Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag(I) Complex as DNA and BSA Binders
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Methods
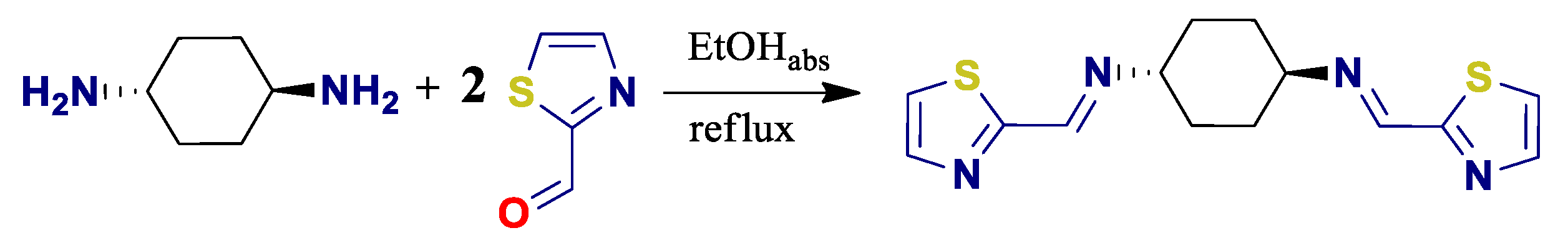
2.2. Synthesis of Ligand L
- 1H NMR (400 MHz, CD3CN-d3): δ(ppm) = 8.51 (s, 1H); 7.90 (d, 1H); 7.55 (d, 1H); 3.47 (m, 1H); 1.83 (m, 2H); 1.71 (m, 2H).
- 13C NMR (75 MHz, DMSO-d6): δ(ppm) = 166.78; 153.66; 144.04; 122.64; 67.04; 31.69 (2C).
- ESI-MS(+) m/z(%): 305 (30) [L+H]+; 327 (10) [L+Na]+.
- Anal. calcd. for (C14H16N4S2): C: 55.23, H: 5.30, N: 18.40, S: 21.07%; found: C: 55.13, H: 5.34, N: 18.48, S: 21.05%.
- FT-IR (KBr): 3446, ν(O−H); 2929, 2854 ν (C−H)aliph; 1640 ν(C=N); 1486 ν(C=C); 1447, 1420 δ(C-H); 1376, 1319 ν(C−N); 1223, 1147 ν(C−S); 947, 893, 780, 699 ν(C-H)arom cm−1.
2.3. Synthesis of Complex [Ag2L2](PF6)2
- 1H NMR (400 MHz, CD3CN–d3): δ(ppm) = 8.58 (s, 1H); 7.97 (d, 1H); 7.77 (d, 1H); 3.48 (m, 1H); 1.91 (m, 2H); 1.69 (m, 2H).
- ESI-MS(+) m/z(%): 412.98 (100) [AgL]+; 717.07 (16) [AgL2]+; 968.93 (2) {[Ag2L2](PF6)}+.
- Anal. calcd. for (C28H32Ag2N8S4P2F12): C: 30.17, H: 2.89, N: 10.05, S: 11.51%; found: C: 30.62, H: 2.72, N: 10.34, S: 11.91%.
- FT-IR (KBr): 3461 ν(O−H); 2930, 2858 ν(C−H)aliph; 1641, 1629 ν(C=N); 1485 ν(C=C); 1382 δ(C-H); 1323 ν(C−N); 1206, 1136 ν(C−S); 1075, 960, 628 ν(C-H)arom; 840 PF6-; 559 ν(Ag−N) cm−1.
2.4. X-ray Crystallography
2.5. DNA Binding Studies
2.5.1. UV-Vis Absorption Titration
2.5.2. Fluorescence Competitive Binding with Ethidium Bromide
2.5.3. Fluorescence Competitive Binding with Hoechst33258
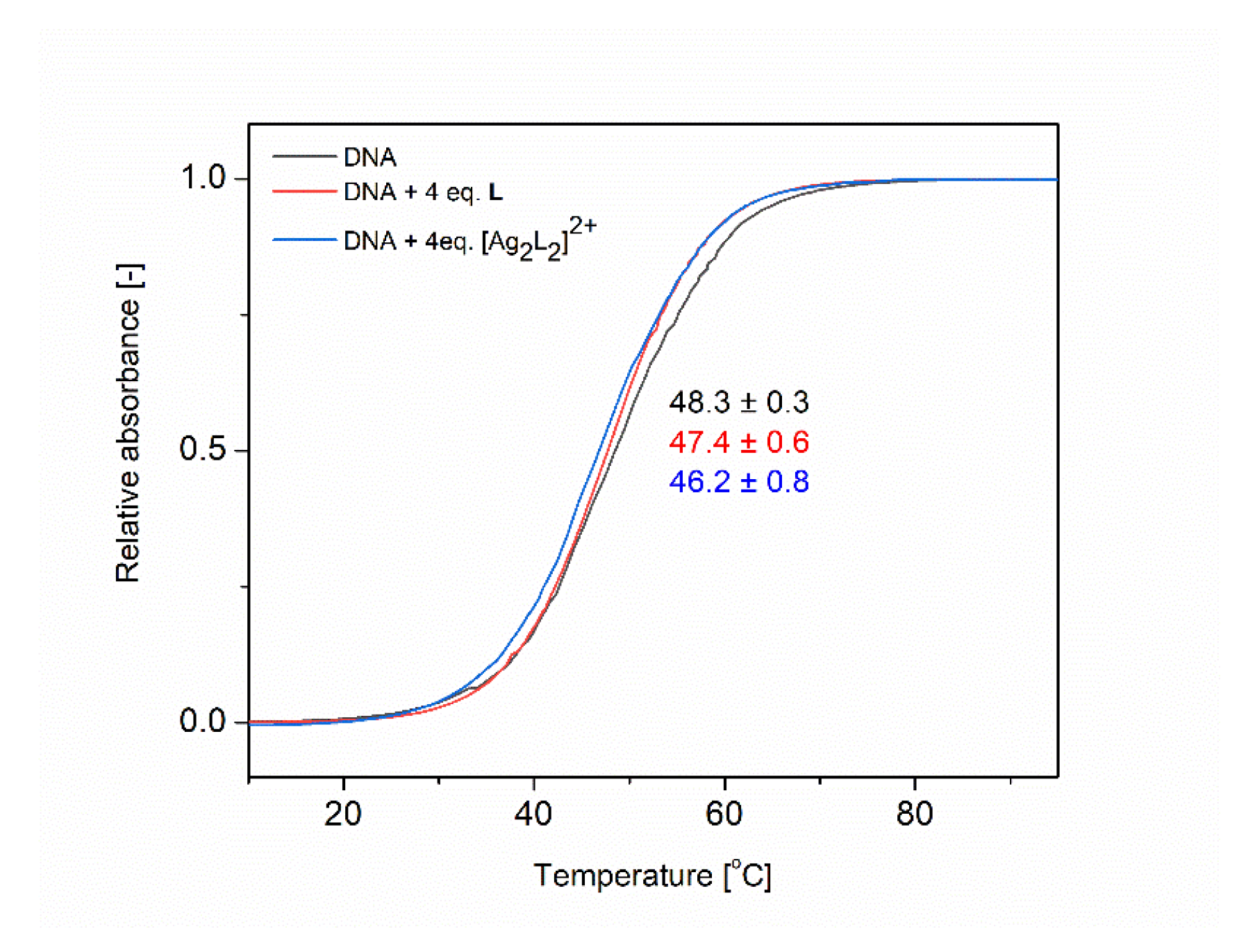
2.5.4. UV Melting Experiments
2.6. Protein Binding Studies
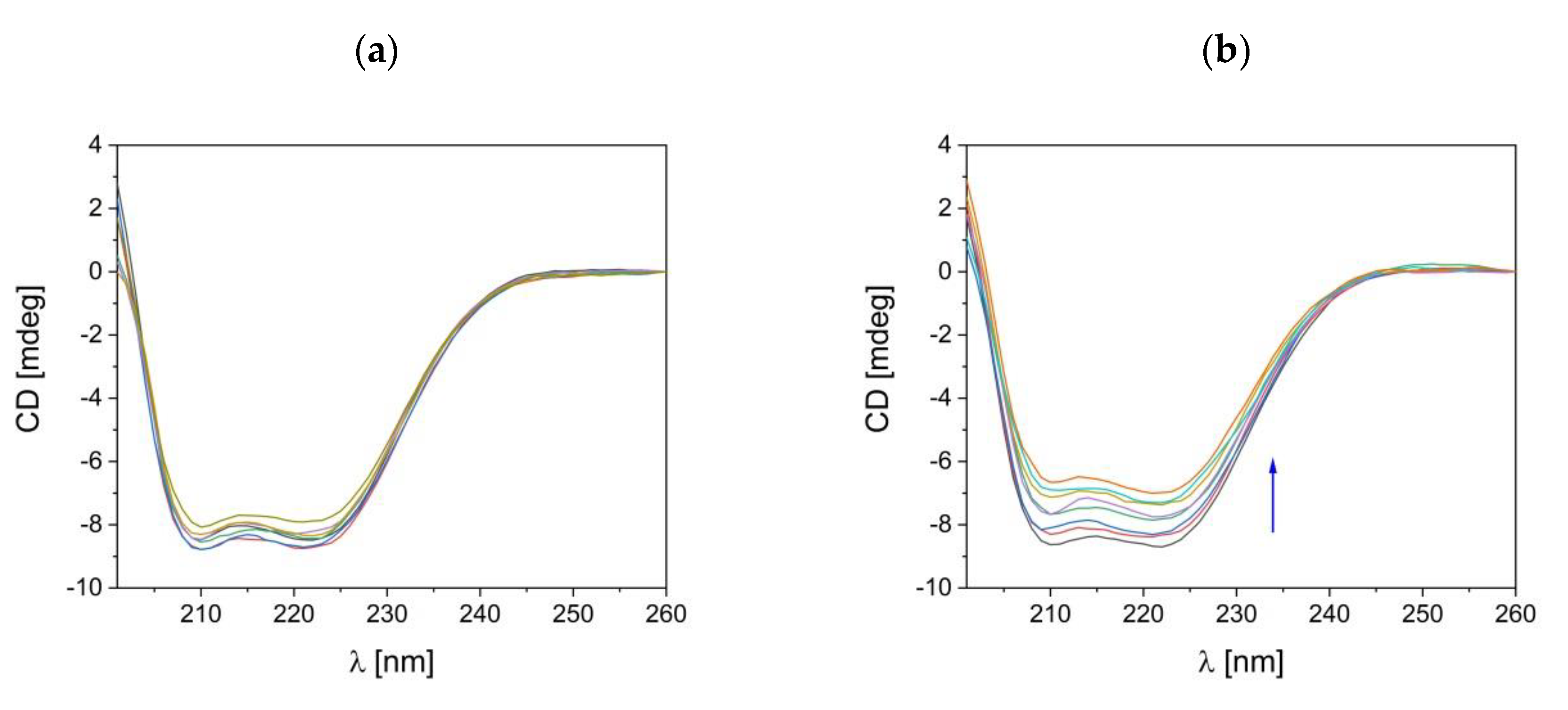
2.6.1. CD Analysis
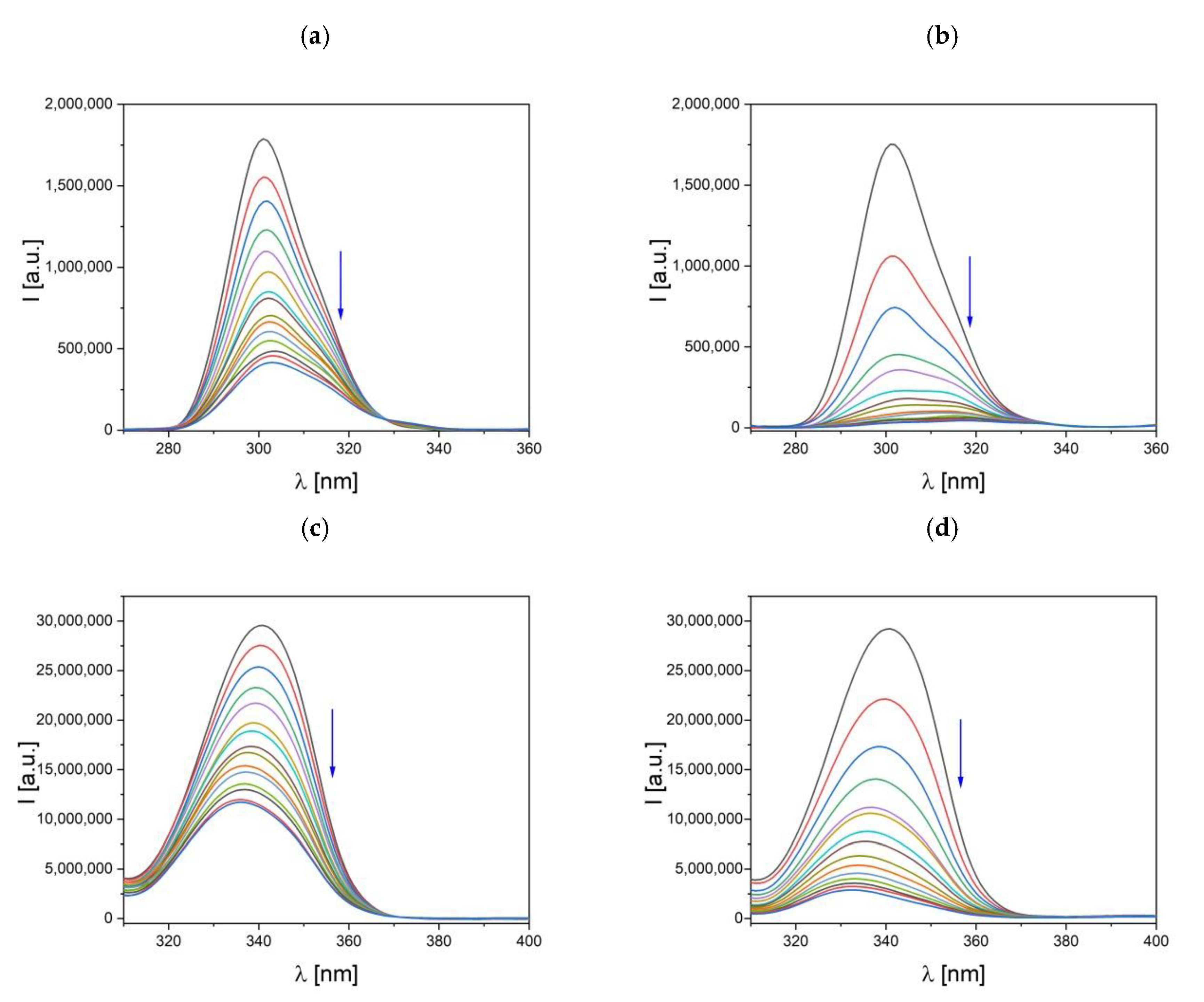
2.6.2. Fluorescence Quenching Studies
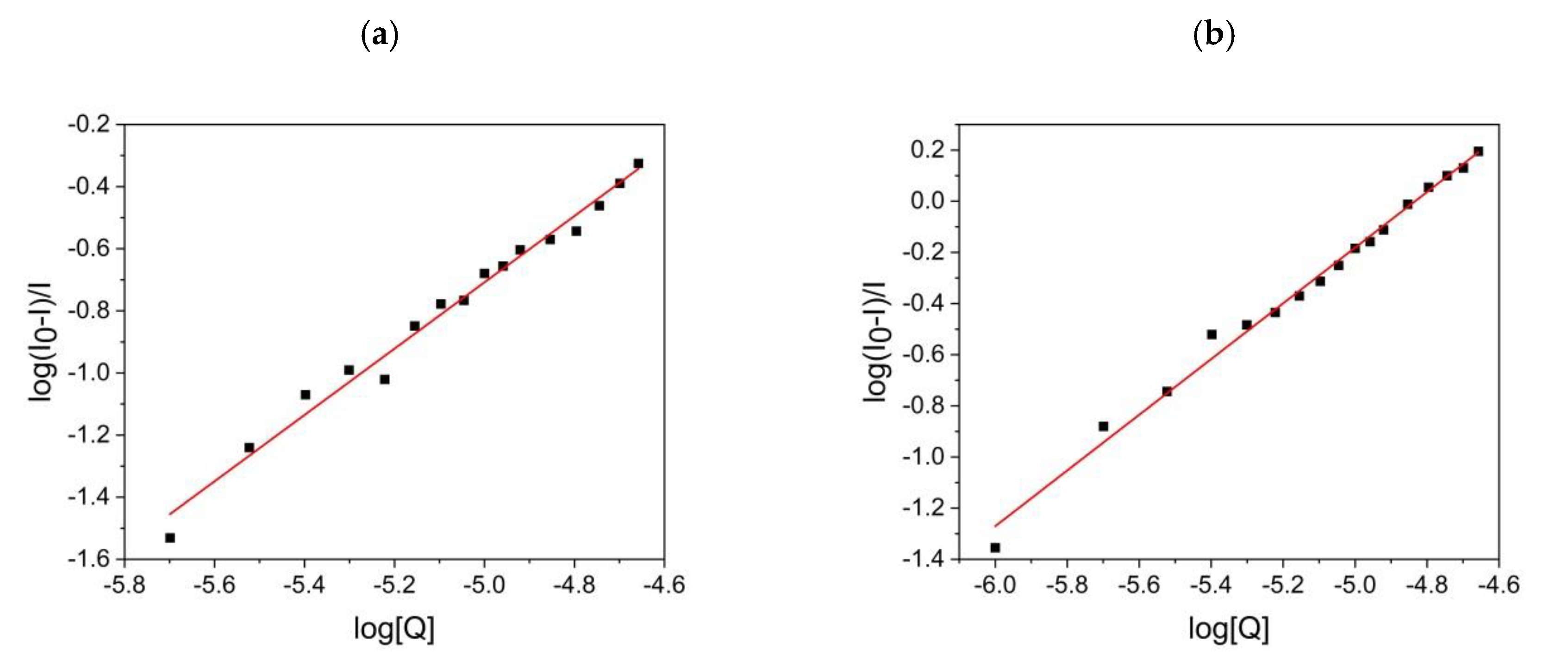
2.6.3. Number of Binding Sites and Binding Constant
2.6.4. Synchronous Fluorescence Studies
3. Results and Discussion
3.1. Design, Synthesis and Characterization
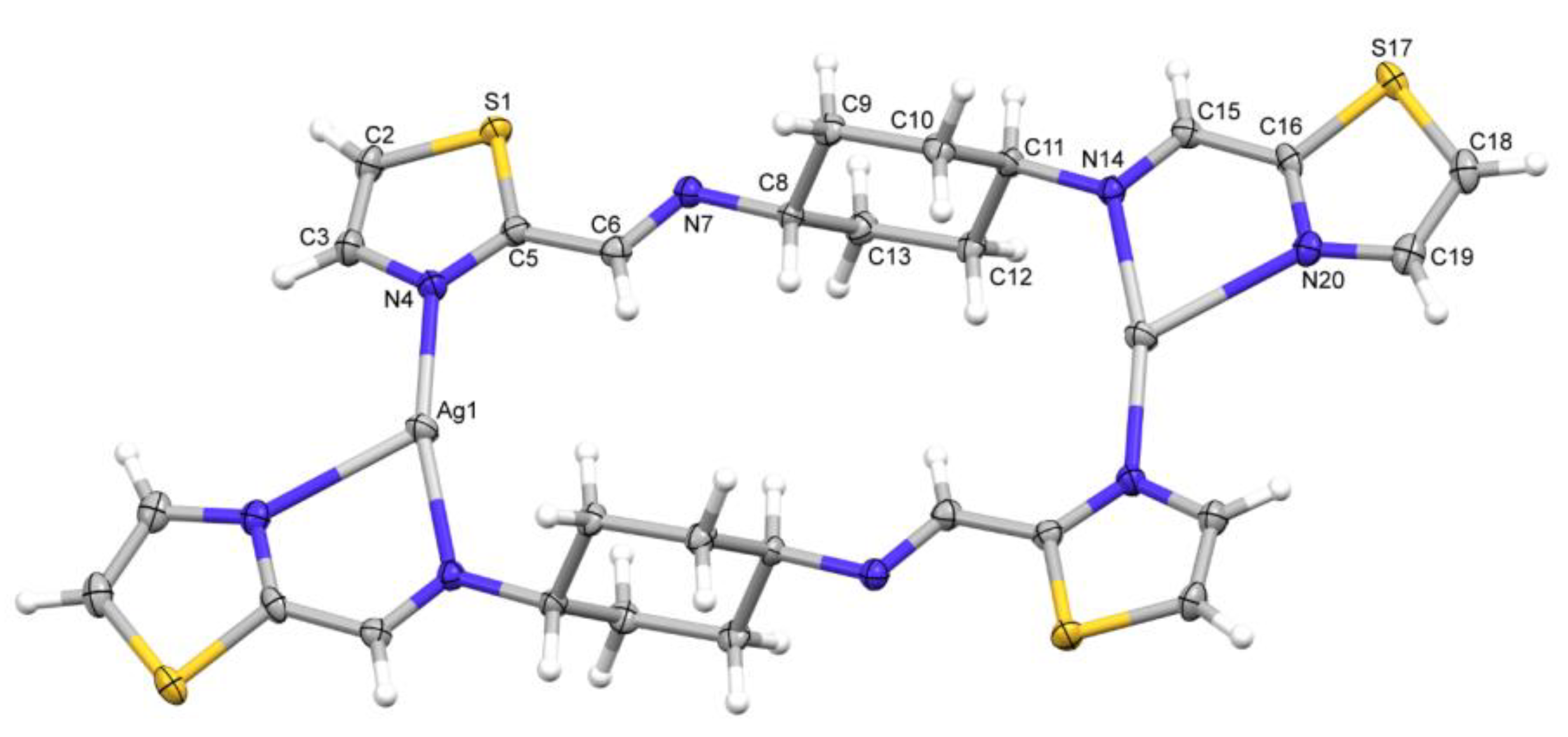
3.2. X-ray Structures
3.3. Interactions with DNA
3.3.1. Electronic Absorption Titration
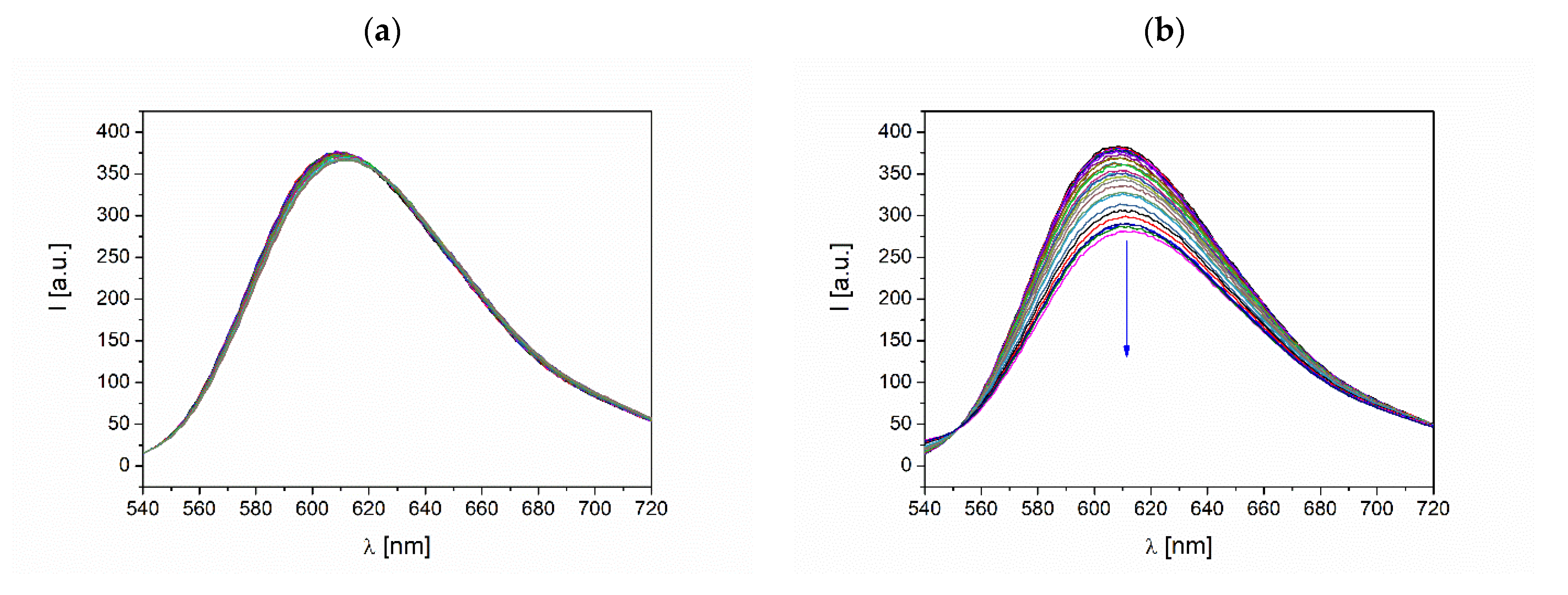
3.3.2. Competitive Studies with Fluorescent Dyes
3.3.3. DNA Melting Studies
3.4. Binding of BSA—A Model Protein
