soc. 2014, 136, 18062–18069. [Google Scholar] [CrossRef]
- Abraham Punnoose, J.; Ma, Y.; Li, Y.; Sakuma, M.; Mandal, S.; Nagasawa, K.; Mao, H. Adaptive and Specific Recognition of Telomeric G-Quadruplexes via Polyvalency Induced Unstacking of Binding Units. J. Am. Chem. Soc. 2017, 139, 7476–7484. [Google Scholar] [CrossRef] [PubMed]
- Jonchhe, S.; Ghimire, C.; Cui, Y.; Sasaki, S.; McCool, M.; Park, S.; Iida, K.; Nagasawa, K.; Sugiyama, H.; Mao, H. Binding of a Telomestatin Derivative Changes the Mechanical Anisotropy of a Human Telomeric G-Quadruplex. Angew. Chem. Int. Ed. Engl. 2019, 58, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Kawamoto, Y.; Yue, Z.; Hashiya, K.; Cui, Y.; Bando, T.; Pandey, S.; Hoque, M.E.; Hossain, M.A.; Sugiyama, H.; et al. Submolecular dissection reveals strong and specific binding of polyamide-pyridostatin conjugates to human telomere interface. Nucleic Acids Res. 2019, 47, 3295–3305. [Google Scholar] [CrossRef]
- Pandey, S.; Li, Y.; Young, M.D.; Mandal, S.; Lu, L.; Shelley, J.T.; Mao, H. Cooperative Heteroligand Interaction with G-Quadruplexes Shows Evidence of Allosteric Binding. Biochemistry 2020, 59, 3438–3446. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Cui, Y.; Koirala, D.; Ghimire, C.; Kushwaha, S.; Yu, Z.; Yangyuoru, P.M.; Mao, H. Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 2013, 41, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Xiao, S.; Dhakal, S.; Tan, Z.; Mao, H. Nascent RNA transcripts facilitate the formation of G-quadruplexes. Nucleic Acids Res. 2014, 42, 7236–7246. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Schonhoft, J.D.; Koirala, D.; Yu, Z.; Basu, S.; Mao, H. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 2010, 132, 8991–8997. [Google Scholar] [CrossRef]
- Cui, Y.; Koirala, D.; Kang, H.; Dhakal, S.; Yangyuoru, P.; Hurley, L.H.; Mao, H. Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL. Nucleic Acids Res. 2014, 42, 5755–5764. [Google Scholar] [CrossRef]
- Sutherland, C.; Cui, Y.; Mao, H.; Hurley, L.H. A Mechanosensor Mechanism Controls the G-Quadruplex/i-Motif Molecular Switch in the MYC Promoter NHE III1. J. Am. Chem. Soc. 2016, 138, 14138–14151. [Google Scholar] [CrossRef]
- Brazda, V.; Haronikova, L.; Liao, J.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef]
- Zhang, X.; Spiegel, J.; Martinez Cuesta, S.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633. [Google Scholar] [CrossRef]
- Pipier, A.; Devaux, A.; Lavergne, T.; Adrait, A.; Coute, Y.; Britton, S.; Calsou, P.; Riou, J.F.; Defrancq, E.; Gomez, D. Constrained G4 structures unveil topology specificity of known and new G4 binding proteins. Sci. Rep. 2021, 11, 13469. [Google Scholar] [CrossRef]
- Zyner, K.G.; Mulhearn, D.S.; Adhikari, S.; Cuesta, S.M.; Di Antonio, M.; Erard, N.; Hannon, G.J.; Tannahill, D.; Balasubramanian, S. Genetic interactions of G-quadruplexes in humans. Elife 2019, 8, e46793. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Audry, J.; Maestroni, L.; Delagoutte, E.; Gauthier, T.; Nakamura, T.M.; Gachet, Y.; Saintome, C.; Geli, V.; Coulon, S. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 2015, 34, 1942–1958. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Lee, W.T.C.; Yin, Y.; Morten, M.J.; Tonzi, P.; Gwo, P.P.; Odermatt, D.C.; Modesti, M.; Cantor, S.B.; Gari, K.; Huang, T.T.; et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021, 12, 2525. [Google Scholar] [CrossRef]
- Tan, J.; Vonrhein, C.; Smart, O.S.; Bricogne, G.; Bollati, M.; Kusov, Y.; Hansen, G.; Mesters, J.R.; Schmidt, C.L.; Hilgenfeld, R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009, 5, e1000428. [Google Scholar] [CrossRef]
- Lavigne, M.; Helynck, O.; Rigolet, P.; Boudria-Souilah, R.; Nowakowski, M.; Baron, B.; Brule, S.; Hoos, S.; Raynal, B.; Guittat, L.; et al. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021, 49, 7695–7712. [Google Scholar] [CrossRef]
- Paeschke, K.; Juranek, S.; Simonsson, T.; Hempel, A.; Rhodes, D.; Lipps, H.J. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008, 15, 598–604. [Google Scholar] [CrossRef]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015, 6, 7643. [Google Scholar] [CrossRef]
- Giraldo, R.; Rhodes, D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994, 13, 2411–2420. [Google Scholar] [CrossRef]
