Modeling Prostate Cancer Treatment Responses in the Organoid Era: 3D Environment Impacts Drug Testing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture and Viability Assays of Metastatic Prostate Cancer Organoid Lines
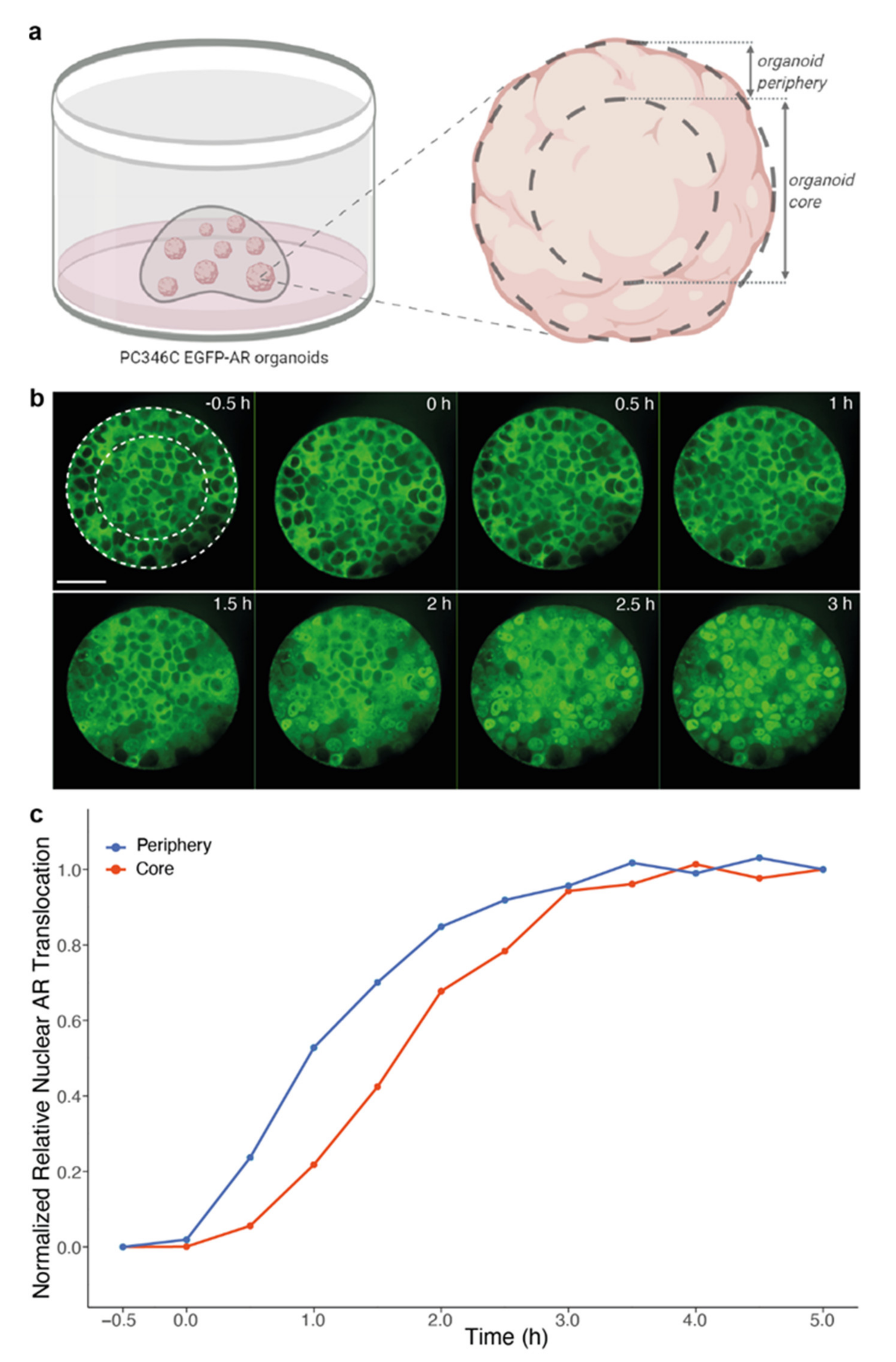
2.2. Live Cell Imaging of Androgen-Induced Nuclear AR Translocation
2.3. Live Cell Imaging of Taxane-Induced Tubulin Stabilization
2.4. Statistical Analysis
3. Results
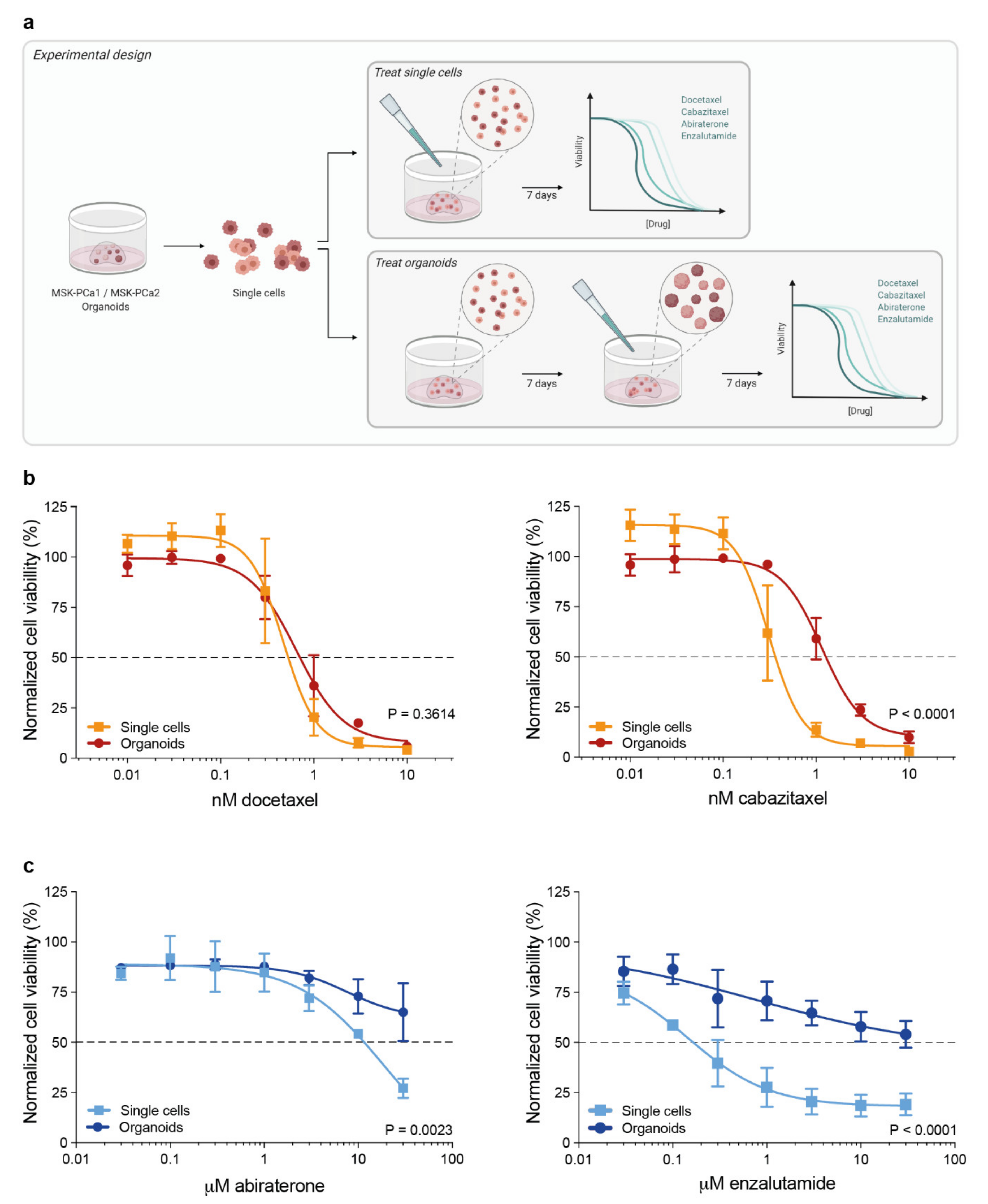
3.1. 3D Organoid Structure Attenuates Sensitivity to Taxane-Based Chemotherapy and Anti-Androgen Treatment
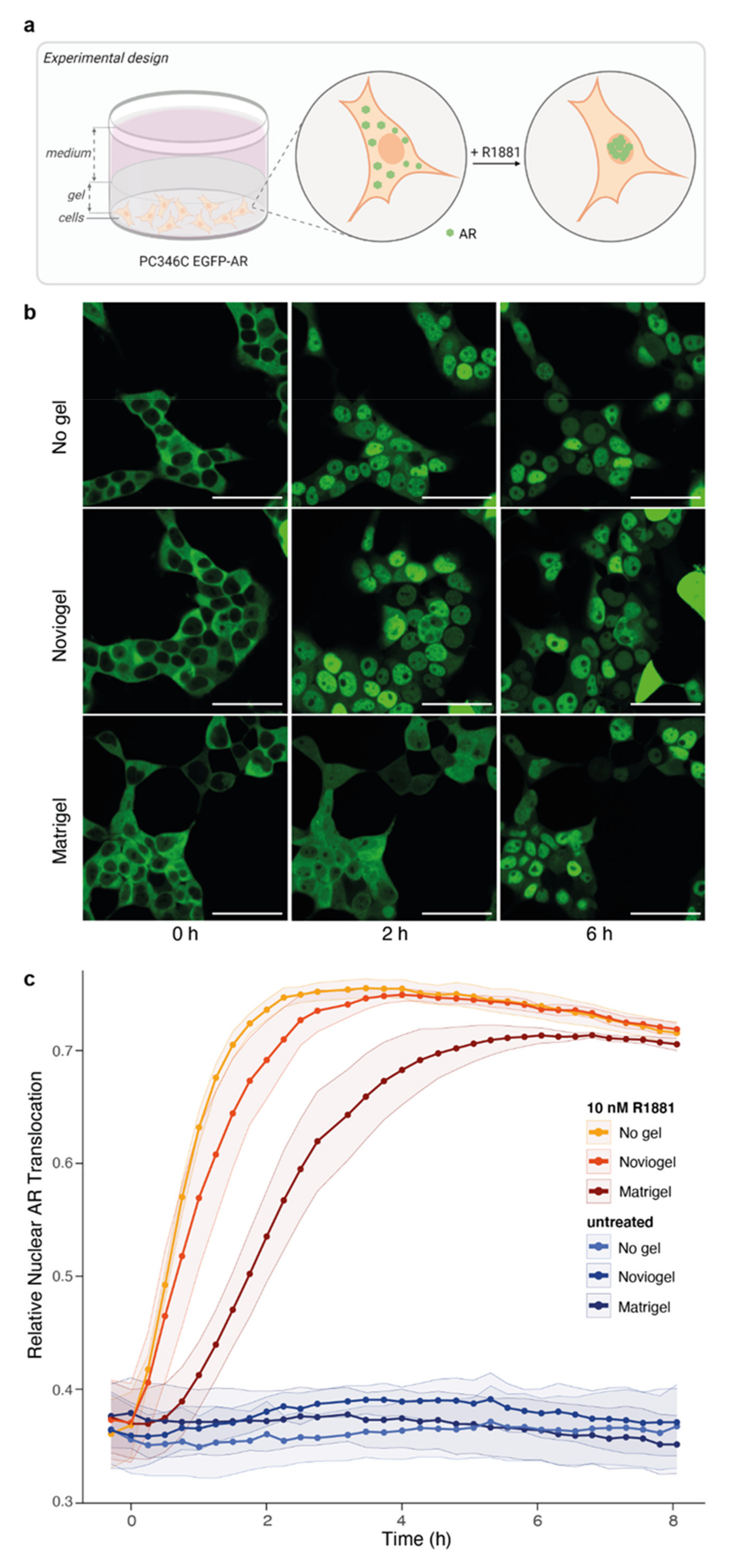
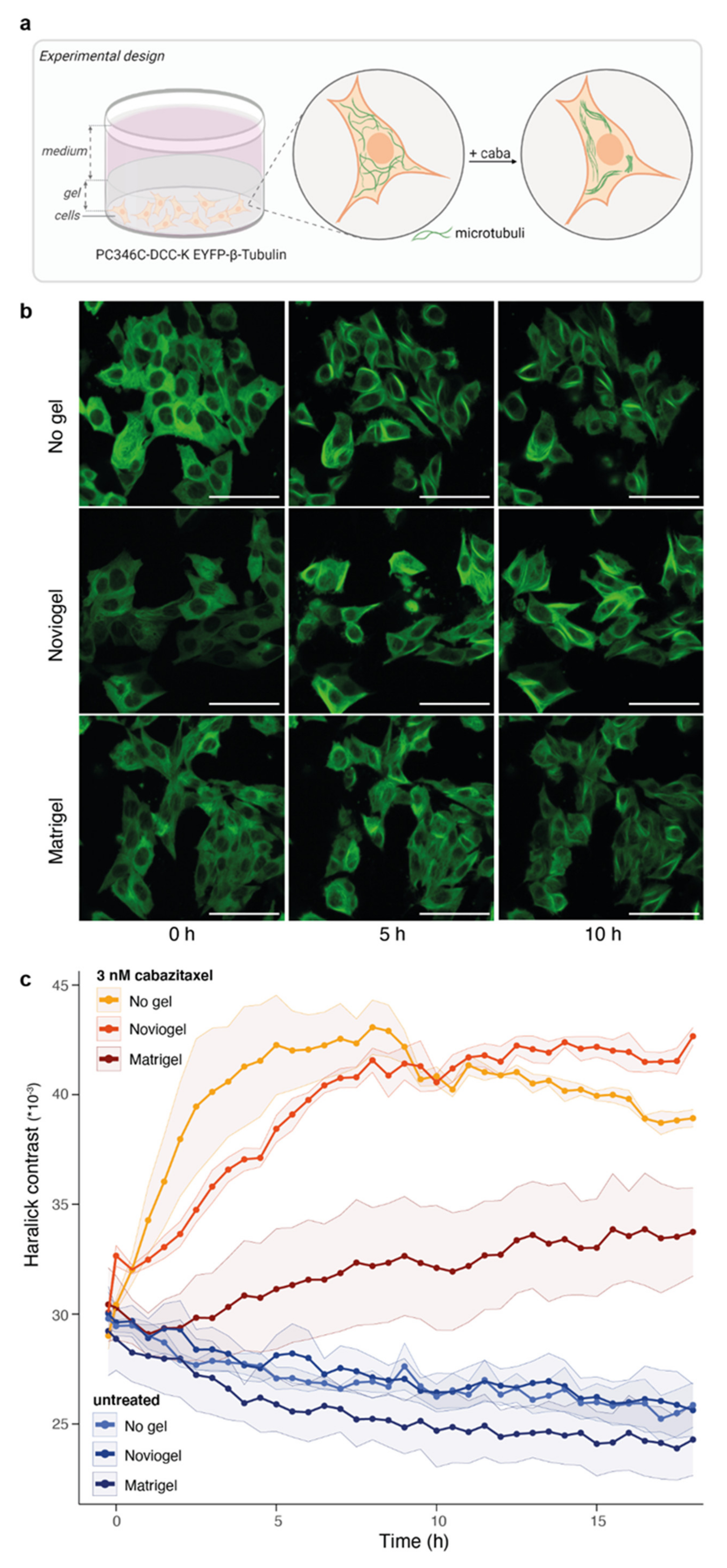
3.2. Extracellular Matrix Affects Androgen-Induced AR Translocation and Chemotherapy Effectiveness
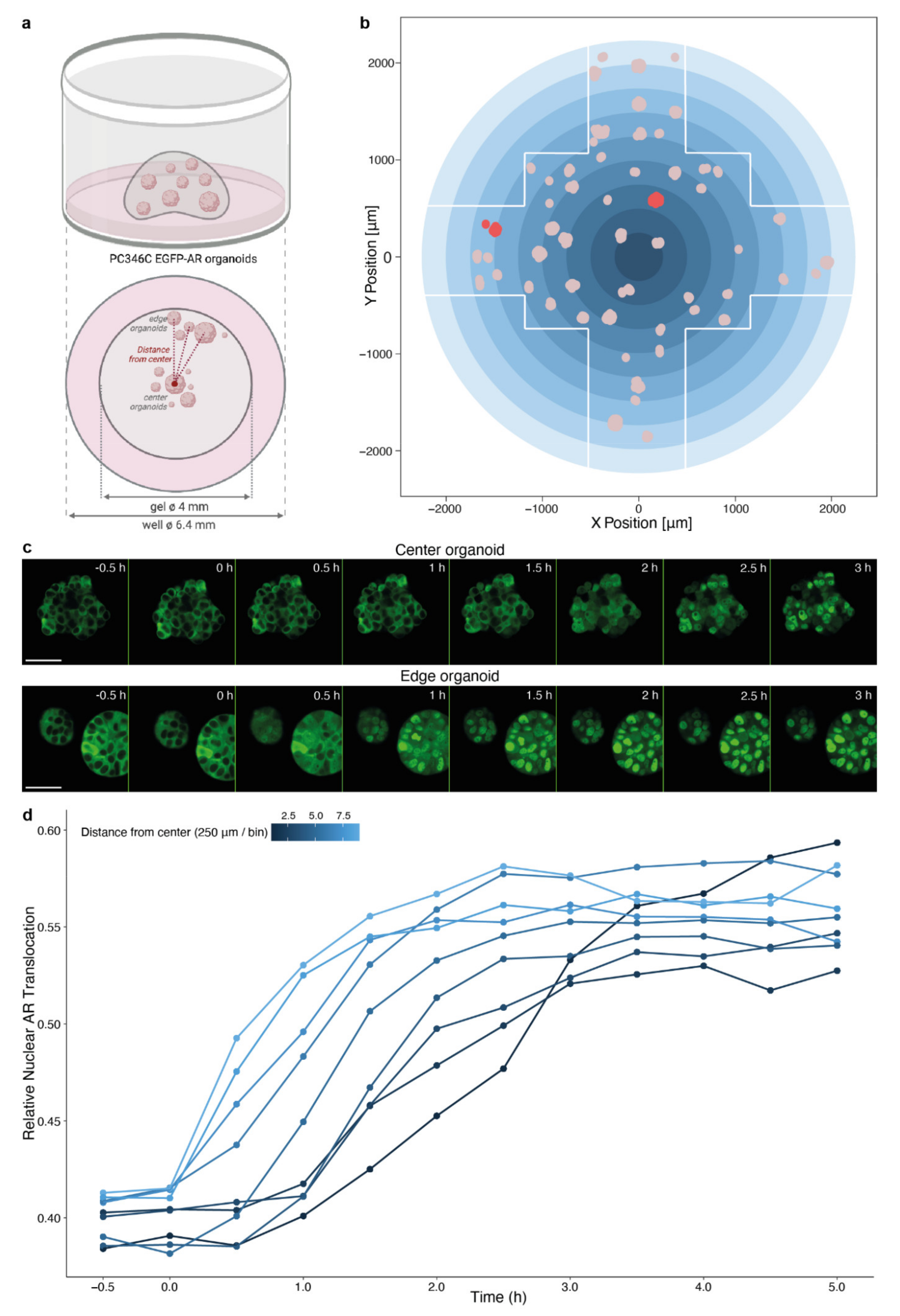
3.3. Spatial Distribution of Organoids Impacts Compound Effectivity
3.4. Impact of Organoid Size on Drug Effectivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.M.; Kannan, M.; Langer, L.F.; Leibowitz, B.D.; Bentaieb, A.; Cancino, A.; Dolgalev, I.; Drummond, B.E.; Dry, J.R.; Ho, C.S.; et al. A pan-cancer organoid platform for precision medicine. Cell Rep. 2021, 36, 109429. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [Green Version]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernandez-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puca, L.; Bareja, R.; Prandi, D.; Shaw, R.; Benelli, M.; Karthaus, W.R.; Hess, J.; Sigouros, M.; Donoghue, A.; Kossai, M.; et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 2018, 9, 2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mout, L.; van Dessel, L.F.; Kraan, J.; de Jong, A.C.; Neves, R.P.L.; Erkens-Schulze, S.; Beaufort, C.M.; Sieuwerts, A.M.; van Riet, J.; Woo, T.L.C.; et al. Generating human prostate cancer organoids from leukapheresis enriched circulating tumour cells. Eur. J. Cancer 2021, 150, 179–189. [Google Scholar] [CrossRef]
- Karkampouna, S.; La Manna, F.; Benjak, A.; Kiener, M.; De Menna, M.; Zoni, E.; Grosjean, J.; Klima, I.; Garofoli, A.; Bolis, M.; et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat. Commun. 2021, 12, 1117. [Google Scholar] [CrossRef]
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.; et al. A pdx/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin. Cancer Res. 2018, 24, 4332–4345. [Google Scholar] [CrossRef] [Green Version]
- Jansson, K.H.; Tucker, J.B.; Stahl, L.E.; Simmons, J.K.; Fuller, C.; Beshiri, M.L.; Agarwal, S.; Fang, L.; Hynes, P.G.; Alilin, A.N.; et al. High-throughput screens identify hsp90 inhibitors as potent therapeutics that target inter-related growth and survival pathways in advanced prostate cancer. Sci. Rep. 2018, 8, 17239. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wei, T.; Ye, Z.; Orme, J.J.; Lin, D.; Sheng, H.; Fazli, L.; Jeffrey Karnes, R.; Jimenez, R.; Wang, L.; et al. A noncanonical ar addiction drives enzalutamide resistance in prostate cancer. Nat. Commun. 2021, 12, 1521. [Google Scholar] [CrossRef] [PubMed]
- Welti, J.; Sharp, A.; Yuan, W.; Dolling, D.; Nava Rodrigues, D.; Figueiredo, I.; Gil, V.; Neeb, A.; Clarke, M.; Seed, G.; et al. Targeting bromodomain and extra-terminal (bet) family proteins in castration-resistant prostate cancer (crpc). Clin. Cancer Res. 2018, 24, 3149–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoarau-Vechot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2d and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [Green Version]
- Velasco, V.; Shariati, S.A.; Esfandyarpour, R. Microtechnology-based methods for organoid models. Microsyst. Nanoeng. 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Djomehri, S.I.; Burman, B.; Gonzalez, M.E.; Takayama, S.; Kleer, C.G. A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer. J. Cell Commun. Signal. 2019, 13, 129–143. [Google Scholar] [CrossRef]