3.4.1. Conformational Changes Detected by CD
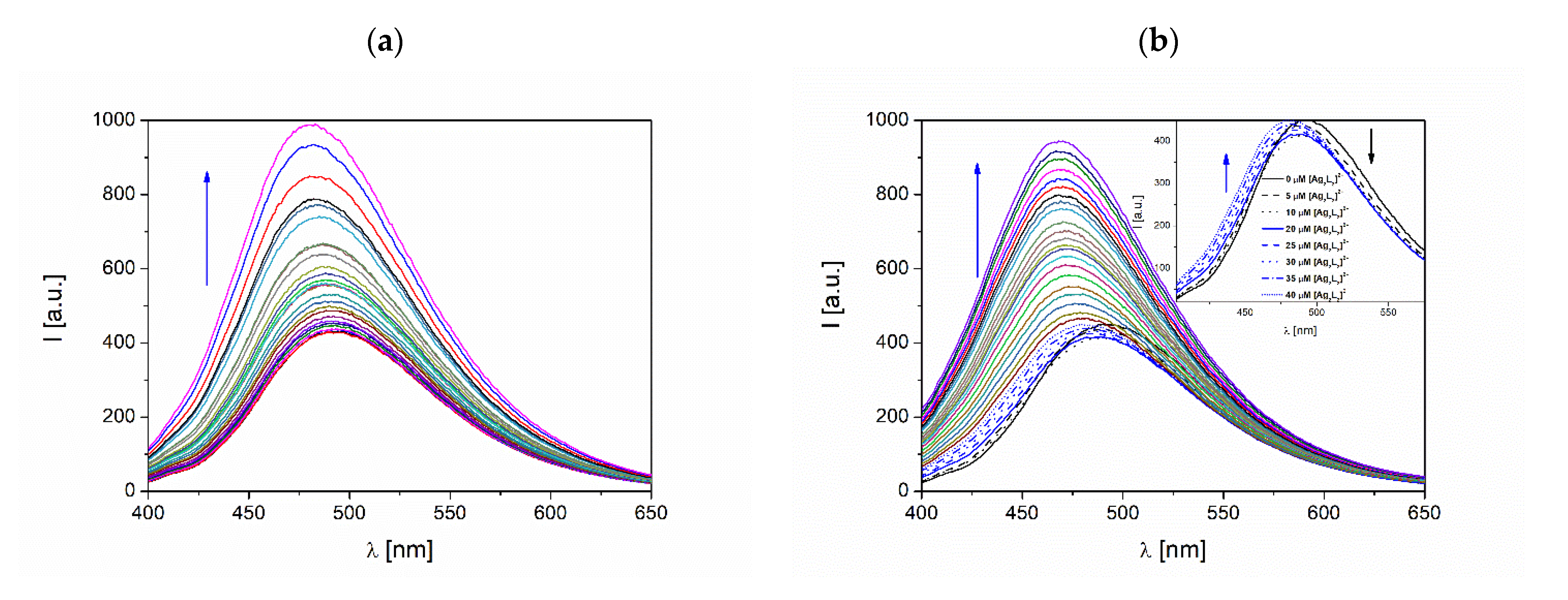
3.4.2. Fluorescence Quenching Studies
3.4.3. Binding Parameters
3.4.4. Synchronous Fluorescence Spectroscopic Studies of BSA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Crisponi, G.; Nurchi, V.M.; Lachowicz, J.I.; Remelli, M.; Zoroddu, M.A. Silver coordination compounds: A new horizon in medicine. Coord. Chem. Rev. 2016, 327–328, 349–359. [Google Scholar] [CrossRef]
- Medvetz, D.A.; Hindi, K.M.; Panzner, M.J.; Ditto, A.J.; Yun, Y.H.; Youngs, W.J. Anticancer Activity of Ag(I) N-Heterocyclic Carbene Complexes Derived from 4,5-Dichloro-1H-Imidazole. Met. Based Drugs 2008, 2008, 384010. [Google Scholar] [CrossRef] [PubMed]
- Hostýnek, J.J.; Hinz, R.S.; Lorence, C.R.; Price, M.; Guy, R.H. Metals and the Skin. Crit. Rev. Toxicol. 1993, 23, 171–235. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T.; Silver, G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 2006, 33, 627–634. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Lachowicz, J.I.; Crisponi, G.; Zoroddu, M.A. Noble metals in medicine: Latest advances. Coord. Chem. Rev. 2015, 284, 329–350. [Google Scholar] [CrossRef]
- Tsyba, I.; Mui, B.B.; Bau, R.; Noguchi, R.; Nomiya, K. Synthesis and Structure of a Water-Soluble Hexanuclear Silver(I) Nicotinate Cluster Comprised of a “Cyclohexane-Chair”-Type of Framework, Showing Effective Antibacterial and Antifungal Activities: Use of “Sparse Matrix” Techniques for Growing Crystals of Water-Soluble Inorganic Complexes. Inorg. Chem. 2003, 42, 8028–8032. [Google Scholar]
- Johnson, A.; Iffland, L.; Singh, K.; Apfel, U.-P.; Suntharalingam, K. A dithiacyclam-coordinated silver(I) polymer with anti-cancer stem cell activity. Dalton Trans. 2021, 50, 5779–5783. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, V.T.; Gocmen, E.; Icsel, C.; Cengiz, M.; Susluer, S.Y.; Buyukgungor, O. Synthesis, crystal structures, in vitro DNA binding, antibacterial and cytotoxic activities of new di- and polynuclear silver(I) saccharinate complexes with tertiary monophosphanes. J. Photochem. Photobiol. B 2014, 131, 31–42. [Google Scholar] [CrossRef]
