Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Mouse Genotyping
2.3. Neural Stem and Progenitor Cell Cultures
2.4. Proliferation Assay
2.5. Self-Renewal Capacity Assay
2.6. Differentiation Assay
2.7. Western Blot
2.8. Statistical Analysis
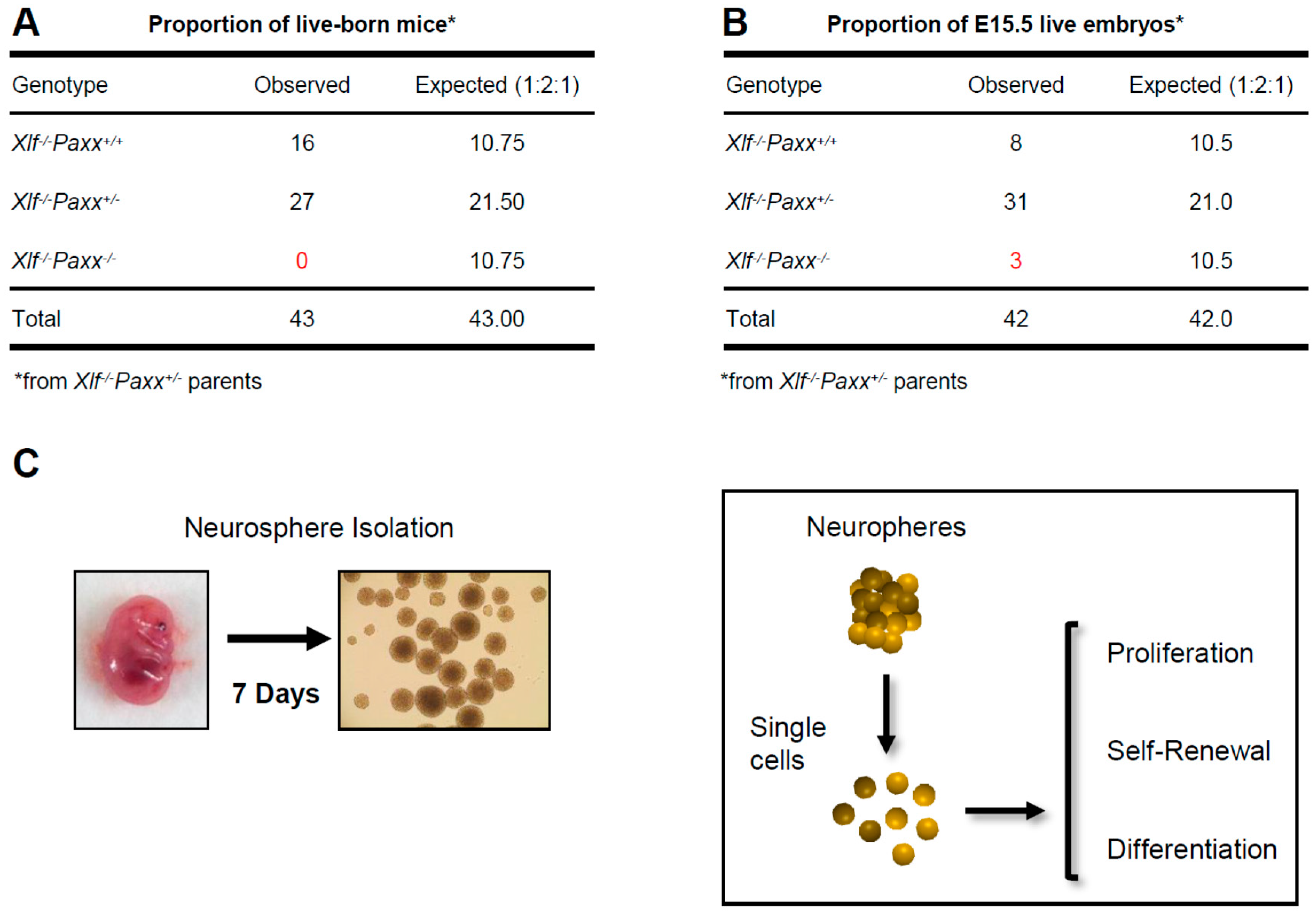
3. Results
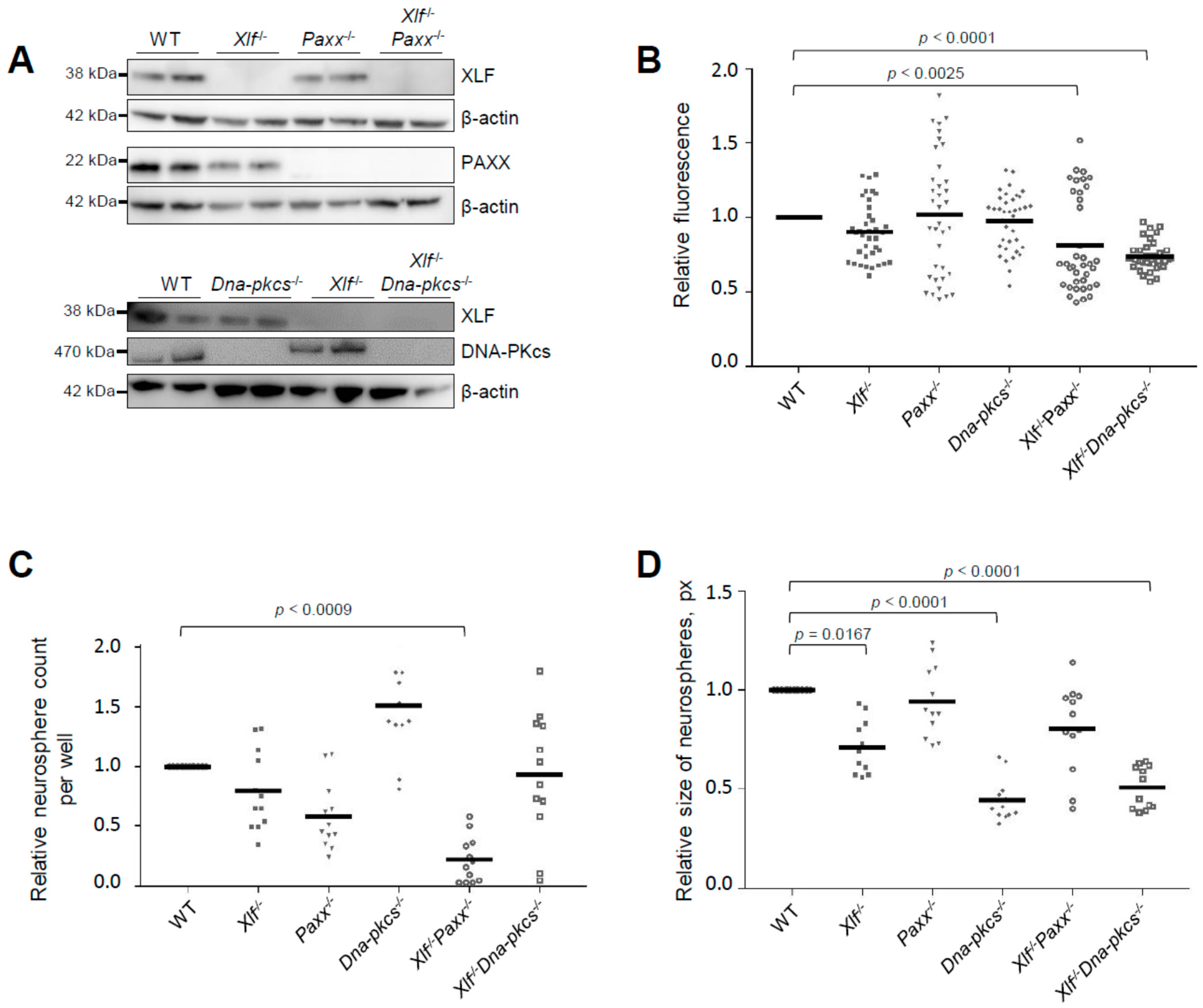
3.1. Impact of XLF, PAXX, and DNA-PKcs on Proliferation and Self-Renewal Capacity of Neural Stem and Progenitor Cells
3.2. Impact of XLF, PAXX, and DNA-PKcs on Differentiation Capacity of Neural Stem and Progenitor Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castañeda-Zegarra, S.; Fernandez-Berrocal, M.; Tkachev, M.; Yao, R.; Upfold, N.L.E.; Oksenych, V. Genetic interaction between the non-homologous end joining factors during B and T lymphocyte development: In Vivo mouse models. Scand. J. Immunol. 2020, 92, e12936. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Zegarra, S.; Zhang, Q.; Alirezaylavasani, A.; Fernandez-Berrocal, M.; Yao, R.; Oksenych, V. Leaky severe combined immunodeficiency in mice lacking non-homologous end joining factors XLF and MRI. Aging 2020, 12, 12. [Google Scholar] [CrossRef]
- Kumar, V.; Alt, F.W.; Oksenych, V. Functional overlaps between XLF and the ATM-dependent DNA double strand break response. DNA Repair 2014, 16, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Lee, B.J.; Zha, S. The recent advances in non-homologous end-joining through the lens of lymphocyte development. DNA Repair 2020, 94, 102874. [Google Scholar] [CrossRef] [PubMed]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Boboila, C.; Oksenych, V.; Gostissa, M.; Wang, J.H.; Zha, S.; Zhang, Y.; Chai, H.; Lee, C.-S.; Jankovic, M.; Saez, L.-M.A.; et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc. Natl. Acad. Sci. USA 2012, 109, 2473–2478. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, Y.; Frank, K.M.; Dikkes, P.; Fujiwara, Y.; Seidl, K.J.; Sekiguchi, J.M.; Rathbun, G.A.; Swat, W.; Wang, J.; et al. A Critical Role for DNA End-Joining Proteins in Both Lymphogenesis and Neurogenesis. Cell 1998, 95, 891–902. [Google Scholar] [CrossRef]
