Extraction and Chemical Characterization of Functional Phenols and Proteins from Coffee (Coffea arabica) By-Products
Abstract
:1. Introduction
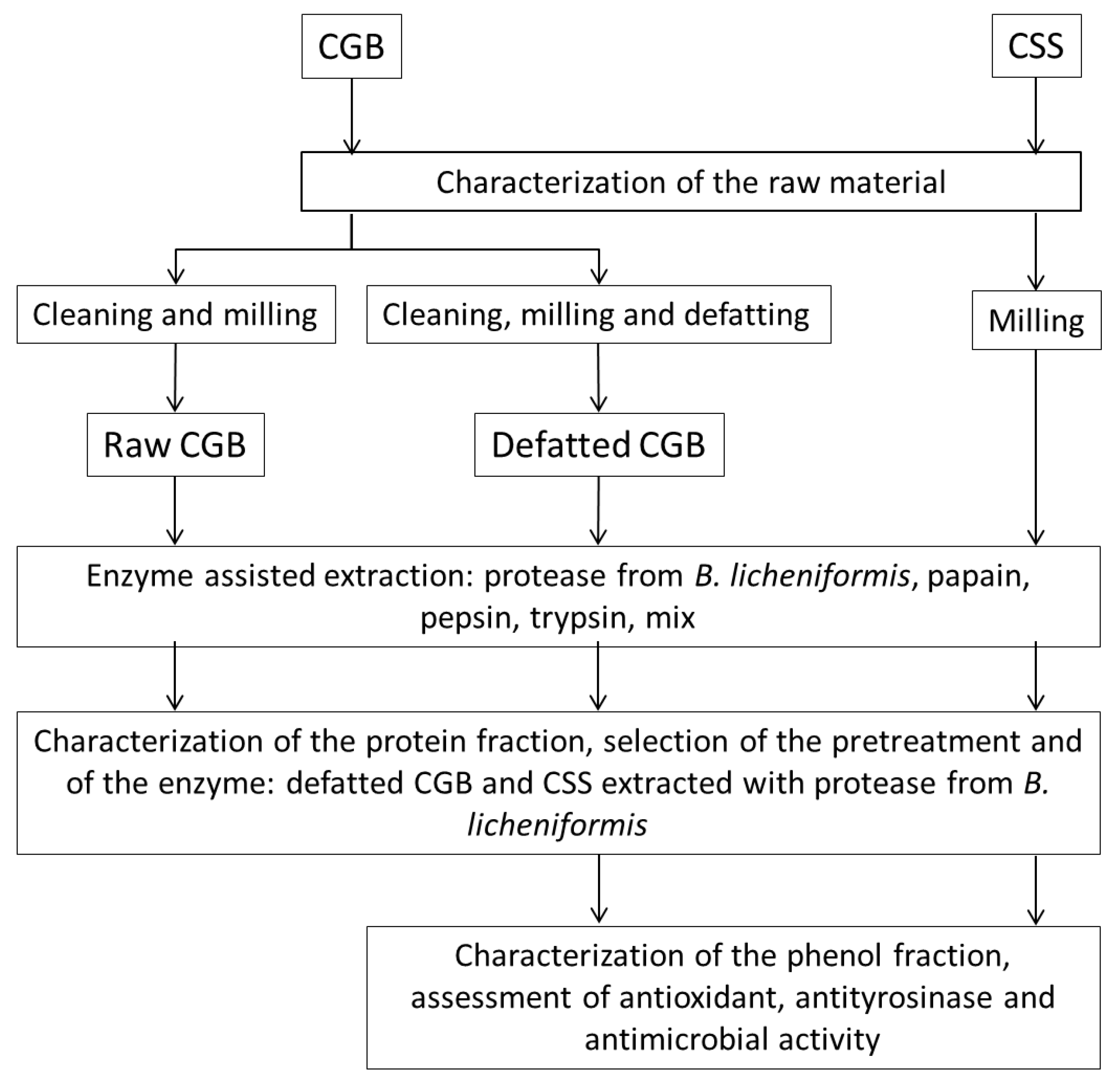
2. Materials and Methods
2.1. Sampling of Raw Materials
2.2. Proximal Analysis of Raw Materials
2.3. Phenolic Compounds and Caffeine Content of Raw Materials
2.4. Pretreatments of CGB
2.5. Protease-Assisted Extraction from the Raw Materials
- Reaction medium: 10 mM phosphate buffer (for protease from B. licheniformis, papain, trypsin, and mix) or 10 mM hydrochloric acid (pepsin).