- Liu, J.J.; Galettis, P.; Farr, A.; Maharaj, L.; Samarasinha, H.; McGechan, A.C.; Baguley, B.C.; Bowen, R.J.; Berners-Price, S.J.; McKeage, M.J. In vitro antitumour and hepatotoxicity profiles of Au(I) and Ag(I) bidentate pyridyl phosphine complexes and relationships to cellular uptake. J. Inorg. Biochem. 2008, 102, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Fik, M.A.; Gorczyński, A.; Kubicki, M.; Hnatejko, Z.; Fedoruk-Wyszomirska, A.; Wyszko, E.; Giel-Pietraszuk, M.; Patroniak, V. 6,6″-Dimethyl-2,2′:6′,2″-terpyridine revisited: New fluorescent silver(I) helicates with in vitro antiproliferative activity via selective nucleoli targeting. Eur. J. Med. Chem. 2014, 86, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Achar, G.; Shahini, C.R.; Patil, S.A.; Małecki, J.G.; Budagumpi, S. Coumarin-substituted 1,2,4-triazole-derived silver(I) and gold(I) complexes: Synthesis, characterization and anticancer studies. New J. Chem. 2019, 43, 1216–1229. [Google Scholar] [CrossRef]
- Adamski, A.; Fik, M.A.; Kubicki, M.; Hnatejko, Z.; Gurda, D.; Fedoruk-Wyszomirska, A.; Wyszko, E.; Kruszka, D.; Dutkiewicz, Z.; Patroniak, V. Full characterization and cytotoxic activity of new silver(I) and copper(I) helicates with quaterpyridine. N. J. Chem. 2016, 40, 7943–7957. [Google Scholar] [CrossRef]
- Szymańska, M.; Insińska-Rak, M.; Dutkiewicz, G.; Roviello, G.N.; Fik-Jaskółka, M.A.; Patroniak, V. Thiophene-benzothiazole dyad ligand and its Ag(I) complex–Synthesis, characterization, interactions with DNA and BSA. J. Mol. Liq. 2020, 319, 114182. [Google Scholar] [CrossRef]
- Caruso, U.; Panunzi, B.; Roviello, G.N.; Roviello, G.; Tingoli, M.; Tuzi, A. Synthesis, structure and reactivity of amino-benzodifurane derivatives. C. R. Chim. 2009, 12, 622–634. [Google Scholar] [CrossRef]
- Padnya, P.; Shibaeva, K.; Arsenyev, M.; Baryshnikova, S.; Terenteva, O.; Shiabiev, I.; Khannanov, A.; Boldyrev, A.; Gerasimov, A.; Grishaev, D.; et al. Catechol-Containing Schiff Bases on Thiacalixarene: Synthesis, Copper(II) Recognition, and Formation of Organic-Inorganic Copper-Based Materials. Molecules 2021, 26, 2334. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, G.; Musumeci, D.; Bucci, E.M.; Pedone, C. Dakin–West reaction on 1-thyminyl acetic acid for the synthesis of 1,3-bis(1-thyminyl)-2-propanone, a heteroaromatic compound with nucleopeptide-binding properties. Amino Acids 2012, 43, 1615–1623. [Google Scholar] [CrossRef]