- Frank, K.M.; Sekiguchi, J.M.; Seidl, K.J.; Swat, W.; Rathbun, G.A.; Cheng, H.-L.; Davidson, L.; Kangaloo, L.; Alt, F.W. Late embryonic lethality and impaired V (D)J recombination in mice lacking DNA ligase IV. Nat. Cell Biol. 1998, 396, 173–177. [Google Scholar] [CrossRef]
- Frank, K.M.; Sharpless, N.E.; Gao, Y.; Sekiguchi, J.M.; Ferguson, D.O.; Zhu, C.; Manis, J.P.; Horner, J.; DePinho, R.A.; Alt, F.W. DNA Ligase IV Deficiency in Mice Leads to Defective Neurogenesis and Embryonic Lethality via the p53 Pathway. Mol. Cell 2000, 5, 993–1002. [Google Scholar] [CrossRef]
- Gao, Y.; Ferguson, D.O.; Xie, W.; Manis, J.P.; Sekiguchi, J.; Frank, K.M.; Chaudhuri, J.; Horner, J.W.; Depinho, R.A.; Alt, F.W. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nat. Cell Biol. 2000, 404, 897–900. [Google Scholar] [CrossRef]
- Gu, Y.; Seidl, K.J.; Rathbun, G.A.; Zhu, C.; Manis, J.P.; Van Der Stoep, N.; Davidson, L.; Cheng, H.-L.; Sekiguchi, J.M.; Frank, K.; et al. Growth Retardation and Leaky SCID Phenotype of Ku70-Deficient Mice. Immunity 1997, 7, 653–665. [Google Scholar] [CrossRef]
- Nussenzweig, A.; Chen, C.; Soares, V.D.C.; Sanchez, M.; Sokol, K.; Nussenzweig, M.C.; Li, G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nat. Cell Biol. 1996, 382, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chaudhuri, J.; Zhu, C.; Davidson, L.; Weaver, D.T.; Alt, F.W. A Targeted DNA-PKcs-Null Mutation Reveals DNA-PK-Independent Functions for KU in V(D)J Recombination. Immunity 1998, 9, 367–376. [Google Scholar] [CrossRef]
- Li, G.; Alt, F.W.; Cheng, H.-L.; Brush, J.W.; Goff, P.H.; Murphy, M.M.; Franco, S.; Zhang, Y.; Zha, S. Lymphocyte-Specific Compensation for XLF/Cernunnos End-Joining Functions in V(D)J Recombination. Mol. Cell 2008, 31, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Vera, G.; Rivera-Munoz, P.; Abramowski, V.; Malivert, L.; Lim, A.; Bole-Feysot, C.; Martin, C.; Florkin, B.; Latour, S.; Revy, P.; et al. Cernunnos Deficiency Reduces Thymocyte Life Span and Alters the T Cell Repertoire in Mice and Humans. Mol. Cell. Biol. 2012, 33, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Abramowski, V.; Etienne, O.; Elsaid, R.; Yang, J.; Berland, A.; Kermasson, L.; Roch, B.; Musilli, S.; Moussu, J.-P.; Lipson-Ruffert, K.; et al. PAXX and Xlf interplay revealed by impaired CNS development and immunodeficiency of double KO mice. Cell Death Differ. 2017, 25, 444–452. [Google Scholar] [CrossRef]