- Giraldo, R.; Suzuki, M.; Chapman, L.; Rhodes, D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: A circular dichroism study. Proc. Natl. Acad. Sci. USA 1994, 91, 7658–7662. [Google Scholar] [CrossRef]
- Traczyk, A.; Liew, C.W.; Gill, D.J.; Rhodes, D. Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1. Nucleic Acids Res. 2020, 48, 4562–4571. [Google Scholar] [CrossRef]
- Fang, G.; Cech, T.R. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell 1993, 74, 875–885. [Google Scholar] [CrossRef]
- Horvath, M.P.; Schultz, S.C. DNA G-quartets in a 1.86 A resolution structure of an Oxytricha nova telomeric protein-DNA complex. J. Mol. Biol. 2001, 310, 367–377. [Google Scholar] [CrossRef]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef]
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef]
- Hwang, H.; Buncher, N.; Opresko, P.L.; Myong, S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure 2012, 20, 1872–1880. [Google Scholar] [CrossRef]
- Salas, T.R.; Petruseva, I.; Lavrik, O.; Bourdoncle, A.; Mergny, J.L.; Favre, A.; Saintome, C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006, 34, 4857–4865. [Google Scholar] [CrossRef] [PubMed]
- Law, M.J.; Lower, K.M.; Voon, H.P.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Enokizono, Y.; Matsugami, A.; Uesugi, S.; Fukuda, H.; Tsuchiya, N.; Sugimura, T.; Nagao, M.; Nakagama, H.; Katahira, M. Destruction of quadruplex by proteins, and its biological implications in replication and telomere maintenance. Nucleic Acids Res. Suppl. 2003, 3, 231–232. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Membrino, A.; Cogoi, S.; Fukuda, H.; Nakagama, H.; Xodo, L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 2009, 37, 2841–2853. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tang, M.L.; Zeng, Z.X.; Wu, R.Y.; Xue, Y.; Hao, Y.H.; Pang, D.W.; Zhao, Y.; Tan, Z. Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20413–20418. [Google Scholar] [CrossRef]
- Scalabrin, M.; Frasson, I.; Ruggiero, E.; Perrone, R.; Tosoni, E.; Lago, S.; Tassinari, M.; Palu, G.; Richter, S.N. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Gonzalez, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef]
- Tosoni, E.; Frasson, I.; Scalabrin, M.; Perrone, R.; Butovskaya, E.; Nadai, M.; Palu, G.; Fabris, D.; Richter, S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015, 43, 8884–8897. [Google Scholar] [CrossRef]
- Thakur, R.K.; Kumar, P.; Halder, K.; Verma, A.; Kar, A.; Parent, J.L.; Basundra, R.; Kumar, A.; Chowdhury, S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009, 37, 172–183. [Google Scholar] [CrossRef]
- Borgognone, M.; Armas, P.; Calcaterra, N.B. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-Myc, promotes the formation of parallel G-quadruplexes. Biochem. J. 2010, 428, 491–498. [Google Scholar] [CrossRef]
- Niu, K.; Xiang, L.; Jin, Y.; Peng, Y.; Wu, F.; Tang, W.; Zhang, X.; Deng, H.; Xiang, H.; Li, S.; et al. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019, 47, 7306–7320. [Google Scholar] [CrossRef]
- Cogoi, S.; Paramasivam, M.; Membrino, A.; Yokoyama, K.K.; Xodo, L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010, 285, 22003–22016. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Williams, P.; Ren, W.; Wang, M.Y.; Gao, Z.; Miao, W.; Huang, M.; Song, J.; Wang, Y. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2021, 17, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Soldatenkov, V.A.; Vetcher, A.A.; Duka, T.; Ladame, S. First evidence of a functional interaction between DNA quadruplexes and poly(ADP-ribose) polymerase-1. ACS Chem. Biol. 2008, 3, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.D.; Marecki, J.C.; Byrd, A.K.; Gao, J.; Raney, K.D. G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 2021, 49, 416–431. [Google Scholar] [CrossRef]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef]
- Wu, Y.; Shin-ya, K.; Brosh, R.M., Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef]
- Sun, H.; Karow, J.K.; Hickson, I.D.; Maizels, N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998, 273, 27587–27592. [Google Scholar] [CrossRef]
- Fry, M.; Loeb, L.A. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999, 274, 12797–12802. [Google Scholar] [CrossRef]
- Mohaghegh, P.; Karow, J.K.; Brosh, R.M.; Bohr, V.A.; Hickson, I.D. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001, 29, 2843–2849. [Google Scholar] [CrossRef]