- Vicidomini, C.; Cioffi, F.; Broersen, K.; Roviello, V.; Riccardi, C.; Montesarchio, D.; Capasso, D.; Di Gaetano, S.; Musumeci, D.; Roviello, G.N. Benzodifurans for biomedical applications: BZ4, a selective anti-proliferative and anti-amyloid lead compound. Future Med. Chem. 2019, 11, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Aragón-Muriel, A.; Liscano, Y.; Upegui, Y.; Robledo, S.M.; Ramírez-Apan, M.T.; Morales-Morales, D.; Oñate-Garzón, J.; Polo-Cerón, D. In Vitro Evaluation of the Potential Pharmacological Activity and Molecular Targets of New Benzimidazole-Based Schiff Base Metal Complexes. Antibiotics 2021, 10, 728. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, A.A.; Zamisa, S.J.; Islam, M.S.; Olofinsan, K.; Salau, V.F.; Mocktar, C.; Omondi, B. Quinoline Functionalized Schiff Base Silver (I) Complexes: Interactions with Biomolecules and In Vitro Cytotoxicity, Antioxidant and Antimicrobial Activities. Molecules 2021, 26, 1205. [Google Scholar] [CrossRef]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. MedChemComm 2018, 9, 409–436. [Google Scholar] [CrossRef]
- Paul, A.; Singh, P.; Kuznetsov, M.L.; Karmakar, A.; Guedes da Silva, M.F.C.; Koch, B.; Pombeiro, A.J.L. Influence of anchoring moieties on new benzimidazole-based Schiff base copper(II) complexes towards estrogen dependent breast cancer cells. Dalton Trans. 2021, 50, 3701–3716. [Google Scholar] [CrossRef]
- Banti, C.N.; Giannoulis, A.D.; Kourkoumelis, N.; Owczarzak, A.M.; Kubicki, M.; Hadjikakou, S.K. Novel metallo-therapeutics of the NSAID naproxen. Interaction with intracellular components that leads the cells to apoptosis. Dalton Trans. 2014, 43, 6848–6863. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G. Hard and Soft Acids and Bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Letelier, M.E.; Faúndez, M.; Jara-Sandoval, J.; Molina-Berríos, A.; Cortés-Troncoso, J.; Aracena-Parks, P.; Marín-Catalán, R. Mechanisms underlying the inhibition of the cytochrome P450 system by copper ions. J. Appl. Toxicol. 2009, 29, 695–702. [Google Scholar] [CrossRef]