- Enzyme/substrate ratio: 1% (w/w or v/w).
- Hydraulic module: 1 to 5 (1 part of sample and 5 parts of reaction medium) for CGB, 1 to 10 for CSS.
- Hydrolysis time: 2 h.
- pH and temperature for each enzyme:
- ○
- Protease from B. licheniformis: pH 6.5–8.5, T 60 °C.
- ○
- Trypsin: pH 7–9, T 37 °C.
- ○
- Pepsin: pH 2–4, T 37 °C.
- ○
- Papain: pH 6–7, T 65 °C.
- ○
- Mix of protease from B. licheniformis and papain: pH 6.5–7, T 62.5 °C.
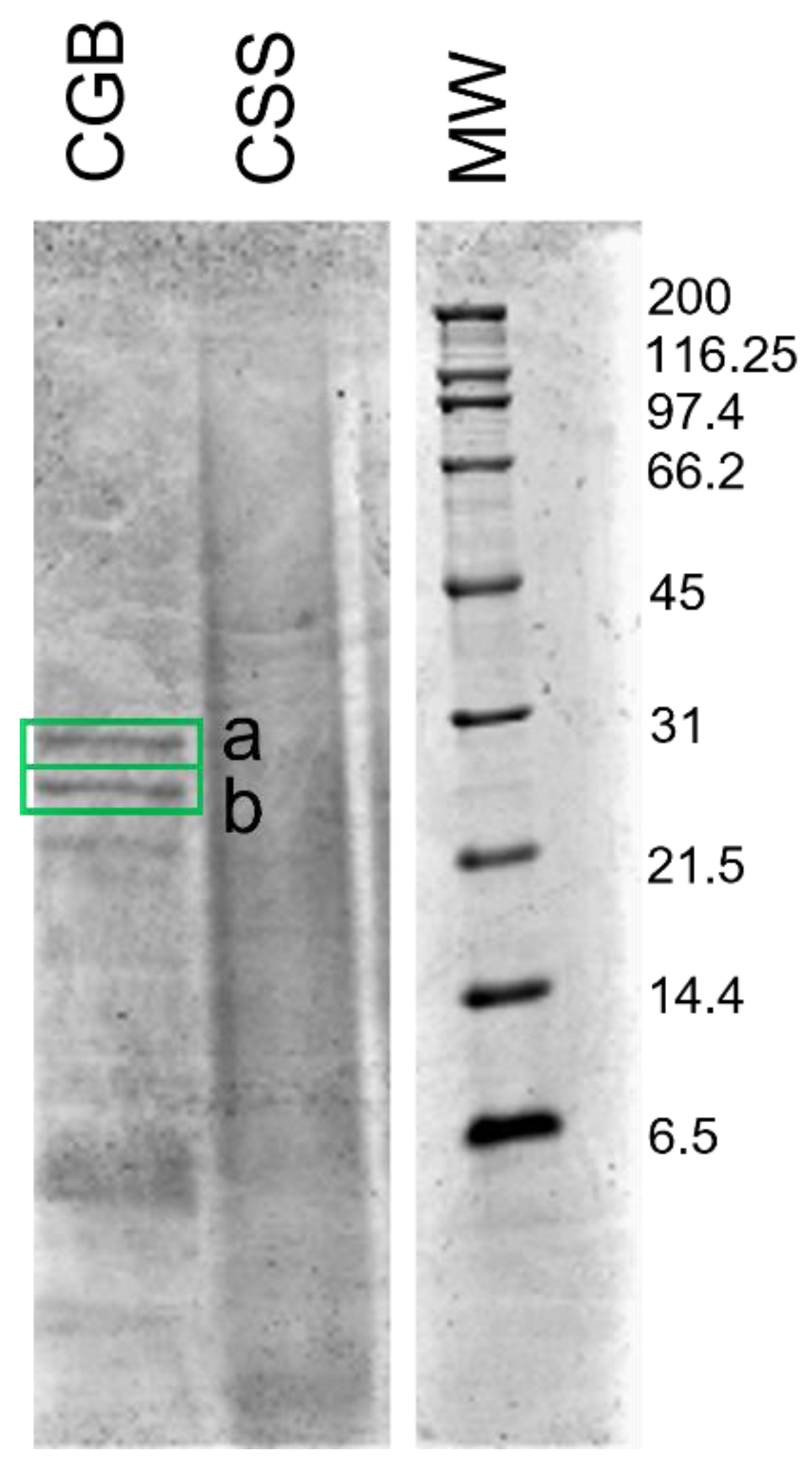