- Eder, T.; Eder, I.E. 3D hanging drop culture to establish prostate cancer organoids. Methods Mol. Biol. 2017, 1612, 167–175. [Google Scholar]
- Marsee, A.; Roos, F.J.M.; Verstegen, M.M.A.; Consortium, H.P.B.O.; Gehart, H.; de Koning, E.; Lemaigre, F.; Forbes, S.J.; Peng, W.C.; Huch, M.; et al. Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 2021, 28, 816–832. [Google Scholar] [CrossRef]
- Van Hemelryk, A.; van Weerden, W.M. Novel patient-derived 3D culture models to guide clinical decision-making in prostate cancer. Curr. Opin. Endocr. Metab. Res. 2020, 10, 7–15. [Google Scholar] [CrossRef]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Kaur, I.; Rawal, P.; Tripathi, D.M.; Vasudevan, A. Non-matrigel scaffolds for organoid cultures. Cancer Lett. 2021, 504, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Wu, A.; Min, S.; Shin, Y.C.; Fleming, R.Y.D.; Eckhardt, S.G.; Kim, H.J. Spatiotemporal gradient and instability of wnt induce heterogeneous growth and differentiation of human intestinal organoids. iScience 2020, 23, 101372. [Google Scholar] [CrossRef]
- Kopper, O.; de Witte, C.J.; Lohmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Obinata, D.; Sandhu, S.; Selth, L.A.; Wong, S.Q.; Porter, L.H.; Lister, N.; Pook, D.; Pezaro, C.J.; Goode, D.L.; et al. Patient-derived models of abiraterone- and enzalutamide-resistant prostate cancer reveal sensitivity to ribosome-directed therapy. Eur. Urol. 2018, 74, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Choo, N.; Ramm, S.; Luu, J.; Winter, J.M.; Selth, L.A.; Dwyer, A.R.; Frydenberg, M.; Grummet, J.; Sandhu, S.; Hickey, T.E.; et al. High-throughput imaging assay for drug screening of 3D prostate cancer organoids. SLAS Discov. 2021, 26, 1107–1124. [Google Scholar] [PubMed]
- Gulculer Balta, G.S.; Monzel, C.; Kleber, S.; Beaudouin, J.; Balta, E.; Kaindl, T.; Chen, S.; Gao, L.; Thiemann, M.; Wirtz, C.R.; et al. 3D cellular architecture modulates tyrosine kinase activity, thereby switching cd95-mediated apoptosis to survival. Cell Rep. 2019, 29, 2295–2306.e6. [Google Scholar] [CrossRef] [Green Version]