- Che, C.-M.; Siu, F.-M. Metal complexes in medicine with a focus on enzyme inhibition. Curr. Opin. Chem. Biol. 2010, 14, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, S.; Brunetto, P.S.; Gagnon, J.; Priebe, M.; Giese, B.; Fromm, K.M. Nanobio Silver: Its Interactions with Peptides and Bacteria, and Its Uses in Medicine. Chem. Rev. 2013, 113, 4708–4754. [Google Scholar] [CrossRef] [PubMed]
- Ašanin, D.P.; Skaro Bogojevic, S.; Perdih, F.; Andrejević, T.P.; Milivojevic, D.; Aleksic, I.; Nikodinovic-Runic, J.; Glišić, B.Đ.; Turel, I.; Djuran, M.I. Structural Characterization, Antimicrobial Activity and BSA/DNA Binding Affinity of New Silver(I) Complexes with Thianthrene and 1,8-Naphthyridine. Molecules 2021, 26, 1871. [Google Scholar] [CrossRef]
- Banti, C.N.; Giannoulis, A.D.; Kourkoumelis, N.; Owczarzak, A.M.; Poyraz, M.; Kubicki, M.; Charalabopoulos, K.; Hadjikakou, S.K. Mixed ligand–silver(I) complexes with anti-inflammatory agents which can bind to lipoxygenase and calf-thymus DNA, modulating their function and inducing apoptosis. Metallomics 2012, 4, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, H.; Neault, J.F.; Tajmir-Riahi, H.A. Silver(I) Complexes with DNA and RNA Studied by Fourier Transform Infrared Spectroscopy and Capillary Electrophoresis. Biophys. J. 2001, 81, 1580–1587. [Google Scholar] [CrossRef]
- Kasyanenko, N.; Qiushi, Z.; Bakulev, V.; Osolodkov, M.; Sokolov, P.; Demidov, V. DNA Binding with Acetate Bis(1,10-phenanthroline)silver(I) Monohydrate in a Solution and Metallization of Formed Structures. Polymers 2017, 9, 211. [Google Scholar] [CrossRef]
- Đurić, S.Ž.; Vojnovic, S.; Andrejević, T.P.; Stevanović, N.L.; Savić, N.D.; Nikodinovic-Runic, J.; Glišić, B.Đ.; Djuran, M.I. Antimicrobial Activity and DNA/BSA Binding Affinity of Polynuclear Silver(I) Complexes with 1,2-Bis(4-pyridyl)ethane/ethene as Bridging Ligands. Bioinorg. Chem. Appl. 2020, 2020, 3812050. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Shih, W.-C.; Chang, H.C.; Kuo, Y.-Y.; Hung, W.-C.; Ong, T.-G.; Li, W.-S. Preparation and Characterization of Amino-Linked Heterocyclic Carbene Palladium, Gold, and Silver Complexes and Their Use as Anticancer Agents That Act by Triggering Apoptotic Cell Death. J. Med. Chem. 2011, 54, 5245–5249. [Google Scholar] [CrossRef]