- Balmus, G.; Barros, A.C.; Wijnhoven, P.W.; Lescale, C.; Hasse, H.L.; Boroviak, K.; Le Sage, C.; Doe, B.; Speak, A.O.; Galli, A.; et al. Synthetic lethality between PAXX and XLF in mammalian development. Genes Dev. 2016, 30, 2152–2157. [Google Scholar] [CrossRef]
- Gago-Fuentes, R.; Xing, M.; Saeterstad, S.; Sarno, A.; Dewan, A.; Beck, C.; Bradamante, S.; Bjørås, M.; Oksenych, V. Normal development of mice lacking PAXX, the paralogue of XRCC4 and XLF. FEBS Open Bio 2018, 8, 426–434. [Google Scholar] [CrossRef]
- Liu, X.; Shao, Z.; Jiang, W.; Lee, B.J.; Zha, S. PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 2017, 8, 13816. [Google Scholar] [CrossRef]
- Castañeda-Zegarra, S.; Xing, M.; Gago-Fuentes, R.; Sæterstad, S.; Oksenych, V. Synthetic lethality between DNA repair factors Xlf and Paxx is rescued by inactivation of Trp53. DNA Repair 2019, 73, 164–169. [Google Scholar] [CrossRef]
- Hung, P.J.; Johnson, B.; Chen, B.-R.; Byrum, A.K.; Bredemeyer, A.L.; Yewdell, W.T.; Johnson, T.E.; Lee, B.J.; Deivasigamani, S.; Hindi, I.; et al. MRI Is a DNA Damage Response Adaptor during Classical Non-homologous End Joining. Mol. Cell 2018, 71, 332–342.e8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Crowe, J.L.; Liu, X.; Nakajima, S.; Wang, Y.; Li, C.; Lee, B.J.; Dubois, R.L.; Liu, C.; Yu, X.; et al. Differential Phosphorylation of DNA-PKcs Regulates the Interplay between End-Processing and End-Ligation during Nonhomologous End-Joining. Mol. Cell 2015, 58, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs kinases-the lessons from the mouse models: Inhibition not equal deletion. Cell Biosci. 2020, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.E.; Stamp, G.; Rosewell, I.; Denzel, A.; Lindahl, T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998, 8, 1395–1398. [Google Scholar] [CrossRef]