- Chambers, K.F.; Mosaad, E.M.; Russell, P.J.; Clements, J.A.; Doran, M.R. 3D cultures of prostate cancer cells cultured in a novel high-throughput culture platform are more resistant to chemotherapeutics compared to cells cultured in monolayer. PLoS ONE 2014, 9, e111029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santo, V.E.; Rebelo, S.P.; Estrada, M.F.; Alves, P.M.; Boghaert, E.; Brito, C. Drug screening in 3D in vitro tumor models: Overcoming current pitfalls of efficacy read-outs. Biotechnol. J. 2017, 12, 1–18. [Google Scholar] [CrossRef]
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016, 11, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Marques, R.B.; van Weerden, W.M.; Erkens-Schulze, S.; de Ridder, C.M.; Bangma, C.H.; Trapman, J.; Jenster, G. The human pc346 xenograft and cell line panel: A model system for prostate cancer progression. Eur. Urol. 2006, 49, 245–257. [Google Scholar] [CrossRef]
- van Soest, R.J.; van Royen, M.E.; de Morree, E.S.; Moll, J.M.; Teubel, W.; Wiemer, E.A.; Mathijssen, R.H.; de Wit, R.; van Weerden, W.M. Cross-resistance between taxanes and new hormonal agents abiraterone and enzalutamide may affect drug sequence choices in metastatic castration-resistant prostate cancer. Eur. J. Cancer 2013, 49, 3821–3830. [Google Scholar] [CrossRef]