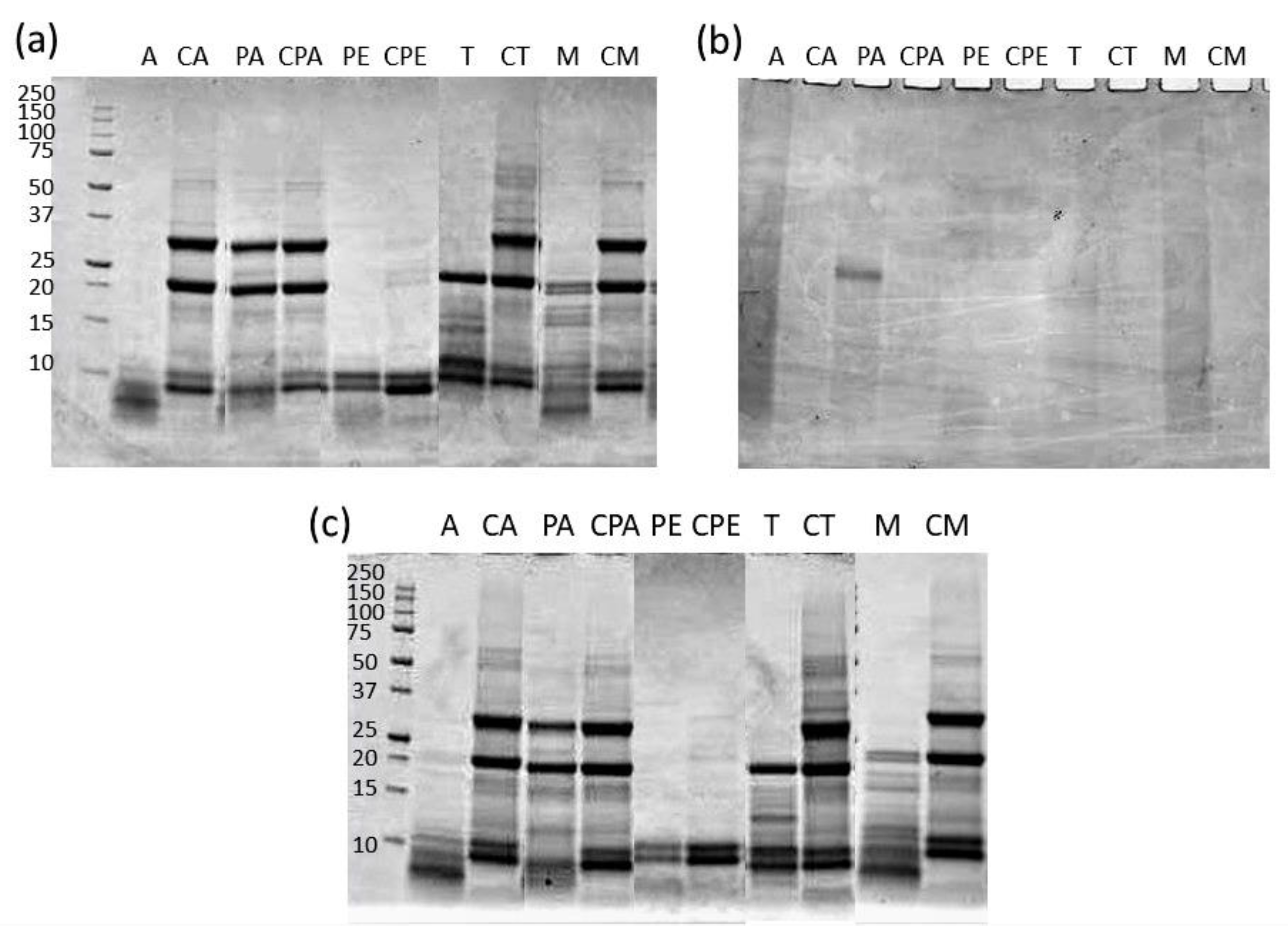
2.6. SDS-PAGE of Extracts
2.7. Analysis of the Total and Free Amino Acid of the Extracts
2.8. Degree of Racemization of the Amino Acids of the Extracts
2.9. Protein Identification
2.10. Biological Activities of the Extracts
2.11. Antimicrobial Activity of the Extracts
2.12. Statistical Analysis
3. Results and Discussion
3.1. Molecular Characterization of By-Product Composition
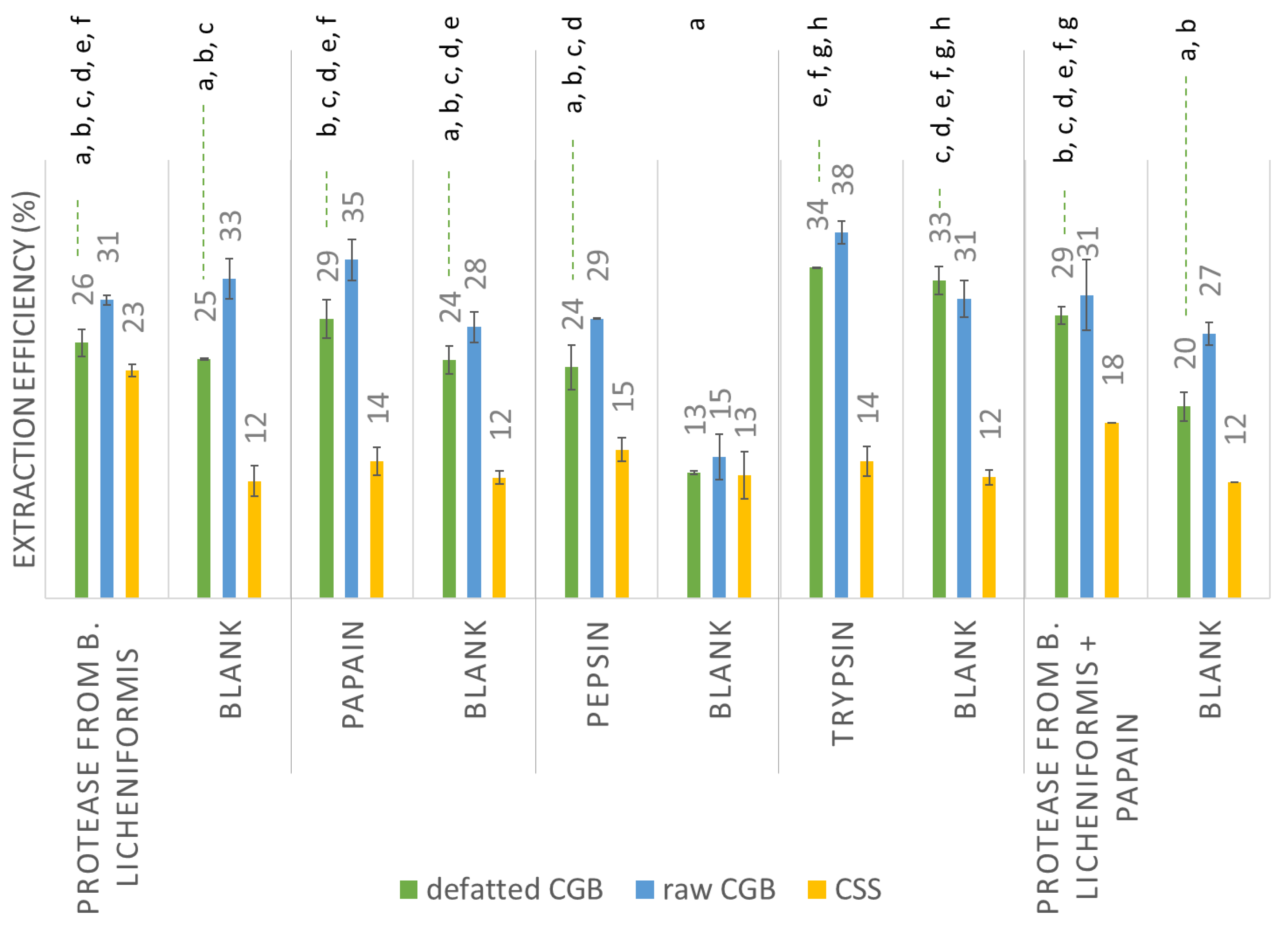
3.2. CGB and CSS Pretreatment
3.3. Protein Extraction
3.4. Characterization of Proteins in the Extracts
3.5. Protein and Amino Acid Content of the Enzymatic Extracts
3.6. Polyphenol Content and Biofunctional Properties of the Enzymatic Extracts
3.7. Antimicrobial Activity of the Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICO (International Coffee Organization) and UN Comtrade Data. Available online: www.statista.com (accessed on 27 March 2020).
