Cdc42-Specific GTPase-Activating Protein Rga1 Squelches Crosstalk between the High-Osmolarity Glycerol (HOG) and Mating Pheromone Response MAPK Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Plasmids and Growth Conditions
2.2. Genetic Selection for Mutants that Result in Crosstalk
2.3. Microscopy of Fluorescent Proteins in Yeast Cells
2.4. Cell Extracts and Immunoblotting
2.5. Protein Purification and In Vitro Protein Kinase Assays
2.6. Immunoprecipitation and Calf Intestinal Phosphatase Treatment of HA-Rga1
3. Results
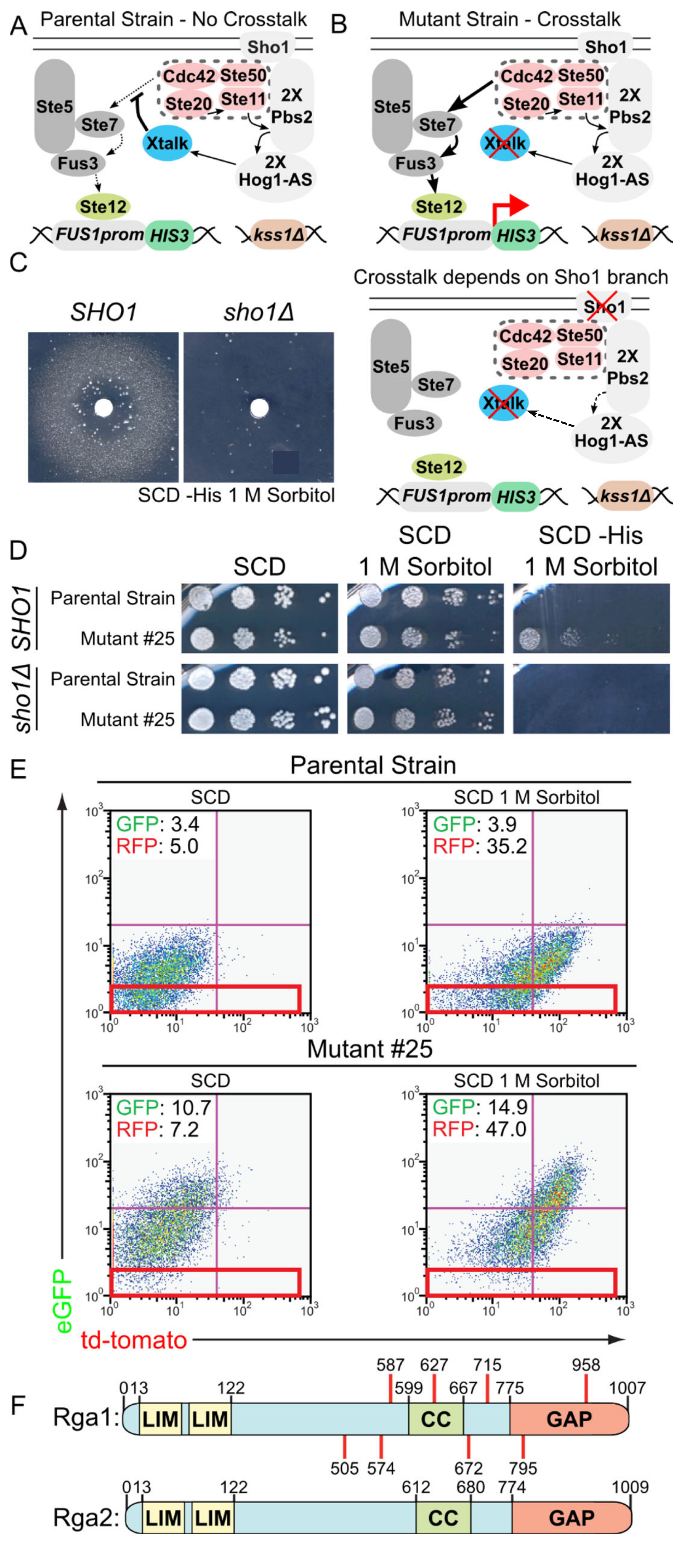
3.1. A Genetic Selection to Recover Mutants that Allow Mating Pathway Activation under Hypertonic Conditions in the Presence of Active Hog1
3.2. A Role for Rga1 in Blocking Crosstalk
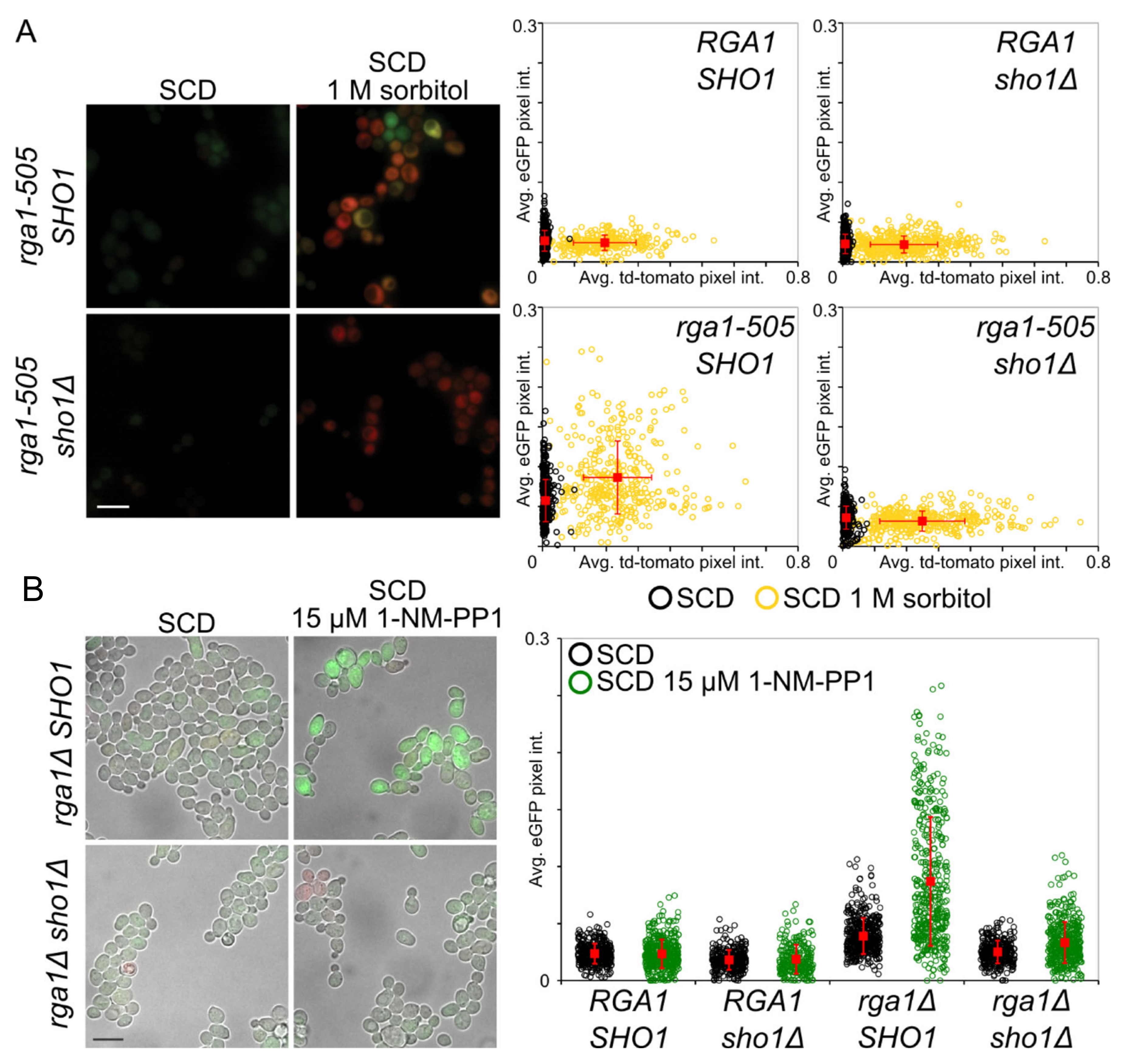
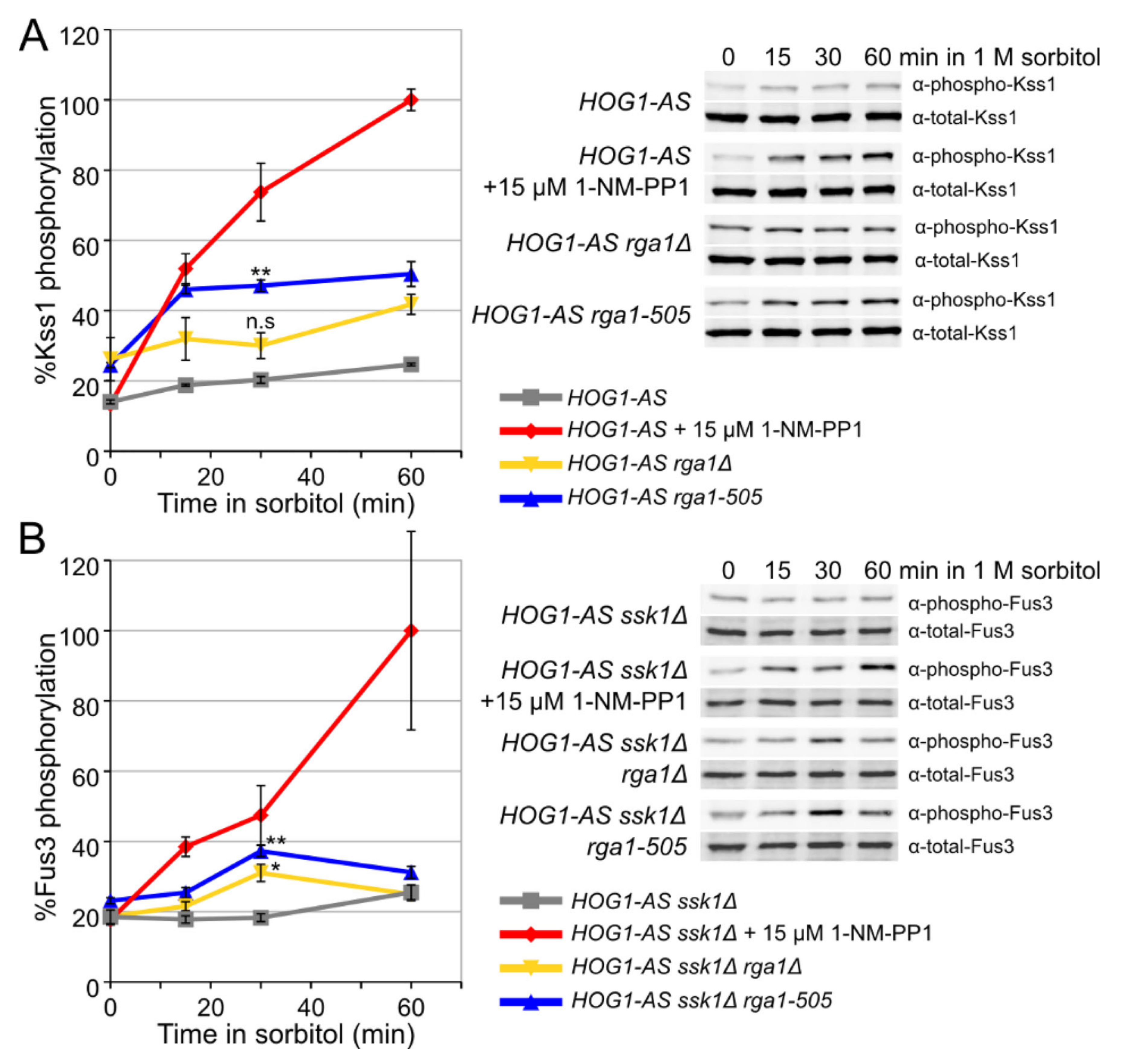
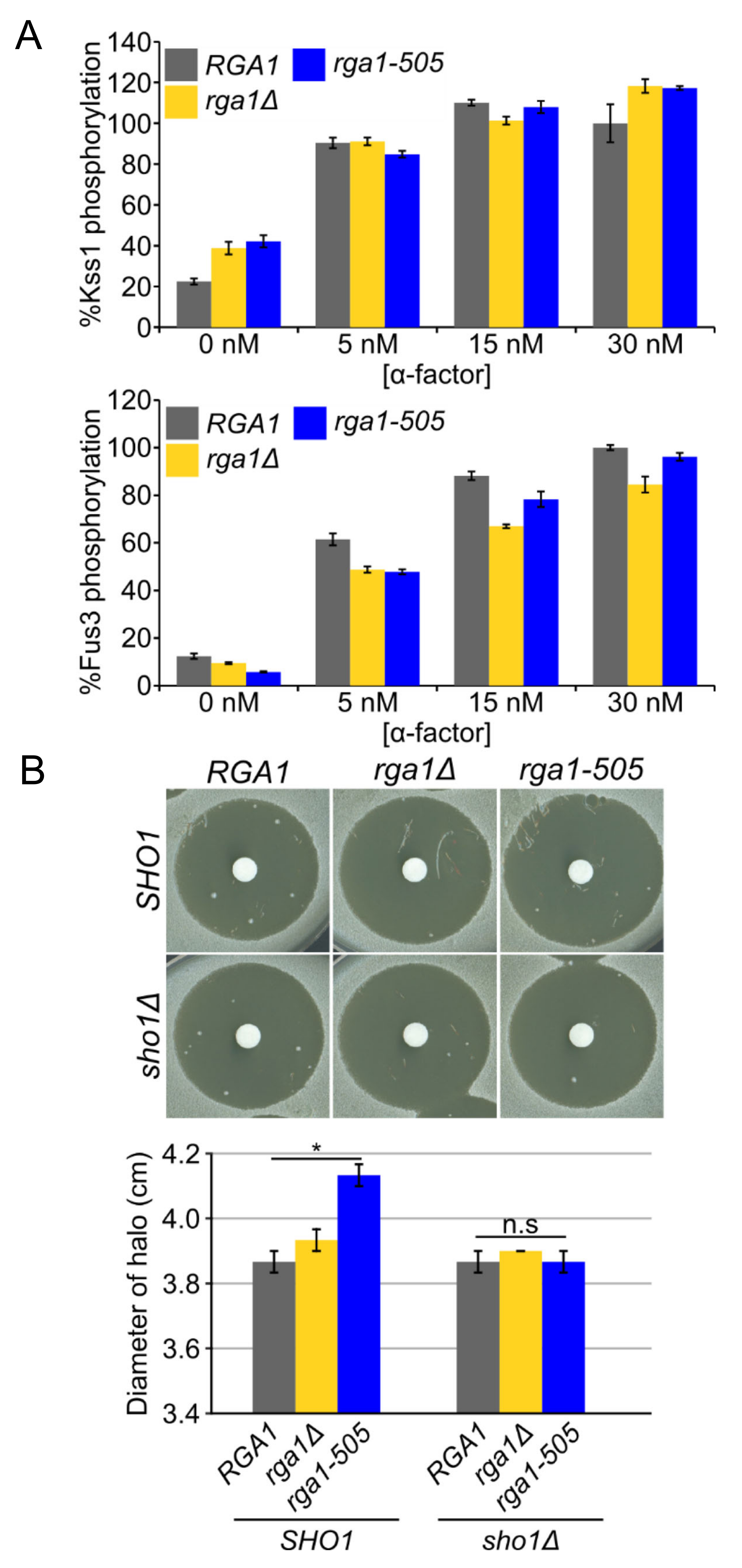
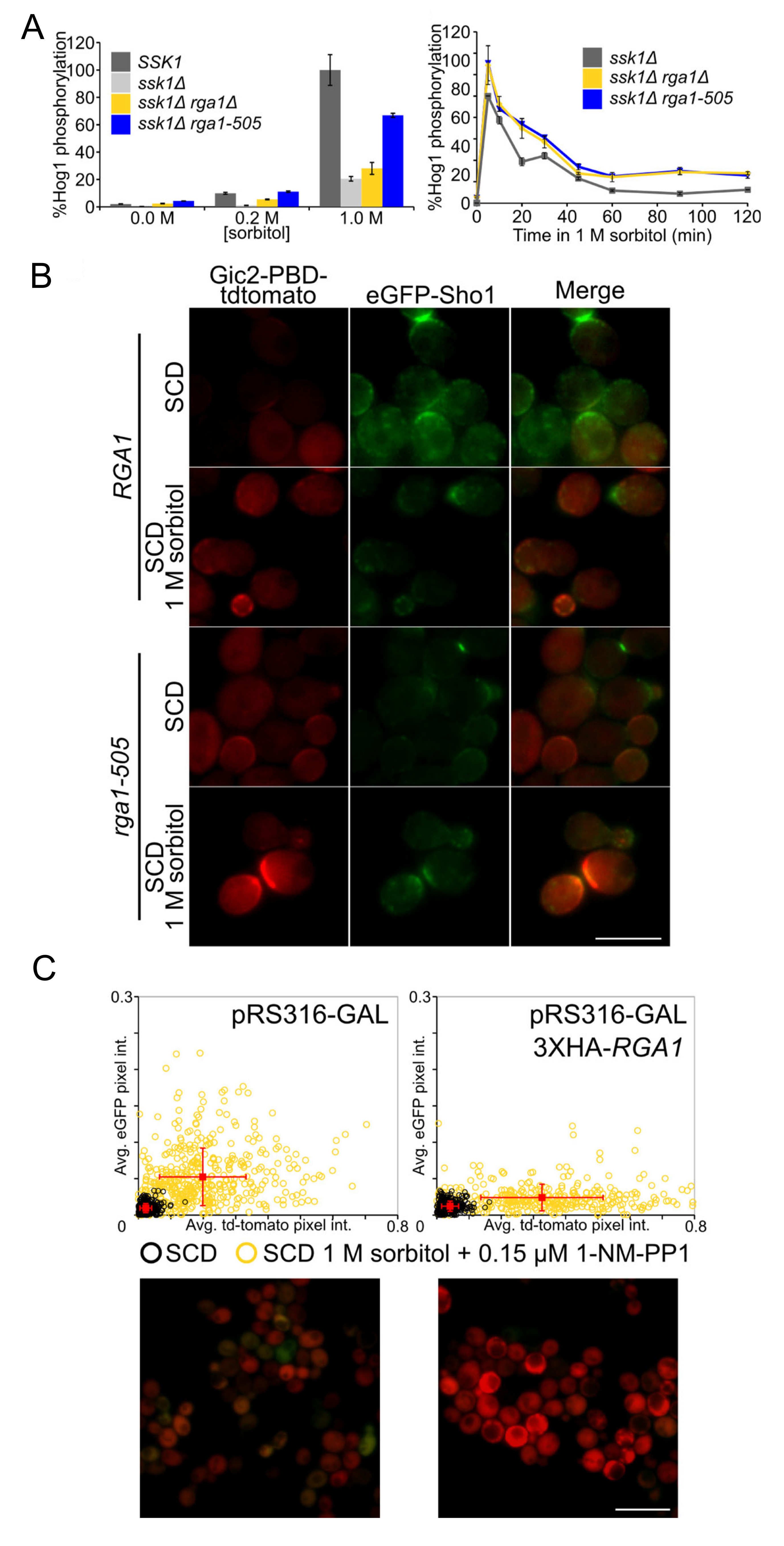
3.3. Rga1 Negatively Regulates the SHO1 Branch of the HOG Pathway
3.4. Rga1 Is a Hog1 Substrate In Vitro
3.5. Hog1 Phosphorylation of Rga1 Is Not Required to Prevent Crosstalk
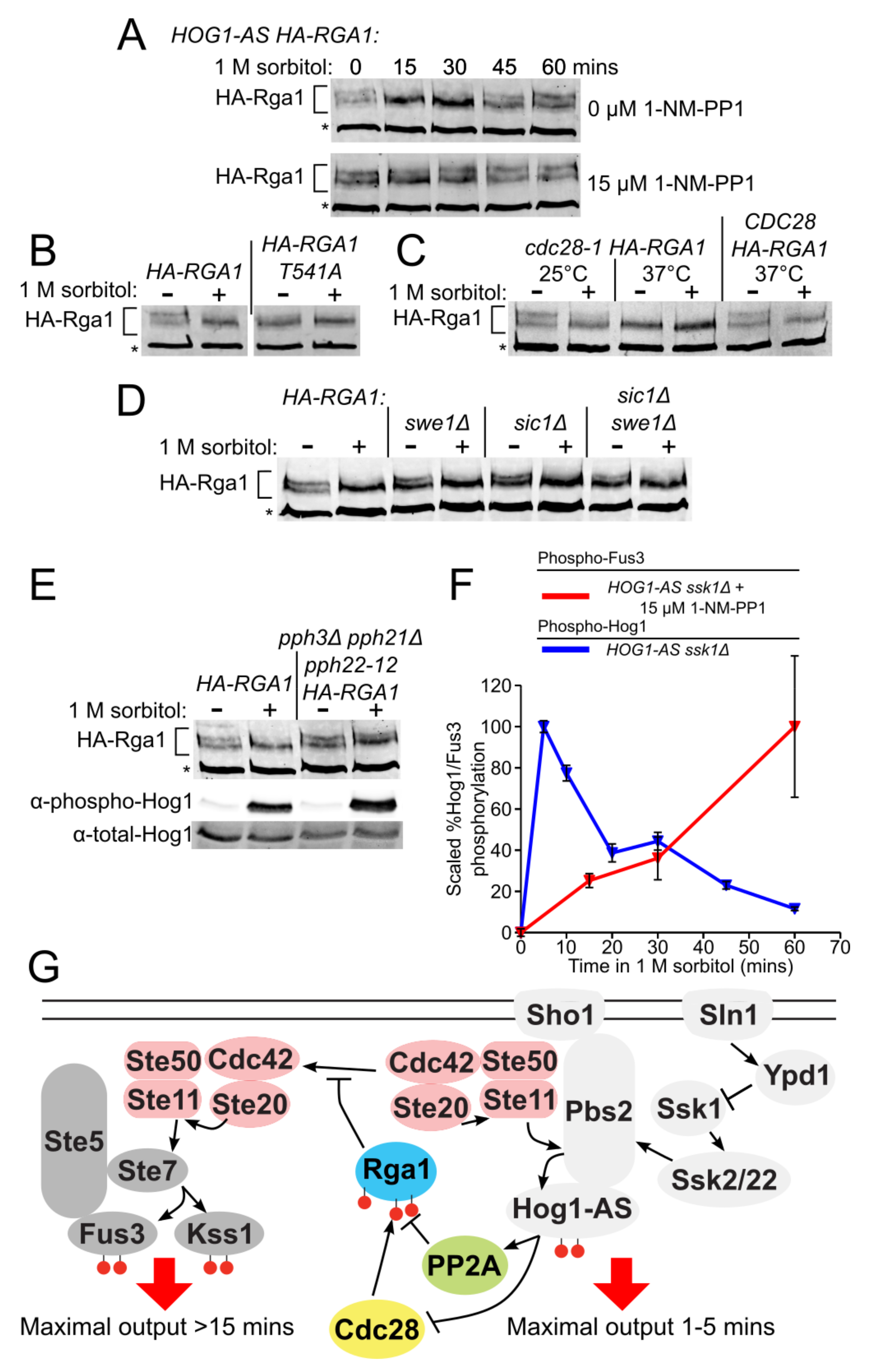
3.6. Rga1 Undergoes Cell Cycle-Dependent Cdc28/Cdk1-Mediated Phosphorylation
3.7. Phosphorylation of Rga1 by Cdk1 Is Regulated Indirectly by Hog1
3.8. PP2A Is the Phosphatase that Counteracts the Cdk1-Dependent Phosphorylation of Rga1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.E.; Thorner, J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA) Bioenerg. 2007, 1773, 1311–1340. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Chen, W.; Cobb, M. Differential regulation and properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Saito, H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 2010, 13, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Witzel, F.; Maddison, L.E.; Blüthgen, N. How scaffolds shape MAPK signaling: What we know and opportunities for systems approaches. Front. Physiol. 2012, 3, 475. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Hohmann, S. Synthetic biology: Lessons from engineering yeast MAPK signalling pathways. Mol. Microbiol. 2013, 88, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Peti, W.; Page, R. Molecular basis of MAP kinase regulation. Protein Sci. 2013, 22, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.-J. Compromised MAPK signaling in human diseases: An update. Arch. Toxicol. 2015, 89, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Van Drogen, F.; Dard, N.; Pelet, S.; Lee, S.S.; Mishra, R.; Srejić, N.; Peter, M. Crosstalk and spatiotemporal regulation between stress-induced MAP kinase pathways and pheromone signaling in budding yeast. Cell Cycle 2020, 19, 1707–1715. [Google Scholar] [CrossRef]
- Vázquez-Ibarra, A.; Rodríguez-Martínez, G.; Guerrero-Serrano, G.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. Negative feedback-loop mechanisms regulating HOG- and pheromone-MAPK signaling in yeast. Curr. Genet. 2020, 66, 867–880. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to hyperosmotic stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef]