- Marques, R.B.; Erkens-Schulze, S.; de Ridder, C.M.; Hermans, K.G.; Waltering, K.; Visakorpi, T.; Trapman, J.; Romijn, J.C.; van Weerden, W.M.; Jenster, G. Androgen receptor modifications in prostate cancer cells upon long-termandrogen ablation and antiandrogen treatment. Int. J. Cancer 2005, 117, 221–229. [Google Scholar] [CrossRef]
- Mout, L.; Moll, J.M.; Chen, M.; de Morree, E.S.; de Ridder, C.M.A.; Gibson, A.; Stuurman, D.; Aghai, A.; Erkens-Schulze, S.; Mathijssen, R.H.J.; et al. Androgen receptor signalling impairs docetaxel efficacy in castration-resistant prostate cancer. Br. J. Cancer 2020, 123, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Mout, L.; van Royen, M.E.; de Ridder, C.M.A.; Stuurman, D.; van de Geer, W.S.; Marques, R.B.; French, P.J.; van de Werken, H.J.G.; Mathijssen, R.H.J.; de Wit, R.; et al. Continued androgen signaling inhibition improves cabazitaxel efficacy in prostate cancer. EBioMedicine 2021. [Google Scholar]
- Berges, N.; Arens, K.; Kreusch, V.; Fischer, R.; Di Fiore, S. Toward discovery of novel microtubule targeting agents: A snap-tag-based high-content screening assay for the analysis of microtubule dynamics and cell cycle progression. SLAS Discov. 2017, 22, 387–398. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 18 October 2021).