- Bicho, N.C.; Lidon, F.C.; Ramalho, J.C.; Leitao, A.E. Quality assessment of Arabica and Robusta green and roasted coffees—A review. Emir. J. Food Agric. 2013, 25, 945–950. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, L.S.; Franca, A.S.; Mendonça, J.C.F.; Barros-Júnior, M.C. Proximate composition and fatty acids profile of green and roasted defective coffee beans. LWT 2006, 39, 235–239. [Google Scholar] [CrossRef]
- Malara, A.; Paone, E.; Frontera, P.; Bonaccorsi, L.; Panzera, G.; Mauriello, F. Sustainable exploitation of coffee silverskin in water remediation. Sustainability 2018, 10, 3547. [Google Scholar] [CrossRef] [Green Version]
- Murthy, P.S.; Naidu, M.M. Sustainable management of coffee industry by-products and value addition—a review. Resour. Conserv. Recycl. 2012, 66, 45–58. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Machado, E.M.S.; Martins, S.; Teixeira, J.A. Production, composition, and application of coffee and its industrial residues. Food Bioproc. Tech. 2011, 4, 661. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, E.R.; da Silva, R.F.; Santos, P.R.; Queiroz, F. An investigation into green coffee press cake as a renewable source of bioactive compounds. Int. J. Food Sci. Technol. 2019, 54, 1187–1196. [Google Scholar] [CrossRef]
- Narita, Y.; Inouye, K. Review on utilization and composition of coffee silverskin. Food Res. Int. 2014, 61, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Zhang, Z.H.; Zhao, M.; Senthamaraikannan, R.; Padamati, R.B.; Sun, D.W.; Tiwari, B.K. Green extraction of soluble dietary fibre from coffee silverskin: Impact of ultrasound/microwave-assisted extraction. Int. J. Food Sci. Technol. 2020, 55, 2242–2250. [Google Scholar] [CrossRef]
- Iriondo-DeHond, A.; Rios, M.B.; Herrera, T.; Rodriguez-Bertos, A.; Nunez, F.; San Andres, M.I.; Sanchez-Fortun, S.; del Castillo, M.D. Coffee silverskin extract: Nutritional value, safety and effect on key biological functions. Nutrients 2019, 11, 2693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regazzoni, L.; Saligari, F.; Marinello, C.; Rossoni, G.; Aldini, G.; Carini, M.; Orioli, M. Coffee silver skin as a source of polyphenols: High resolution mass spectrometric profiling of components and antioxidant activity. J. Funct. Foods 2016, 20, 472–485. [Google Scholar] [CrossRef]
- Kusumocahyo, S.P.; Tangguh, P.; Annelies, C.D.; Sutanto, H. Utilization of coffee silverskin by-product from coffee roasting industry through extraction process for the development of antioxidant skin gel. J. Cosmet Sci. 2019, 70, 313–325. [Google Scholar]
- Rebollo-Hernanz, M.; Zhang, Q.Z.; Aguilera, Y.; Martin-Cabrejas, M.A.; de Mejia, E.G. Relationship of the phytochemicals from coffee and cocoa by-products with their potential to modulate biomarkers of metabolic syndrome in vitro. Antioxidants 2019, 8, 279. [Google Scholar] [CrossRef] [Green Version]
- Rebollo-Hernanz, M.; Zhang, Q.Z.; Aguilera, Y.; Martin-Cabrejas, M.A.; de Mejia, E.G. Phenolic compounds from coffee by-products modulate adipogenesis-related inflammation, mitochondrial dysfunction, and insulin resistance in adipocytes, via insulin/PI3K/AKT signaling pathways. Food Chem. Toxicol. 2019, 132, 110672. [Google Scholar] [CrossRef] [PubMed]
- Perez-Miguez, R.; Marina, M.L.; Castro-Puyana, M. High resolution liquid chromatography tandem mass spectrometry for the separation and identification of peptides in coffee silverskin protein hydrolysates. Microchem J. 2019, 149, 103951. [Google Scholar] [CrossRef]