- Brewster, J.L.; Gustin, M. Hog1: 20 years of discovery and impact. Sci. Signal. 2014, 7, re7. [Google Scholar] [CrossRef] [PubMed]
- Tatebayashi, K.; Yamamoto, K.; Tomida, T.; Nishimura, A.; Takayama, T.; Oyama, M.; Kozuka-Hata, H.; Adachi-Akahane, S.; Tokunaga, Y.; Saito, H. Osmostress enhances activating phosphorylation of Hog1 MAP kinase by mono-phosphorylated Pbs2 MAP 2K. EMBO J. 2020, 39, e103444. [Google Scholar] [CrossRef] [PubMed]
- Tatebayashi, K.; Yamamoto, K.; Nagoya, M.; Takayama, T.; Nishimura, A.; Sakurai, M.; Momma, T.; Saito, H. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat. Commun. 2015, 6, 6975. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Tatebayashi, K.; Nishimura, A.; Yamamoto, K.; Yang, H.-Y.; Saito, H. Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci. Signal. 2014, 7, ra21. [Google Scholar] [CrossRef]
- Takayama, T.; Yamamoto, K.; Saito, H.; Tatebayashi, K. Interaction between the transmembrane domains of Sho1 and Opy2 enhances the signaling efficiency of the Hog1 MAP kinase cascade in Saccharomyces cerevisiae. PLoS ONE 2019, 14, e0211380. [Google Scholar] [CrossRef]
- Chiou, J.G.; Balasubramanian, M.K.; Lew, D.J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 2017, 33, 77–101. [Google Scholar] [CrossRef]
- Basu, S.; González, B.; Li, B.; Kimble, G.; Kozminski, K.G.; Cullen, P.J. Functions for Cdc42p BEM adaptors in regulating a differentiation-type MAP kinase pathway. Mol. Biol. Cell 2020, 31, 491–510. [Google Scholar] [CrossRef]
- Miller, K.E.; Kang, P.J.; Park, H.-O. Regulation of Cdc42 for polarized growth in budding yeast. Microb. Cell 2020, 7, 175–189. [Google Scholar] [CrossRef]
- Truckses, D.M.; Bloomekatz, J.E.; Thorner, J. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Jansen, G.; Zhang, J.; Thomas, D.; Whiteway, M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006, 20, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.J.; Warner, N.; Maini, J.; Tung, K.W.C.; Zakaria, H.; Pawson, T.; Donaldson, L.W. Saccharomyces cerevisiae Ste50 binds the MAPKKK Ste11 through a head-to-tail SAM domain interaction. J. Mol. Biol. 2006, 356, 142–154. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Yamamoto, K.; Tanaka, K.; Tomida, T.; Maruoka, T.; Kasukawa, E.; Saito, H. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006, 25, 3033–3044. [Google Scholar] [CrossRef]
- Tatebayashi, K.; Tanaka, K.; Yang, H.-Y.; Yamamoto, K.; Matsushita, Y.; Tomida, T.; Imai, M.; Saito, H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007, 26, 3521–3533. [Google Scholar] [CrossRef] [PubMed]
- Ekiel, I.; Sulea, T.; Jansen, G.; Kowalik, M.; Minailiuc, O.; Cheng, J.; Harcus, D.; Cygler, M.; Whiteway, M.; Wu, C. Binding the atypical RA domain of Ste50p to the unfolded Opy2p cytoplasmic tail is essential for the high-osmolarity glycerol pathway. Mol. Biol. Cell 2009, 20, 5117–5126. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Tatebayashi, K.; Tanaka, K.; Saito, H. Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol. Cell 2010, 40, 87–98. [Google Scholar] [CrossRef]