- Jiang, W.; Estes, V.M.; Wang, X.S.; Shao, Z.; Lee, B.J.; Lin, X.; Crowe, J.L.; Zha, S. Phosphorylation at S2053 in Murine (S2056 in Human) DNA-PKcs Is Dispensable for Lymphocyte Development and Class Switch Recombination. J. Immunol. 2019, 203, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, H.; Shiraishi, K.; Sakaguchi, K.; Nakahama, T.; Kodama, S. Repair kinetics of DNA double-strand breaks and incidence of apoptosis in mouse neural stem/progenitor cells and their differentiated neurons exposed to ionizing radiation. J. Radiat. Res. 2018, 59, 261–271. [Google Scholar] [CrossRef]
- Oksenych, V.; Kumar, V.; Liu, X.; Guo, C.; Schwer, B.; Zha, S.; Alt, F.W. Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining. Proc. Natl. Acad. Sci. USA 2013, 110, 2234–2239. [Google Scholar] [CrossRef]
- Xing, M.; Bjørås, M.; Daniel, J.A.; Alt, F.W.; Oksenych, V. Synthetic lethality between murine DNA repair factors XLF and DNA-PKcs is rescued by inactivation of Ku70. DNA Repair 2017, 57, 133–138. [Google Scholar] [CrossRef]
- Castañeda-Zegarra, S.; Huse, C.; Røsand, Ø.; Sarno, A.; Xing, M.; Gago-Fuentes, R.; Zhang, Q.; Alirezaylavasani, A.; Werner, J.; Ji, P.; et al. Generation of a Mouse Model Lacking the Non-Homologous End-Joining Factor Mri/Cyren. Biomolecules 2019, 9, 798. [Google Scholar] [CrossRef]
- Wang, W.; Osenbroch, P.; Skinnes, R.; Esbensen, Y.; Bjørås, M.; Eide, L. Mitochondrial DNA Integrity Is Essential For Mitochondrial Maturation During Differentiation of Neural Stem Cells. Stem Cells 2010, 28, 2195–2204. [Google Scholar] [CrossRef]
- Xing, M.; Oksenych, V. Genetic interaction between DNA repair factors PAXX, XLF, XRCC4 and DNA-PKcs in human cells. FEBS Open Bio 2019, 9, 1315–1326. [Google Scholar] [CrossRef]
- Dewan, A.; Xing, M.; Lundbaek, M.B.; Gago-Fuentes, R.; Beck, C.; Aas, P.A.; Liabakk, N.-B.; Saeterstad, S.; Chau, K.T.P.; Kavli, B.M.; et al. Robust DNA repair in PAXX-deficient mammalian cells. FEBS Open Bio 2018, 8, 442–448. [Google Scholar] [CrossRef]
- Ragunathan, K.; Upfold, N.L.E.; Oksenych, V. Interaction between Fibroblasts and Immune Cells Following DNA Damage Induced by Ionizing Radiation. Int. J. Mol. Sci. 2020, 21, 8635. [Google Scholar] [CrossRef]