- Totaro, G.; Sisti, L.; Fiorini, M.; Lancellotti, I.; Andreola, F.N.; Saccani, A. Formulation of green particulate composites from PLA and PBS matrix and wastes deriving from the coffee production. J. Polym. Environ. 2019, 27, 1488–1496. [Google Scholar] [CrossRef]
- Gigante, V.; Seggiani, M.; Cinelli, P.; Signori, F.; Vania, A.; Navarini, L.; Amato, G.; Lazzeri, A. Utilization of coffee silverskin in the production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) biopolymer-based thermoplastic biocomposites for food contact applications. Compos. Part A Appl. Sci. Manuf. 2021, 140, 106172. [Google Scholar] [CrossRef]
- Jooste, T.; García-Aparicio, M.P.; Brienzo, M.; van Zyl, W.H.; Görgens, J.F. Enzymatic hydrolysis of spent coffee ground. Appl. Biochem. Biotechnol. 2013, 169, 2248–2262. [Google Scholar] [CrossRef]
- Almeida, F.S.; Gancalves Dias, F.F.; Kawazoe Sato, A.C.; Nobrega de Moura Bell, J.M.L. From solvent extraction to the concurrent extraction of lipids and proteins from green coffee: An eco-friendly approach to improve process feasibility. Food Bioprod Process. 2021, 129, 144–156. [Google Scholar] [CrossRef]
- Full, G.; Lonzarich, V.; Suggi-Liverani, F. Differences in chemical composition of electronically sorted green coffee beans. In Proceedings of the 18th International Scientific Colloquium on Coffee, Helsinki, Finland, 1 January 1999. [Google Scholar]
- Prandi, B.; Zurlini, C.; Cicognini, I.M.; Cutroneo, S.; Di Massimo, M.; Bondi, M.; Brutti, A.; Sforza, S.; Tedeschi, T. Targeting the nutritional value of proteins from legumes by-products through mild extraction technologies. Front. Nutr. 2021, 8, 695793. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Method Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Janming, W. The determination of flavonoid in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- McMurrough, I.; McDowell, J. Chromatographic separation and automated analysis of flavanols. Anal. Biochem. 1978, 91, 92–100. [Google Scholar] [CrossRef]
- Porter, L.J.; Hrstich, L.N.; Chan, B.G. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry 1986, 25, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Belay, A.; Ture, K.; Redi, M.; Asfaw, A. Measurement of caffeine in coffee beans with UV/vis spectrometer. Food Chem. 2008, 108, 310–315. [Google Scholar] [CrossRef]
- Prandi, B.; Lambertini, F.; Varani, M.; Faccini, A.; Suman, M.; Leporati, A.; Tedeschi, T.; Sforza, S. Assessment of enzymatic improvers in flours using LC−MS/MS detection of marker tryptic peptides. J. Am. Mass Spectrom. 2020, 31, 240–248. [Google Scholar] [CrossRef]
- Gasparini, A.; Van Gool, M.P.; Bultsma, M.; Cutroneo, S.; Sforza, S.; Tedeschi, T. Modifications induced by controlled storage conditions on whey protein concentrates: Effects on whey protein lactosylation and solubility. Int. Dairy J. 2020, 109, 104765. [Google Scholar] [CrossRef]
- Prandi, B.; Faccini, A.; Lambertini, F.; Bencivenni, M.; Jorba, M.; Van Droogenbroek, B.; Bruggeman, G.; Schöber, J.; Petrusan, J.; Elst, K.; et al. Food wastes from agrifood industry as possible sources of proteins: A detailed molecular view on the composition of the nitrogen fraction, amino acid profile and racemisation degree of 39 food waste streams. Food Chem. 2019, 286, 567–575. [Google Scholar] [CrossRef]
- Ferri, M.; Gianotti, A.; Tassoni, A. Optimisation of assay conditions for the determination of antioxidant capacity and polyphenols in cereal food components. J. Food Comp. Anal. 2013, 30, 94–101. [Google Scholar] [CrossRef]