- Nishimura, A.; Yamamoto, K.; Oyama, M.; Kozuka-Hata, H.; Saito, H.; Tatebayashi, K. Scaffold protein Ahk1, which associates with Hkr1, Sho1, Ste11, and Pbs2, inhibits cross talk signaling from the Hkr1 osmosensor to the Kss1 mitogen-activated protein kinase. Mol. Cell. Biol. 2016, 36, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Lamson, R.E.; Winters, M.J.; Pryciak, P.M. Cdc42 regulation of kinase activity and signaling by the Yyeast p21-activated kinase Ste20. Mol. Cell. Biol. 2002, 22, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Pryciak, P.M. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell 2007, 18, 4945–4956. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.; Andrianopoulos, A. Ste20-related kinases: Effectors of signaling and morphogenesis in fungi. Trends Microbiol. 2011, 19, 400–410. [Google Scholar] [CrossRef]
- van Drogen, F.; O’Rourke, S.M.; Stucke, V.M.; Jaquenoud, M.; Neiman, A.; Peter, M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000, 10, 630–639. [Google Scholar] [CrossRef]
- Raitt, D.C.; Posas, F.; Saito, H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000, 19, 4623–4631. [Google Scholar] [CrossRef]
- Maeda, T.; Takekawa, M.; Saito, H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 1995, 269, 554–558. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Bhattacharyya, R.P.; Nittler, M.; A Lim, W. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 2004, 14, 825–832. [Google Scholar] [CrossRef]
- Posas, F.; Saito, H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science 1997, 276, 1702–1705. [Google Scholar] [CrossRef]
- Escoté, X.; Zapater, M.; Clotet, J.; Posas, F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004, 6, 997–1002. [Google Scholar] [CrossRef]
- Chang, Y.L.; Tseng, S.F.; Huang, Y.C.; Shen, Z.J.; Hsu, P.H.; Hsieh, M.H.; Yang, C.W.; Tognetti, S.; Canal, B.; Subirana, L.; et al. Yeast Cip1 is activated by environmental stress to inhibit Cdk1-G1 cyclins via Mcm1 and Msn2/4. Nat. Commun. 2017, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Yaakov, G.; Duch, A.; García-Rubio, M.; Clotet, J.; Jimenez, J.; Aguilera, A.; Posas, F. The stress-activated protein kinase Hog1 mediates S phase delay in response to osmostress. Mol. Biol. Cell 2009, 20, 3572–3582. [Google Scholar] [CrossRef] [PubMed]
- Clotet, J.; Escoté, X.; Adrover, M.À.; Yaakov, G.; Gari, E.; Aldea, M.; de Nadal, E.; Posas, F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006, 25, 2338–2346. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, S.; Jiménez, J.; Viganò, M.; Duch, A.; Queralt, E.; de Nadal, E.; Posas, F. Hog1 activation delays mitotic exit via phosphorylation of Net1. Proc. Natl. Acad. Sci. USA 2020, 117, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015, 61, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Westfall, P.J.; Patterson, J.C.; Chen, R.E.; Thorner, J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl. Acad. Sci. USA 2008, 105, 12212–12217. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jeschke, G.; Roelants, F.M.; Thorner, J.; Turk, B.E. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 2012, 32, 4705–4717. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.; Roelants, F.M.; Timmons, G.; Leskoske, K.L.; Thorner, J. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. Elife 2015, 4, e09336. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, C.G.; Thorner, J. Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J. Biol. Chem. 2016, 291, 7788–7795. [Google Scholar] [CrossRef] [PubMed]
- Frawley, D.; Bayram, Ö. The pheromone response module, a mitogen-activated protein kinase pathway implicated in the regulation of fungal development, secondary metabolism and pathogenicity. Fungal Genet. Biol. 2020, 144, 103469. [Google Scholar] [CrossRef]
- Leeuw, T.; Wu, C.; Schrag, J.D.; Whiteway, M.; Thomas, D.Y.; Leberer, E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 1998, 391, 191–195. [Google Scholar] [CrossRef]
- Butty, A.-C.; Pryciak, P.M.; Huang, L.S.; Herskowitz, I.; Peter, M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 1998, 282, 1511–1516. [Google Scholar] [CrossRef]
- Gartner, A.; Jovanović, A.; Jeoung, D.-I.; Bourlat, S.; Cross, F.R.; Ammerer, G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol. Cell. Biol. 1998, 18, 3681–3691. [Google Scholar] [CrossRef]
- Doncic, A.; Atay, O.; Valk, E.; Grande, A.; Bush, A.; Vasen, G.; Colman-Lerner, A.; Loog, M.; Skotheim, J.M. Compartmentalization of a bistable switch enables memory to cross a feedback-driven transition. Cell 2015, 160, 1182–1195. [Google Scholar] [CrossRef]
- Elion, E.A. The Ste5p scaffold. J. Cell Sci. 2001, 114, 3967–3978. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F. The regulation of filamentous growth in yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Sette, C.; Inouye, C.J.; Stroschein, S.L.; Iaquinta, P.J.; Thorner, J.; Ramm, G.; Slot, J.W.; James, D.E.; Stoorvogel, W. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 2000, 11, 4033–4049. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Good, M.; Tang, G.; Singleton, J.; Reményi, A.; Lim, W.A. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 2009, 136, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.G. Molecular mechanisms of chemotropism and cell fusion in unicellular fungi. J. Cell Sci. 2019, 132, jcs230706. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.C.; Klimenko, E.S.; Thorner, J. Single-cell analysis reveals that insulation maintains signaling specificity between two yeast MAPK pathways with common components. Sci. Signal. 2010, 3, ra75. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, S.M.; Herskowitz, I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998, 12, 2874–2886. [Google Scholar] [CrossRef]
- Westfall, P.J.; Thorner, J. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: Use of an analog-sensitive HOG1 allele. Eukaryot. Cell 2006, 5, 1215–1228. [Google Scholar] [CrossRef]