- Ketkar, A.; Voehler, M.; Mukiza, T.; Eoff, R.L. Residues in the RecQ C-terminal Domain of the Human Werner Syndrome Helicase Are Involved in Unwinding G-quadruplex DNA. J. Biol. Chem. 2017, 292, 3154–3163. [Google Scholar] [CrossRef]
- Vaughn, J.P.; Creacy, S.D.; Routh, E.D.; Joyner-Butt, C.; Jenkins, G.S.; Pauli, S.; Nagamine, Y.; Akman, S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005, 280, 38117–38120. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Tippana, R.; Demeshkina, N.A.; Murat, P.; Balasubramanian, S.; Myong, S.; Ferre-D’Amare, A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Hidaka, K.; Tran, P.L.T.; Mergny, J.L.; Gorelick, R.J.; Sugiyama, H. HIV-1 Nucleocapsid Proteins as Molecular Chaperones for Tetramolecular Antiparallel G-Quadruplex Formation. J. Am. Chem. Soc. 2013, 135, 18575–18585. [Google Scholar] [CrossRef] [PubMed]
- Arimondo, P.B.; Riou, J.F.; Mergny, J.L.; Tazi, J.; Sun, J.S.; Garestier, T.; Helene, C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000, 28, 4832–4838. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Padmanabhan, K.P.; Ferrara, J.D.; Sadler, J.E.; Tulinsky, A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993, 268, 17651–17654. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Tulinsky, A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 272–282. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144. [Google Scholar] [CrossRef]
- Guo, J.U.; Bartel, D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353, 6306. [Google Scholar] [CrossRef]
- Chen, W.F.; Rety, S.; Guo, H.L.; Dai, Y.X.; Wu, W.Q.; Liu, N.N.; Auguin, D.; Liu, Q.W.; Hou, X.M.; Dou, S.X.; et al. Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model. Structure 2018, 26, 403–415. [Google Scholar] [CrossRef]
- Armas, P.; Nasif, S.; Calcaterra, N.B. Cellular nucleic acid binding protein binds G-rich single-stranded nucleic acids and may function as a nucleic acid chaperone. J. Cell Biochem. 2008, 103, 1013–1036. [Google Scholar] [CrossRef]
- Lee, C.Y.; McNerney, C.; Ma, K.; Zhao, W.; Wang, A.; Myong, S. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat. Commun. 2020, 11, 3392. [Google Scholar] [CrossRef]
- Lim, G.; Hohng, S. Single-molecule fluorescence studies on cotranscriptional G-quadruplex formation coupled with R-loop formation. Nucleic Acids Res. 2020, 48, 9195–9203. [Google Scholar] [CrossRef]
- De Cian, A.; Cristofari, G.; Reichenbach, P.; De Lemos, E.; Monchaud, D.; Teulade-Fichou, M.P.; Shin-Ya, K.; Lacroix, L.; Lingner, J.; Mergny, J.L. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. USA 2007, 104, 17347–17352. [Google Scholar] [CrossRef]
- Jansson, L.I.; Hentschel, J.; Parks, J.W.; Chang, T.R.; Lu, C.; Baral, R.; Bagshaw, C.R.; Stone, M.D. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc. Natl. Acad. Sci. USA 2019, 116, 9350–9359. [Google Scholar] [CrossRef]
- Crabbe, L.; Cesare, A.J.; Kasuboski, J.M.; Fitzpatrick, J.A.; Karlseder, J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012, 2, 1521–1529. [Google Scholar] [CrossRef]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef]
- Mcrae, E.K.S.; Booy, E.P.; Padilla-Meier, G.P.; McKenna, S.A. On Characterizing the Interactions between Proteins and Guanine Quadruplex Structures of Nucleic Acids. J. Nucleic Acids 2017, 2017, 9675348. [Google Scholar] [CrossRef]
- Hossain, K.A.; Jurkowski, M.; Czub, J.; Kogut, M. Mechanism of recognition of parallel G-quadruplexes by DEAH/RHAU helicase DHX36 explored by molecular dynamics simulations. Comput. Struct. Biotec. 2021, 19, 2526–2536. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Zhang, Y.; You, H. Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy. Biomolecules 2021, 11, 1579. https://doi.org/10.3390/biom11111579
Cheng Y, Zhang Y, You H. Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy. Biomolecules. 2021; 11(11):1579. https://doi.org/10.3390/biom11111579
Chicago/Turabian StyleCheng, Yuanlei, Yashuo Zhang, and Huijuan You. 2021. "Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy" Biomolecules 11, no. 11: 1579. https://doi.org/10.3390/biom11111579
APA StyleCheng, Y., Zhang, Y., & You, H. (2021). Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy. Biomolecules, 11(11), 1579. https://doi.org/10.3390/biom11111579