- Shahabadi, N.; Maghsudi, M. Multi-spectroscopic and molecular modeling studies on the interaction of antihypertensive drug; methyldopa with calf thymus DNA. Mol. BioSyst. 2014, 10, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Matesanz, A.I.; Albacete, P.; Perles, J.; Souza, P. A structural and biological study on the new 3,5-diacetyl-1,2,4-triazol bis(p-chlorophenylthiosemicarbazone) ligand and its bimetallic complexes. Inorg. Chem. Front. 2015, 2, 75–84. [Google Scholar] [CrossRef][Green Version]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Lansdown, A.B.G. Silver in Health Care: Antimicrobial Effects and Safety in Use. Curr. Probl. Dermatol. 2006, 33, 17–34. [Google Scholar]
- Lansdown, A.B.G. Silver in Healthcare: Its Antimicrobial Efficacy and Safety in Use; Royal Society of Chemistry Publishing: Cambridge, UK, 2010. [Google Scholar]
- Ricketts, C.R.; Lowbury, E.J.L.; Lawrence, J.C.; Hall, M.; Wilkins, M.D. Mechanism of Prophylaxis by Silver Compounds against Infection of Burns. Br. Med. J. 1970, 2, 444–446. [Google Scholar] [CrossRef] [PubMed]
- Banti, C.N.; Hadjikakou, S.K. Anti-proliferative and anti-tumor activity of silver(I) compounds. Metallomics 2013, 5, 569–596. [Google Scholar] [CrossRef] [PubMed]
- Jover, J.; Bosque, R.; Sales, J. Quantitative Structure−Property Relationship Estimation of Cation Binding Affinity of the Common Amino Acids. J. Phys. Chem. A 2009, 113, 3703–3708. [Google Scholar] [CrossRef] [PubMed]
- Jover, J.; Bosque, R.; Sales, J. A comparison of the binding affinity of the common amino acids with different metal cations. Dalton Trans. 2008, 45, 6441–6453. [Google Scholar] [CrossRef]
- Glusker, J.P. Structural Aspects of Metal Liganding to Functional Groups in Proteins. Adv. Protein Chem. 1991, 42, 1–76. [Google Scholar] [PubMed]
- Lansdown, A.B.G. A Pharmacological and Toxicological Profile of Silver as an Antimicrobial Agent in Medical Devices. Adv. Pharmacol. Sci. 2010, 2010, 910686. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Pothiraj, K.; Baskaran, T. DNA interaction, antimicrobial, electrochemical and spectroscopic studies of metal(II) complexes with tridentate heterocyclic Schiff base derived from 2′-methylacetoacetanilide. J. Mol. Struct. 2011, 1000, 135–144. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Diffraction, R.O. CrysAlis PRO (Version 1.171.38.46) 2015. Available online: https://www.rigaku.com/zh-hans/products/smc/crysalis (accessed on 30 September 2021).