- Nagiec, M.J.; Dohlman, H.G. Checkpoints in a yeast differentiation pathway coordinate signaling during hyperosmotic stress. PLoS Genet. 2012, 8, e1002437. [Google Scholar] [CrossRef]
- Bardwell, L. Mechanisms of MAPK signalling specificity. Biochem. Soc. Trans. 2006, 34, 837–841. [Google Scholar] [CrossRef]
- Del Vecchio, D.; Ninfa, A.J.; Sontag, E.D. Modular cell biology: Retroactivity and insulation. Mol. Syst. Biol. 2008, 4, 161. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Ballon, D.R.; Flanary, P.L.; Gladue, D.; Konopka, J.; Dohlman, H.G.; Thorner, J. DEP-domain-mediated regulation of GPCR signaling responses. Cell 2006, 126, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.Y.; Pinkham, J.L. Tightly regulated, beta-estradiol dose-dependent expression system for yeast. Biotechniques 2000, 29, 1226–1231. [Google Scholar] [CrossRef]
- McIsaac, R.S.; Silverman, S.J.; McClean, M.; Gibney, P.A.; Macinskas, J.; Hickman, M.; Petti, A.A.; Botstein, D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 2011, 22, 4447–4459. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Boeke, J.D.; La Croute, F.; Fink, G.R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Genet. Genom. 1984, 197, 345–346. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Tkach, J.M.; Yimit, A.; Lee, A.Y.; Riffle, M.; Costanzo, M.; Jaschob, D.; Hendry, J.A.; Ou, J.; Moffat, J.; Boone, C.; et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012, 14, 966–976. [Google Scholar] [CrossRef]
- Baum, P.; Thorner, J.; Honig, L. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1978, 75, 4962–4966. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.J.; Li, M.; Brinkworth, C.S.; Paulson, J.L.; Wang, D.; Hübner, A.; Chou, W.-H.; Davis, R.J.; Burlingame, A.L.; Messing, R.; et al. A semisynthetic epitope for kinase substrates. Nat. Methods 2007, 4, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Horecka, J.; Sprague, G.F. Use of imidazoleglycerolphosphate dehydratase (His3) as a biological reporter in yeast. Methods Enzymol. 2000, 326, 107–119. [Google Scholar] [CrossRef]
- Jauert, P.A.; Jensen, L.E.; Kirkpatrick, D.T. A novel yeast genomic DNA library on a geneticin-resistance vector. Yeast 2005, 22, 653–657. [Google Scholar] [CrossRef]
- Smith, G.R.; Givan, S.A.; Cullen, P.; Sprague, G.F., Jr. GTPase-activating proteins for Cdc42. Eukaryot. Cell 2002, 1, 469–480. [Google Scholar] [CrossRef]
- Pérez, P.; Rincón, S.A. Rho GTPases: Regulation of cell polarity and growth in yeasts. Biochem. J. 2010, 426, 243–253. [Google Scholar] [CrossRef]
- Jonasson, E.M.; Rossio, V.; Hatakeyama, R.; Abe, M.; Ohya, Y.; Yoshida, S. Zds1/Zds2–PP2ACdc55 complex specifies signaling output from Rho1 GTPase. J. Cell Biol. 2016, 212, 51–61. [Google Scholar] [CrossRef]