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Vrignaud, P.; Semiond, D.; Lejeune, P.; Bouchard, H.; Calvet, L.; Combeau, C.; Riou, J.F.; Commercon, A.; Lavelle, F.; Bissery, M.C. Preclinical antitumor activity of cabazitaxel, a semisynthetic taxane active in taxane-resistant tumors. Clin. Cancer Res. 2013, 19, 2973–2983. [Google Scholar] [CrossRef] [Green Version]
- Narasimhan, V.; Wright, J.A.; Churchill, M.; Wang, T.; Rosati, R.; Lannagan, T.R.M.; Vrbanac, L.; Richardson, A.B.; Kobayashi, H.; Price, T.; et al. Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin. Cancer Res. 2020, 26, 3662–3670. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef]
- Mosaad, E.; Chambers, K.; Futrega, K.; Clements, J.; Doran, M.R. Using high throughput microtissue culture to study the difference in prostate cancer cell behavior and drug response in 2D and 3D co-cultures. BMC Cancer 2018, 18, 592. [Google Scholar] [CrossRef]
- Muguruma, M.; Teraoka, S.; Miyahara, K.; Ueda, A.; Asaoka, M.; Okazaki, M.; Kawate, T.; Kuroda, M.; Miyagi, Y.; Ishikawa, T. Differences in drug sensitivity between two-dimensional and three-dimensional culture systems in triple-negative breast cancer cell lines. Biochem. Biophys. Res. Commun. 2020, 533, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Close, D.A.; Camarco, D.P.; Johnston, P.A. High-content screening comparison of cancer drug accumulation and distribution in two-dimensional and three-dimensional culture models of head and neck cancer. Assay Drug Dev. Technol. 2018, 16, 27–50. [Google Scholar] [CrossRef]
- Au, J.L.; Jang, S.H.; Zheng, J.; Chen, C.T.; Song, S.; Hu, L.; Wientjes, M.G. Determinants of drug delivery and transport to solid tumors. J. Control. Release 2001, 74, 31–46. [Google Scholar] [CrossRef]
- Edmondson, R.; Adcock, A.F.; Yang, L. Influence of matrices on 3D-cultured prostate cancer cells’ drug response and expression of drug-action associated proteins. PLoS ONE 2016, 11, e0158116. [Google Scholar] [CrossRef]
- Stowers, R.S.; Shcherbina, A.; Israeli, J.; Gruber, J.J.; Chang, J.; Nam, S.; Rabiee, A.; Teruel, M.N.; Snyder, M.P.; Kundaje, A.; et al. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng. 2019, 3, 1009–1019. [Google Scholar] [CrossRef]
- Scuderi, S.; Altobelli, G.G.; Cimini, V.; Coppola, G.; Vaccarino, F.M. Cell-to-cell adhesion and neurogenesis in human cortical development: A study comparing 2D monolayers with 3D organoid cultures. Stem Cell Rep. 2021, 16, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Tanaka, H.; Nakayama, T.; Taniura, N.; Miyake, T.; Tani, M.; Kushima, R.; Yamamoto, G.; Sugihara, H.; Mukaisho, K.I. Similarities and differences in metabolites of tongue cancer cells among two- and three-dimensional cultures and xenografts. Cancer Sci. 2021, 112, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Yang, M.; Fan, Z.; Li, S.; Gao, T.; Fang, Z. Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J. Exp. Clin. Cancer Res. 2015, 34, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Hemelryk, A.; Mout, L.; Erkens-Schulze, S.; French, P.J.; van Weerden, W.M.; van Royen, M.E. Modeling Prostate Cancer Treatment Responses in the Organoid Era: 3D Environment Impacts Drug Testing. Biomolecules 2021, 11, 1572. https://doi.org/10.3390/biom11111572
Van Hemelryk A, Mout L, Erkens-Schulze S, French PJ, van Weerden WM, van Royen ME. Modeling Prostate Cancer Treatment Responses in the Organoid Era: 3D Environment Impacts Drug Testing. Biomolecules. 2021; 11(11):1572. https://doi.org/10.3390/biom11111572
Chicago/Turabian StyleVan Hemelryk, Annelies, Lisanne Mout, Sigrun Erkens-Schulze, Pim J. French, Wytske M. van Weerden, and Martin E. van Royen. 2021. "Modeling Prostate Cancer Treatment Responses in the Organoid Era: 3D Environment Impacts Drug Testing" Biomolecules 11, no. 11: 1572. https://doi.org/10.3390/biom11111572
APA StyleVan Hemelryk, A., Mout, L., Erkens-Schulze, S., French, P. J., van Weerden, W. M., & van Royen, M. E. (2021). Modeling Prostate Cancer Treatment Responses in the Organoid Era: 3D Environment Impacts Drug Testing. Biomolecules, 11(11), 1572. https://doi.org/10.3390/biom11111572