- Sheldrick, G. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. A 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G. Crystal structure refinement with SHELXL. Acta Cryst. C 2015, C71, 3–8. [Google Scholar] [CrossRef]
- Thordarson, P. Determining association constants from titration experiments in supramolecular chemistry. Chem. Soc. Rev. 2011, 40, 1305–1323. [Google Scholar] [CrossRef]
- Brynn Hibbert, D.; Thordarson, P. The death of the Job plot, transparency, open science and online tools, uncertainty estimation methods and other developments in supramolecular chemistry data analysis. Chem. Commun. 2016, 52, 12792–12805. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://supramolecular.org (accessed on 30 September 2021).
- Shiri, F.; Rahimi-Nasrabadi, M.; Ahmadi, F.; Ehrlich, H. Multispectroscopic and molecular modeling studies on the interaction of copper-ibuprofenate complex with bovine serum albumin (BSA). Spectrochim. Acta A 2018, 203, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Feng, X.-Z.; Lin, Z.; Yang, L.-J.; Wang, C.-H.; Bai, C.-I. Investigation of the interaction between acridine orange and bovine serum albumin. Talanta 1998, 47, 1223–1229. [Google Scholar] [CrossRef]
- Gorczyński, A.; Pakulski, D.; Szymańska, M.; Kubicki, M.; Bułat, K.; Łuczak, T.; Patroniak, V. Electrochemical deposition of the new manganese(II) Schiff-base complex on a gold template and its application for dopamine sensing in the presence of interfering biogenic compounds. Talanta 2016, 149, 347–355. [Google Scholar] [CrossRef]
- Mishra, B.B.; Kumar, D.; Singh, A.S.; Tripathi, R.P.; Tiwari, V.K. Chapter 17—Ionic Liquids-Prompted Synthesis of Biologically Relevant Five- and Six-Membered Heterocyclic Skeletons: An Update. In Green Synthetic Approaches for Biologically Relevant Heterocycles; Elsevier: Amsterdam, The Netherlands, 2015; pp. 437–493. [Google Scholar]
- Hamza, M.; Ali, A.; Khan, S.; Ahmed, S.; Attique, Z.; Ur Rehman, S.; Khan, A.; Ali, H.; Rizwan, M.; Munir, A.; et al. nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of Moringa oleifera and hydroxychloroquine. J. Biomol. Struct. Dyn. 2021, 39, 4089–4099. [Google Scholar] [CrossRef]
- Britovsek, G.J.P.; Gibson, V.C.; Spitzmesser, S.K.; Tellmann, K.P.; White, A.J.P.; Williams, D.J. Cationic 2,6-bis(imino)pyridine iron and cobalt complexes: Synthesis, structures, ethylene polymerisation and ethylene/polar monomer co-polymerisation studies. J. Chem. Soc. Dalton Trans. 2002, 6, 1159–1171. [Google Scholar] [CrossRef]
- Ayati, A.; Emami, S.; Asadipour, A.; Shafiee, A.; Foroumadi, A. Recent applications of 1,3-thiazole core structure in the identification of new lead compounds and drug discovery. Eur. J. Med. Chem. 2015, 97, 699–718. [Google Scholar] [CrossRef]
- Mejía, C.; Ortega-Rosales, S.; Ruiz-Azuara, L. Mechanism of Action of Anticancer Metallodrugs. In Biomedical Applications of Metals; Springer International Publishing: Cham, Switzerland, 2018; pp. 213–234. [Google Scholar]
- Roviello, G.N.; Roviello, V.; Autiero, I.; Saviano, M. Solid phase synthesis of TyrT, a thymine–tyrosine conjugate with poly(A) RNA-binding ability. RSC Adv. 2016, 6, 27607–27613. [Google Scholar] [CrossRef]
- Shivakumar, L.; Shivaprasad, K.; Revanasiddappa, H.D. Synthesis, spectroscopic characterization, antimicrobial, DNA binding and oxidative-induced DNA cleavage activities: New oxovanadium(IV) complexes of 2-(2-hydroxybenzylideneamino)isoindoline-1,3-dione. Spectrochim. Acta A 2012, 97, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Montalbano, A.; Barraja, P.; Dattolo, G.; Almerico, A.M. DNA Minor Groove Binders: An Overview on Molecular Modeling and QSAR Approaches. Curr. Med. Chem. 2007, 14, 2136–2160. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Guo, Q.; Dong, J.; Xu, T.; Li, J. DNA binding, DNA cleavage and BSA interaction of a mixed-ligand copper(II) complex with taurine Schiff base and 1,10-phenanthroline. J. Photochem. Photobiol. B 2013, 125, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Thakor, K.P.; Lunagariya, M.V.; Bhatt, B.S.; Patel, M.N. Fluorescence and absorption studies of DNA–Pd(II) complex interaction: Synthesis, spectroanalytical investigations and biological activities. Luminescence 2019, 34, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, J.; Li, J.; Zheng, K.-C.; Huang, X.-M.; Tan, C.-P.; Chen, L.-M.; Ji, L.-N. Synthesis, characterization and DNA-binding of novel chiral complexes Δ- and Λ-[Ru(bpy)2L]2+ (L=o-mopip and p-mopip). J. Inorg. Biochem. 2006, 100, 385–395. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Adeleke, A.A.; Islam, M.S.; Sanni, O.; Mocktar, C.; Zamisa, S.J.; Omondi, B. Aryl variation and anion effect on CT-DNA binding and in vitro biological studies of pyridinyl Ag(I) complexes. J. Inorg. Biochem. 2021, 214, 111266. [Google Scholar] [CrossRef]
- Elsayed, S.A.; El-Gharabawy, H.M.; Butler, I.S.; Atlam, F.M. Novel metal complexes of 3-acetylcoumarin-2-hydrazinobenzothiazole Schiff base: Design, structural characterizations, DNA binding, DFT calculations, molecular docking and biological studies. Appl. Organometal. Chem. 2020, 34, e5643. [Google Scholar] [CrossRef]
- Molphy, Z.; Prisecaru, A.; Slator, C.; Barron, N.; McCann, M.; Colleran, J.; Chandran, D.; Gathergood, N.; Kellett, A. Copper Phenanthrene Oxidative Chemical Nucleases. Inorg. Chem. 2014, 53, 5392–5404. [Google Scholar] [CrossRef]
- Shahabadi, N.; Shiri, F.; Hadidi, S.; Farshadfar, K.; Darbemamieh, M.; Mark Roe, S. The role of both intercalation and groove binding at AT-rich DNA regions in the interaction process of a dinuclear Cu(I) complex probed by spectroscopic and simulation analysis. J. Mol. Liq. 2021, 335, 116290. [Google Scholar] [CrossRef]