- Gingras, R.; Lwin, K.M.; Miller, A.; Bretscher, A. Yeast Rgd3 is a phospho-regulated F-BAR–containing RhoGAP involved in the regulation of Rho3 distribution and cell morphology. Mol. Biol. Cell 2020, 31, 2570–2582. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Abe, M.; Ohya, Y. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 2001, 18, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Fitch, P.G.; Gammie, A.; Lee, D.J.; de Candal, V.B.; Rose, M.D. Lrg1p is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics 2004, 168, 733–746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knaus, M.; Wiget, P.; Shimada, Y.; Peter, M. Control of cell polarity in response to intra- and extracellular signals in budding yeast. Novartis Found. Symp. 2005, 269, 47–54. [Google Scholar]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef]
- Stevenson, B.J.; Ferguson, B.; De Virgilio, C.; Bi, E.; Pringle, J.R.; Ammerer, G.; Sprague, G.F. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 1995, 9, 2949–2963. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F. The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 884–894. [Google Scholar] [CrossRef]
- Pinsky, B.A.; Kotwaliwale, C.V.; Tatsutani, S.Y.; Breed, C.A.; Biggins, S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 2006, 26, 2648–2660. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 2018, 6, 192–205.e3. [Google Scholar] [CrossRef] [PubMed]
- Parmar, J.H.; Bhartiya, S.; Venkatesh, K.V. A model-based study delineating the roles of the two signaling branches of Saccharomyces cerevisiae, Sho1 and Sln1, during adaptation to osmotic stress. Phys. Biol. 2009, 6, 036019. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.L.; Deschenes, R.; Guan, K.L. Differential regulation of Fus3 MAP kinase by tyrosine-specific phosphatases Ptp2/Ptp3 and dual-specificity phosphatase Msg5 in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- González-Rubio, G.; Fernández-Acero, T.; Martín, H.; Molina, M. Mitogen-activated protein kinase phosphatases (MKPs) in fungal signaling: Conservation, function, and regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef]
- Reneke, J.E.; Blumer, K.J.; Courchesne, W.E.; Thorner, J. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell 1988, 55, 221–234. [Google Scholar] [CrossRef]
- O’Rourke, S.M.; Herskowitz, I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 2004, 15, 532–542. [Google Scholar] [CrossRef]
- Brown, J.L.; Jaquenoud, M.; Gulli, M.-P.; Chant, J.; Peter, M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997, 11, 2972–2982. [Google Scholar] [CrossRef]
- Chen, G.-C.; Kim, Y.-J.; Chan, C.S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 2958–2971. [Google Scholar] [CrossRef] [PubMed]
- Mok, J.; Kim, P.M.; Lam, H.Y.K.; Piccirillo, S.; Zhou, X.; Jeschke, G.R.; Sheridan, D.L.; Parker, S.A.; Desai, V.; Jwa, M.; et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 2010, 3, ra12. [Google Scholar] [CrossRef]
- Albuquerque, C.P.; Smolka, M.B.; Payne, S.; Bafna, V.; Eng, J.; Zhou, H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteom. 2008, 7, 1389–1396. [Google Scholar] [CrossRef]
- Holt, L.; Tuch, B.B.; Villen, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef]