- Wei, P.-C.; Chang, A.N.; Kao, J.; Du, Z.; Meyers, R.M.; Alt, F.W.; Schwer, B. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell 2016, 164, 644–655. [Google Scholar] [CrossRef]
- Schwer, B.; Wei, P.-C.; Chang, A.N.; Kao, J.; Du, Z.; Meyers, R.M.; Alt, F.W. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA 2016, 113, 2258–2263. [Google Scholar] [CrossRef]
- Alt, F.W.; Schwer, B. DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair 2018, 71, 158–163. [Google Scholar] [CrossRef]
- Glover, T.W.; Wilson, T.E. Molecular biology: Breaks in the brain. Nature 2016, 532, 46–47. [Google Scholar] [CrossRef]
- Wang, M.; Wei, P.-C.; Lim, C.K.; Gallina, I.S.; Marshall, S.; Marchetto, M.C.; Alt, F.W.; Gage, F.H. Increased Neural Progenitor Proliferation in a hiPSC Model of Autism Induces Replication Stress-Associated Genome Instability. Cell Stem Cell 2020, 26, 221–233.e6. [Google Scholar] [CrossRef]
- Li, J.; Shang, Y.; Wang, L.; Zhao, B.; Sun, C.; Li, J.; Li, S.; Li, C.; Tang, M.; Meng, F.-L.; et al. Genome integrity and neurogenesis of postnatal hippocampal neural stem/progenitor cells require a unique regulator Filia. Sci. Adv. 2020, 6, 1–15. [Google Scholar] [CrossRef]
- Buck, D.; Malivert, L.; De Chasseval, R.; Barraud, A.; Fondanèche, M.-C.; Sanal, O.; Plebani, A.; Stéphan, J.-L.; Hufnagel, M.; Le Deist, F.; et al. Cernunnos, a Novel Nonhomologous End-Joining Factor, Is Mutated in Human Immunodeficiency with Microcephaly. Cell 2006, 124, 287–299. [Google Scholar] [CrossRef]
- Du, L.; Peng, R.; Björkman, A.; De Miranda, N.F.; Rosner, C.; Kotnis, A.; Berglund, M.; Liu, C.; Rosenquist, R.; Enblad, G.; et al. Cernunnos influences human immunoglobulin class switch recombination and may be associated with B cell lymphomagenesis. J. Exp. Med. 2012, 209, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Zha, S.; Guo, C.; Boboila, C.; Oksenych, V.; Cheng, H.-L.; Zhang, Y.; Wesemann, D.R.; Yuen, G.; Patel, H.; Goff, P.H.; et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nat. Cell Biol. 2010, 469, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Castañeda-Zegarra, S.; Huse, C.; Xing, M.; Oksenych, V. Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF. Biomolecules 2019, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, W.; Dubois, R.L.; Yamamoto, K.; Wolner, Z.; Zha, S. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc. Natl. Acad. Sci. USA 2012, 109, 3903–3908. [Google Scholar] [CrossRef]