- Palmucci, J.; Mahmudov, K.T.; Guedes da Silva, M.F.C.; Marchetti, F.; Pettinari, C.; Petrelli, D.; Vitali, L.A.; Quassinti, L.; Bramucci, M.; Lupidi, G.; et al. DNA and BSA binding, anticancer and antimicrobial properties of Co(II), Co(II/III), Cu(II) and Ag(I) complexes of arylhydrazones of barbituric acid. RSC Adv. 2016, 6, 4237–4249. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Gümüş, F.; Eren, G.; Açık, L.; Çelebi, A.; Öztürk, F.; Yılmaz, Ş.; Saǧkan, R.I.; Gür, S.; Özkul, A.; Elmalı, A.; et al. Synthesis, Cytotoxicity, and DNA Interactions of New Cisplatin Analogues Containing Substituted Benzimidazole Ligands. J. Med. Chem. 2009, 52, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Deka, B.; Sarkar, T.; Banerjee, S.; Kumar, A.; Mukherjee, S.; Deka, S.; Saikia, K.K.; Hussain, A. Novel mitochondria targeted copper(II) complexes of ferrocenyl terpyridine and anticancer active 8-hydroxyquinolines showing remarkable cytotoxicity, DNA and protein binding affinity. Dalton Trans. 2017, 46, 396–409. [Google Scholar] [CrossRef]
- Raj Kumar, R.; Ramesh, R. Synthesis, molecular structure and electrochemical properties of nickel(II) benzhydrazone complexes: Influence of ligand substitution on DNA/protein interaction, antioxidant activity and cytotoxicity. RSC Adv. 2015, 5, 101932–101948. [Google Scholar] [CrossRef]
- Roviello, G.N.; Oliviero, G.; Di Napoli, A.; Borbone, N.; Piccialli, G. Synthesis, self-assembly-behavior and biomolecular recognition properties of thyminyl dipeptides. Arab. J. Chem. 2020, 13, 1966–1974. [Google Scholar] [CrossRef]
- Hu, Y.-J.; Ou-Yang, Y.; Dai, C.-M.; Liu, Y.; Xiao, X.-H. Site-Selective Binding of Human Serum Albumin by Palmatine: Spectroscopic Approach. Biomacromolecules 2010, 11, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-H.; Wang, J.; Zhu, Y.-Y.; Chen, J. Characterization of interaction between isoliquiritigenin and bovine serum albumin: Spectroscopic and molecular docking methods. J. Lumin. 2014, 145, 643–650. [Google Scholar] [CrossRef]
- Cao, X.; He, Y.; Liu, D.; He, Y.; Hou, X.; Cheng, Y.; Liu, J. Characterization of interaction between scoparone and bovine serum albumin: Spectroscopic and molecular docking methods. RSC Adv. 2018, 8, 25519–25525. [Google Scholar] [CrossRef]
- Shi, J.-H.; Pan, D.-Q.; Jiang, M.; Liu, T.-T.; Wang, Q. Binding interaction of ramipril with bovine serum albumin (BSA): Insights from multi-spectroscopy and molecular docking methods. J. Photochem. Photobiol. B 2016, 164, 103–111. [Google Scholar] [CrossRef]
- Samari, F.; Hemmateenejad, B.; Shamsipur, M.; Rashidi, M.; Samouei, H. Affinity of Two Novel Five-Coordinated Anticancer Pt(II) Complexes to Human and Bovine Serum Albumins: A Spectroscopic Approach. Inorg. Chem. 2012, 51, 3454–3464. [Google Scholar] [CrossRef]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching of indole and model micelle systems. J. Phys. Chem. 1976, 80, 486–493. [Google Scholar] [CrossRef]
- Hong, M.; Geng, H.; Niu, M.; Wang, F.; Li, D.; Liu, J.; Yin, H. Organotin(IV) complexes derived from Schiff base N’-[(1E)-(2-hydroxy-3-methoxyphenyl)methylidene]pyridine-4-carbohydrazone: Synthesis, in vitro cytotoxicities and DNA/BSA interaction. Eur. J. Med. Chem. 2014, 86, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.B.F.; Evett, I.W. Prediction of peak wavelengths and intensities in synchronously excited fluorescence emission spectra. Anal. Chem. 1977, 49, 1710–1715. [Google Scholar] [CrossRef]
- Jayabharathi, J.; Thanikachalam, V.; Venkatesh Perumal, M. Mechanistic investigation on binding interaction of bioactive imidazole with protein bovine serum albumin—A biophysical study. Spectrochim. Acta A 2011, 79, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.N. Recent advances in molecular luminescence analysis. Proc. Anal. Div. Chem. Soc. 1979, 16, 203–208. [Google Scholar]




| L | [Ag2L2]2+ | |
|---|---|---|
| Ag1-N4 | 2.144(3) | |
| Ag1-N14i | 2.192(3) | |
| Ag1-N20i | 2.479(3) | |
| N4-Ag1-N14i | 166.20(11) | |
| N4-Ag1-N20i | 120.12(10) | |
| N14i-Ag1-N20i | 73.34(10) | |
| C2-S1-C5 | 89.32(5) | 89.82(18) |
| C3-N4-C5 | 109.44(18) | 111.1(3) |
| C16-S17-C18 | 89.30(18) | |
| C16-N20-C19 | 110.3(3) |
| Compound | N | Kbin [M−1] |
|---|---|---|
| L | 1.06 | 4.27 × 104 |
| [Ag2L2]2+ | 1.08 | 1.84 × 105 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymańska, M.; Pospieszna-Markiewicz, I.; Mańka, M.; Insińska-Rak, M.; Dutkiewicz, G.; Patroniak, V.; Fik-Jaskółka, M.A. Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag(I) Complex as DNA and BSA Binders. Biomolecules 2021, 11, 1449. https://doi.org/10.3390/biom11101449
Szymańska M, Pospieszna-Markiewicz I, Mańka M, Insińska-Rak M, Dutkiewicz G, Patroniak V, Fik-Jaskółka MA. Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag(I) Complex as DNA and BSA Binders. Biomolecules. 2021; 11(10):1449. https://doi.org/10.3390/biom11101449
Chicago/Turabian StyleSzymańska, Martyna, Izabela Pospieszna-Markiewicz, Martyna Mańka, Małgorzata Insińska-Rak, Grzegorz Dutkiewicz, Violetta Patroniak, and Marta A. Fik-Jaskółka. 2021. "Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag(I) Complex as DNA and BSA Binders" Biomolecules 11, no. 10: 1449. https://doi.org/10.3390/biom11101449
APA StyleSzymańska, M., Pospieszna-Markiewicz, I., Mańka, M., Insińska-Rak, M., Dutkiewicz, G., Patroniak, V., & Fik-Jaskółka, M. A. (2021). Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag(I) Complex as DNA and BSA Binders. Biomolecules, 11(10), 1449. https://doi.org/10.3390/biom11101449