- Breitkreutz, A.; Choi, H.; Sharom, J.R.; Boucher, L.; Neduva, V.; Larsen, B.; Lin, Z.-Y.; Breitkreutz, B.-J.; Stark, C.; Liu, G.; et al. A Global protein kinase and phosphatase interaction network in yeast. Science 2010, 328, 1043–1046. [Google Scholar] [CrossRef]
- Soulard, A.; Cremonesi, A.; Moes, S.; Schütz, F.; Jenö, P.; Hall, M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 2010, 21, 3475–3486. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villen, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef]
- MacGilvray, M.E.; Shishkova, E.; Place, M.; Wagner, E.R.; Coon, J.J.; Gasch, A.P. Phosphoproteome response to dithiothreitol reveals unique versus shared features of Saccharomyces cerevisiae stress responses. J. Proteome Res. 2020, 19, 3405–3417. [Google Scholar] [CrossRef] [PubMed]
- Lanz, M.C.; Yugandhar, K.; Gupta, S.; Sanford, E.J.; Faça, V.M.; Vega, S.; Joiner, A.M.N.; Fromme, J.C.; Yu, H.; Smolka, M.B. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 2021, 22, e51121. [Google Scholar] [CrossRef] [PubMed]
- Kanshin, E.; Bergeron-Sandoval, L.-P.; Isik, S.S.; Thibault, P.; Michnick, S.W. A cell-signaling network temporally resolves specific versus promiscuous phosphorylation. Cell Rep. 2015, 10, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Janschitz, M.; Romanov, N.; Varnavides, G.; Hollenstein, D.M.; Gérecová, G.; Ammerer, G.; Hartl, M.; Reiter, W. Novel interconnections of HOG signaling revealed by combined use of two proteomic software packages. Cell Commun. Signal. 2019, 17, 1–17. [Google Scholar] [CrossRef]
- Sopko, R.; Huang, D.; Smith, J.C.; Figeys, D.; Andrews, B.J. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007, 26, 4487–4500. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, X.-D.; Howell, A.S.; Bose, I.; Lew, D.J.; Bi, E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein–mediated zone of inhibition. J. Cell Biol. 2007, 179, 1375–1384. [Google Scholar] [CrossRef]
- Ly, N.; Cyert, M.S. Calcineurin, the Ca2+-dependent phosphatase, regulates Rga2, a Cdc42 GTPase-activating protein, to modulate pheromone signaling. Mol. Biol. Cell 2017, 28, 576–586. [Google Scholar] [CrossRef]
- Shulewitz, M.J.; Inouye, C.J.; Thorner, J. Hsl7 localizes to the septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of the protein kinase Cdc28 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 7123–7137. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Evans, D.R.H.; Stark, M. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics 1997, 145, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Behar, M.; Parnell, S.C.; Torres, M.P.; Borchers, C.H.; Elston, T.C.; Dohlman, H.G. A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr. Biol. 2007, 17, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Zeng, Y.; Elston, T.C.; Dohlman, H.G. Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J. Biol. Chem. 2008, 283, 33798–33802. [Google Scholar] [CrossRef] [PubMed]
- Shock, T.R.; Thompson, J.; Yates, J.R., 3rd; Madhani, H.D. Hog1 mitogen-activated protein kinase (MAPK) interrupts signal transduction between the Kss1 MAPK and the Tec1 transcription factor to maintain pathway specificity. Eukaryot. Cell 2009, 8, 606–616. [Google Scholar] [CrossRef]