- Oksenych, V.; Alt, F.W.; Kumar, V.; Schwer, B.; Wesemann, D.R.; Hansen, E.; Patel, H.; Su, A.; Guo, C. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc. Natl. Acad. Sci. USA 2012, 109, 2455–2460. [Google Scholar] [CrossRef]
- Lescale, C.; Abramowski, V.; Bedora-Faure, M.; Murigneux, V.; Vera, G.; Roth, D.B.; Revy, P.; De Villartay, J.-P.; Deriano, L. RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat. Commun. 2016, 7, 10529. [Google Scholar] [CrossRef]
- Graham, T.G.; Walter, J.C.; Loparo, J.J. Two-Stage Synapsis of DNA Ends during Non-homologous End Joining. Mol. Cell 2016, 61, 850–858. [Google Scholar] [CrossRef]
- Carney, S.M.; Moreno, A.T.; Piatt, S.C.; Cisneros-Aguirre, M.; Lopezcolorado, F.W.; Stark, J.M.; Loparo, J.J. XLF acts as a flexible connector during non-homologous end joining. eLife 2020, 9, 9. [Google Scholar] [CrossRef]
- Zhao, B.; Watanabe, G.; Morten, M.J.; Reid, D.A.; Rothenberg, E.; Lieber, M.R. The essential elements for the noncovalent association of two DNA ends during NHEJ synapsis. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Zhao, B.; Rothenberg, E.; Ramsden, D.A.; Lieber, M.R. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 2020, 21, 765–781. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gago-Fuentes, R.; Oksenych, V. Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population. Biomolecules 2021, 11, 20. https://doi.org/10.3390/biom11010020
Gago-Fuentes R, Oksenych V. Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population. Biomolecules. 2021; 11(1):20. https://doi.org/10.3390/biom11010020
Chicago/Turabian StyleGago-Fuentes, Raquel, and Valentyn Oksenych. 2021. "Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population" Biomolecules 11, no. 1: 20. https://doi.org/10.3390/biom11010020
APA StyleGago-Fuentes, R., & Oksenych, V. (2021). Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population. Biomolecules, 11(1), 20. https://doi.org/10.3390/biom11010020