- Masmoto, Y.; Iida, S.; Kubo, M. Inhibitory effect of Chinese crude drugs on tyrosinase. Planta Med. 1980, 40, 361–365. [Google Scholar] [CrossRef]
- Acuña, R.; Bassüner, R.; Beilinson, V.; Cortina, H.; Cadena-Gómez, G.; Montes, V.; Nielsen, N.C. Coffee seeds contain 11S storage proteins. Physiol. Plant. 1999, 105, 122–131. [Google Scholar] [CrossRef]
- Casal, S.; Mendes, E.; Oliveira, M.B.P.P.; Ferreira, M.A. Roast effects on coffee amino acid enantiomers. Food Chem. 2005, 89, 333–340. [Google Scholar] [CrossRef]
- Franca, A.S.; Oliveira, L.S. Chemistry of defective coffee beans. In Food Chemistry Research Developments; Koeffer, E.N., Ed.; Nova Science Publisher, Inc.: Hauppage, NY, USA, 2008; pp. 105–138. [Google Scholar]
- Tripetch, P.; Borompichaichartkul, C. Effect of packaging materials and storage time on changes of colour, phenolic content, chlorogenic acid and antioxidant activity in arabica green coffee beans (Coffea arabica L. cv. Catimor). J. Stored Prod. Res. 2019, 84, 101510. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Zhang, Y.; Ruan, X.; Zhang, R. Purification, characterization, synthesis, in vitro ACE inhibition and in vivo antihypertensive activity of bioactive peptides derived from oil palm kernel glutelin-2 hydrolysates. J. Funct. Foods 2017, 28, 48–58. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Zhang, Y. Purification and identification of antioxidative peptides of palm kernel expeller glutelin-1 hydrolysates. RSC Adv. 2017, 7, 54196. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.-C.; Lee, J.-S.; Lee, J.-H.; Kim, Y.; So, W.-Y. Anti-oxidative and anti-inflammatory activity of Kenya grade AA green coffee bean extracts. Iran. J. Public Health 2019, 48, 2025–2034. [Google Scholar]
- Castro, A.; Oda, F.; Almeida-Cincotto, M.; Davanço, M.; Chiari-Andréo, B.G.; Cicarelli, R.; Peccinini, R.; Zocolo, G.; Ribeiro, P.; Corrêa, M.; et al. Green coffee seed residue: A sustainable source of antioxidant compounds. Food Chem. 2018, 246, 48–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zengin, G.; Sinan, K.I.; Mahomoodally, M.F.; Angeloni, S.; Mustafa, A.M.; Vittori, S.; Maggi, F.; Caprioli, G. Chemical composition, antioxidant and enzyme inhibitory properties of different extracts obtained from spent coffee ground and coffee silverskin. Foods 2020, 9, 713. [Google Scholar] [CrossRef]
- Rudra, S.G.; Nishad, J.; Jakhar, N.; Kaur, C. Food industry waste: Mine of nutraceuticals. Int. J. Sci. Environ. Technol. 2015, 4, 205–222. [Google Scholar]
- Buzanello, E.B.; Pinheiro Machado, G.T.B.; Kuhnen, S.; Mazzarino, L.; Maraschin, M. Nanoemulsions containing oil and aqueous extract of green coffee beans with antioxidant and antimicrobial activities. Nano Express 2020, 1, 010058. [Google Scholar] [CrossRef]
- Getachew, A.T.; Chun, B.S. Influence of hydrothermal process on bioactive compounds extraction from green coffee bean. Innov Food Sci. Emerg. Technol. 2016, 38 Pt A, 24–31. [Google Scholar] [CrossRef]
- Nzekoue, F.K.; Angeloni, S.; Navarini, L.; Angeloni, C.; Freschi, M.; Hrelia, S.; Vitali, L.A.; Sagratini, G.; Vittori, S.; Caprioli, G. Coffee silverskin extracts: Quantification of 30 bioactive compounds by a new HPLC-MS/MS method and evaluation of their antioxidant and antibacterial activities. Food Res. Int. 2020, 133, 109128. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.; Palmeira-de-Oliveira, A.; das Neves, J.; Sarmento, B.; Amaral, M.H.; Oliveira, M.B. Coffee silverskin: A possible valuable cosmetic ingredient. Pharm. Biol. 2015, 53, 386–394. [Google Scholar] [CrossRef] [PubMed]