- Johnson, D.I. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999, 63, 54–105. [Google Scholar] [CrossRef] [PubMed]
- Hopfield, J.J. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA 1974, 71, 4135–4139. [Google Scholar] [CrossRef] [PubMed]
- Reha-Krantz, L.J. Regulation of DNA polymerase exonucleolytic proofreading activity: Studies of bacteriophage T4 “anti-mutator” DNA polymerases. Genetics 1998, 148, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Qian, H. Sensitivity and specificity amplification in signal transduction. Cell Biophys. 2003, 39, 45–60. [Google Scholar] [CrossRef]
- English, J.G.; Shellhammer, J.P.; Malahe, M.; McCarter, P.C.; Elston, T.C.; Dohlman, H.G. MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast. Sci. Signal. 2015, 8, ra5. [Google Scholar] [CrossRef]
- Pelet, S.; Rudolf, F.; Nadal-Ribelles, M.; de Nadal, E.; Posas, F.; Peter, M. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science 2011, 332, 732–735. [Google Scholar] [CrossRef]
- Han, J.; Lee, J.; Bibbs, L.; Ulevitch, R. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef]
- Liu, Q.; Hofmann, P.A. Modulation of protein phosphatase 2A by adenosine A1 receptors in cardiomyocytes: Role for p38 MAPK. Am. J. Physiol. Circ. Physiol. 2003, 285, H97–H103. [Google Scholar] [CrossRef]
- Grethe, S.; Porn-Ares, M.I. p38 MAPK regulates phosphorylation of Bad via PP2A-dependent suppression of the MEK1/2-ERK1/2 survival pathway in TNF-alpha induced endothelial apoptosis. Cell Signal 2006, 18, 531–540. [Google Scholar] [CrossRef]
- Bodenmiller, B.; Campbell, D.; Gerrits, B.; Lam, H.; Jovanovic, M.; Picotti, P.; Schlapbach, R.; Aebersold, R. PhosphoPep—A database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008, 26, 1339–1340. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patterson, J.C.; Goupil, L.S.; Thorner, J. Cdc42-Specific GTPase-Activating Protein Rga1 Squelches Crosstalk between the High-Osmolarity Glycerol (HOG) and Mating Pheromone Response MAPK Pathways. Biomolecules 2021, 11, 1530. https://doi.org/10.3390/biom11101530
Patterson JC, Goupil LS, Thorner J. Cdc42-Specific GTPase-Activating Protein Rga1 Squelches Crosstalk between the High-Osmolarity Glycerol (HOG) and Mating Pheromone Response MAPK Pathways. Biomolecules. 2021; 11(10):1530. https://doi.org/10.3390/biom11101530
Chicago/Turabian StylePatterson, Jesse C., Louise S. Goupil, and Jeremy Thorner. 2021. "Cdc42-Specific GTPase-Activating Protein Rga1 Squelches Crosstalk between the High-Osmolarity Glycerol (HOG) and Mating Pheromone Response MAPK Pathways" Biomolecules 11, no. 10: 1530. https://doi.org/10.3390/biom11101530
APA StylePatterson, J. C., Goupil, L. S., & Thorner, J. (2021). Cdc42-Specific GTPase-Activating Protein Rga1 Squelches Crosstalk between the High-Osmolarity Glycerol (HOG) and Mating Pheromone Response MAPK Pathways. Biomolecules, 11(10), 1530. https://doi.org/10.3390/biom11101530