| Component Analysis | CGB | CSS |
|---|---|---|
| Dry residue (%) | 96.7 ± 0.0 a | 82.5 ± 1.6 a |
| Proteins (% g/100 g DW) | 15.3 ± 0.1 a | 16.3 ± 0.2 a |
| Fibers (% g/100 g DW) | 56.4 ± 0.1 | 69.8 ± 0.2 |
| Sugars (% g/100 g DW) | 8.0 ± 0.8 a | 0.4 ± 0.2 a |
| Lipids (% g/100 g DW) | 13.6 ± 0.2 a | 6.3 ± 0.1 a |
| d-Ala | 2.0 ± 0.3 | 2.8 ± 0.4 |
| d-Asp | 4.7 ± 0.4 | 9.0 ± 0.7 |
| d-Glu | 1.9 ± 1.2 | 5.5 ± 3.5 |
| d-Phe | 1.7 ± 0.3 | 2.9 ± 0.6 |
| d-Lys | 2.3 ± 0.9 | 1.7 ± 0.6 |
| Total polyphenols (mg GA eq/g DW) | 27.22 ± 1.03 a | 5.36 ± 0.33 b |
| Total flavonoids (mg CAT eq/g DW) | 16.29 ± 0.65 a | 4.81 ± 0.66 b |
| Total flavanols (mg CAT eq/g DW) | 5.26 ± 0.49 a | 0.90 ± 0.10 b |
| Tannins (mg/g DW) | 2.46 ± 0.40 a | 2.48 ± 0.14 b |
| Caffeine (mg/g DW) | 15.18 ± 0.49 a | 9.08 ± 0.97 b |
| Extraction Condition | Non-Defatted CGB | Defatted CGB | CSS |
|---|---|---|---|
| EAE—Bacillus licheniformis protease | 17 | 16 | 10 |
| EAE—papain | 17 | 19 | 5 |
| EAE—pepsin | 15 | 15 | 4 |
| EAE—trypsin | 17 | 18 | 5 |
| Extraction Condition | Raw CGB | Defatted CGB | CSS |
|---|---|---|---|
| EAE (Bacillus licheniformis protease) | 18 | 18 | 3 |
| EAE (papain) | 17 | 16 | 11 |
| EAE (pepsin) | 21 | 20 | 6 |
| EAE (trypsin) | 15 | 12 | 4 |
| EAE (Bacillus licheniformis protease + papain) | 18 | 18 | 4 |
| Coffee By-Product | Total Polyphenols (mg GA eq/g DW) | Antioxidant Activity (mg AA eq/g DW) | Antityrosinase Activity (mg KA eq/g DW) |
|---|---|---|---|
| CGB—EAE-(Bacillus licheniformis protease) | 99.02 ± 10.13 a | 78.74 ± 4.90 a | 2.25 ± 0.16 a |
| CSS—EAE-(Bacillus licheniformis protease) | 12.63 ± 0.13 b | 18.95 ± 0.31 b | 2.85 ± 0.31 b |
| Sample | Kill off E. coli F4 (%) | Kill off S. suis (%) | ||
|---|---|---|---|---|
| 1.25 kg/MT | 12.5 kg/MT | 1.25 kg/MT | 12.5 kg/MT | |
| Defatted CGB—EAE (alcalase) | 51 | 65 | 55.5 | 64.2 |
| CSS—EAE (alcalase) | 4.7 | 58.8 | 2.8 | 22.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prandi, B.; Ferri, M.; Monari, S.; Zurlini, C.; Cigognini, I.; Verstringe, S.; Schaller, D.; Walter, M.; Navarini, L.; Tassoni, A.; et al. Extraction and Chemical Characterization of Functional Phenols and Proteins from Coffee (Coffea arabica) By-Products. Biomolecules 2021, 11, 1571. https://doi.org/10.3390/biom11111571
Prandi B, Ferri M, Monari S, Zurlini C, Cigognini I, Verstringe S, Schaller D, Walter M, Navarini L, Tassoni A, et al. Extraction and Chemical Characterization of Functional Phenols and Proteins from Coffee (Coffea arabica) By-Products. Biomolecules. 2021; 11(11):1571. https://doi.org/10.3390/biom11111571
Chicago/Turabian StylePrandi, Barbara, Maura Ferri, Stefania Monari, Chiara Zurlini, Ilaria Cigognini, Stefanie Verstringe, Dennis Schaller, Martha Walter, Luciano Navarini, Annalisa Tassoni, and et al. 2021. "Extraction and Chemical Characterization of Functional Phenols and Proteins from Coffee (Coffea arabica) By-Products" Biomolecules 11, no. 11: 1571. https://doi.org/10.3390/biom11111571
APA StylePrandi, B., Ferri, M., Monari, S., Zurlini, C., Cigognini, I., Verstringe, S., Schaller, D., Walter, M., Navarini, L., Tassoni, A., Sforza, S., & Tedeschi, T. (2021). Extraction and Chemical Characterization of Functional Phenols and Proteins from Coffee (Coffea arabica) By-Products. Biomolecules, 11(11), 1571. https://doi.org/10.3390/biom11111571







