Loss of ADAM9 Leads to Modifications of the Extracellular Matrix Modulating Tumor Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Cells and Reagents, Preparation of Conditioned Media
2.2. Proliferation Assays
2.3. Preparation of Recombinant ADAM9
2.4. Western Blot Analysis
2.5. Zymographic and In Vitro Enzymatic Assays
2.6. RNA Isolation, RT-PCR and Real-Time PCR
2.7. Tumor Growth Assay In Vivo
2.8. Mass Spectrometry
2.9. Histology
2.10. Statistical Analysis
3. Results
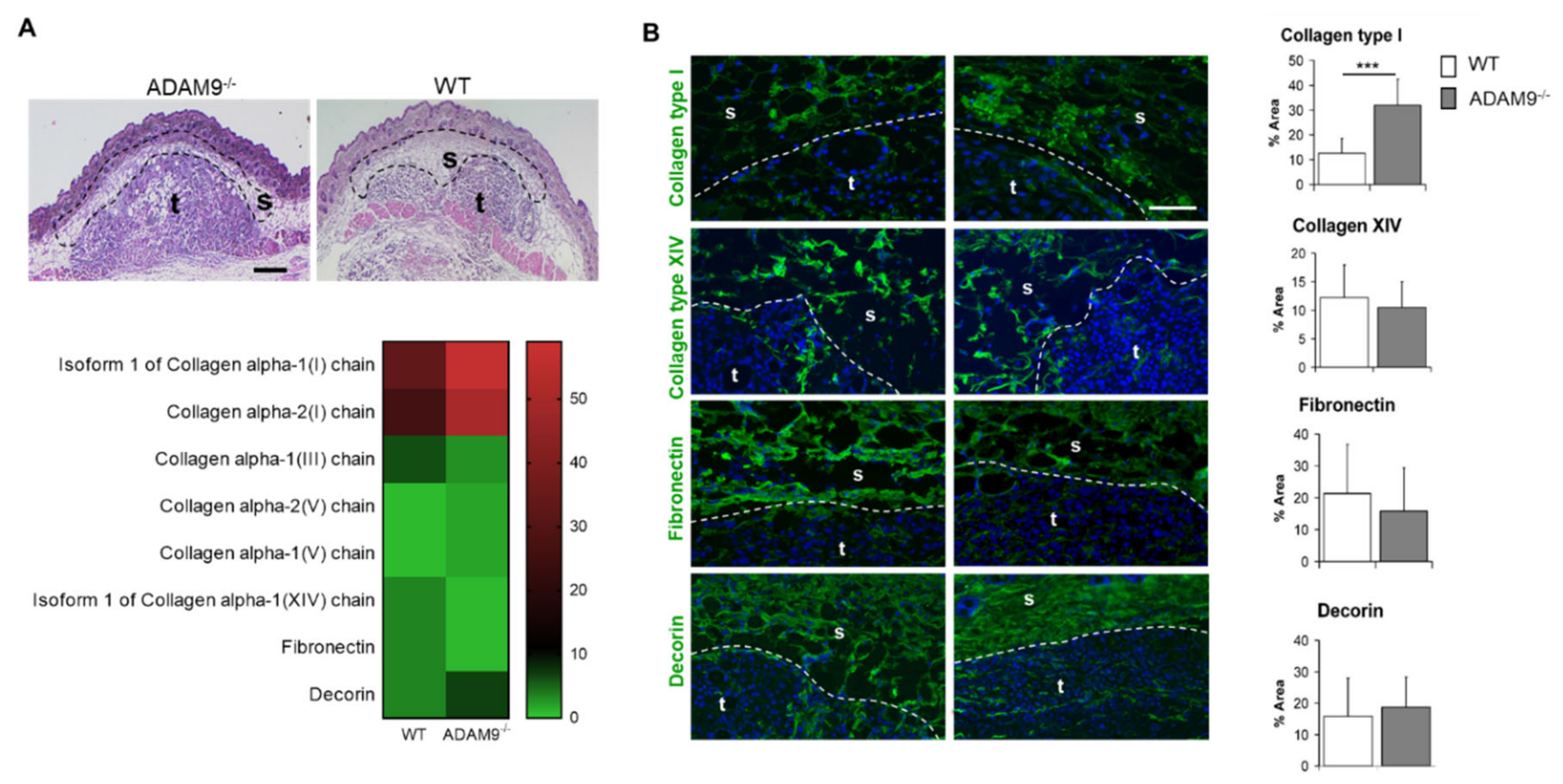
3.1. ECM Protein Expression at the Tumor-Stroma Border of B16F1 Melanomas
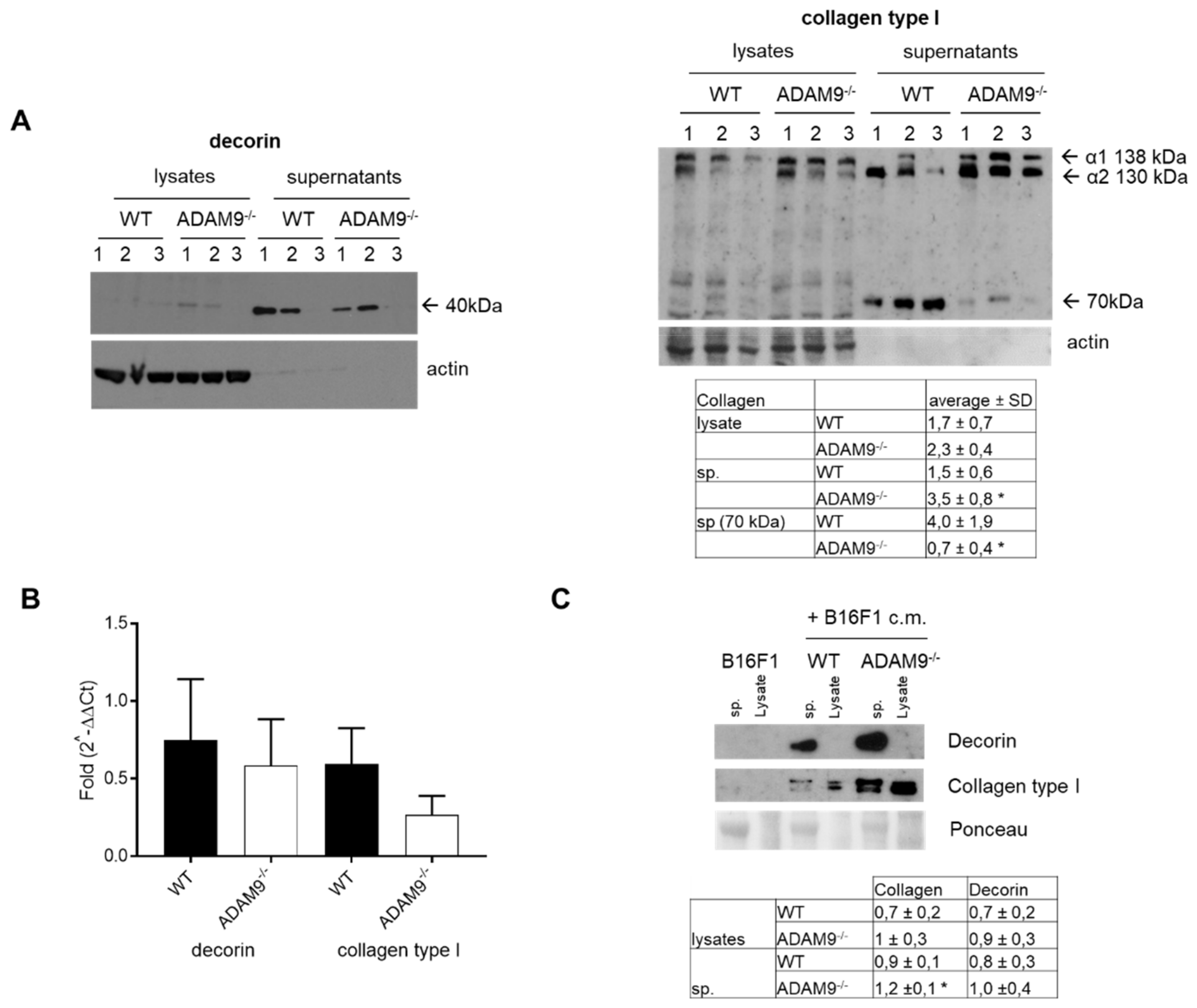
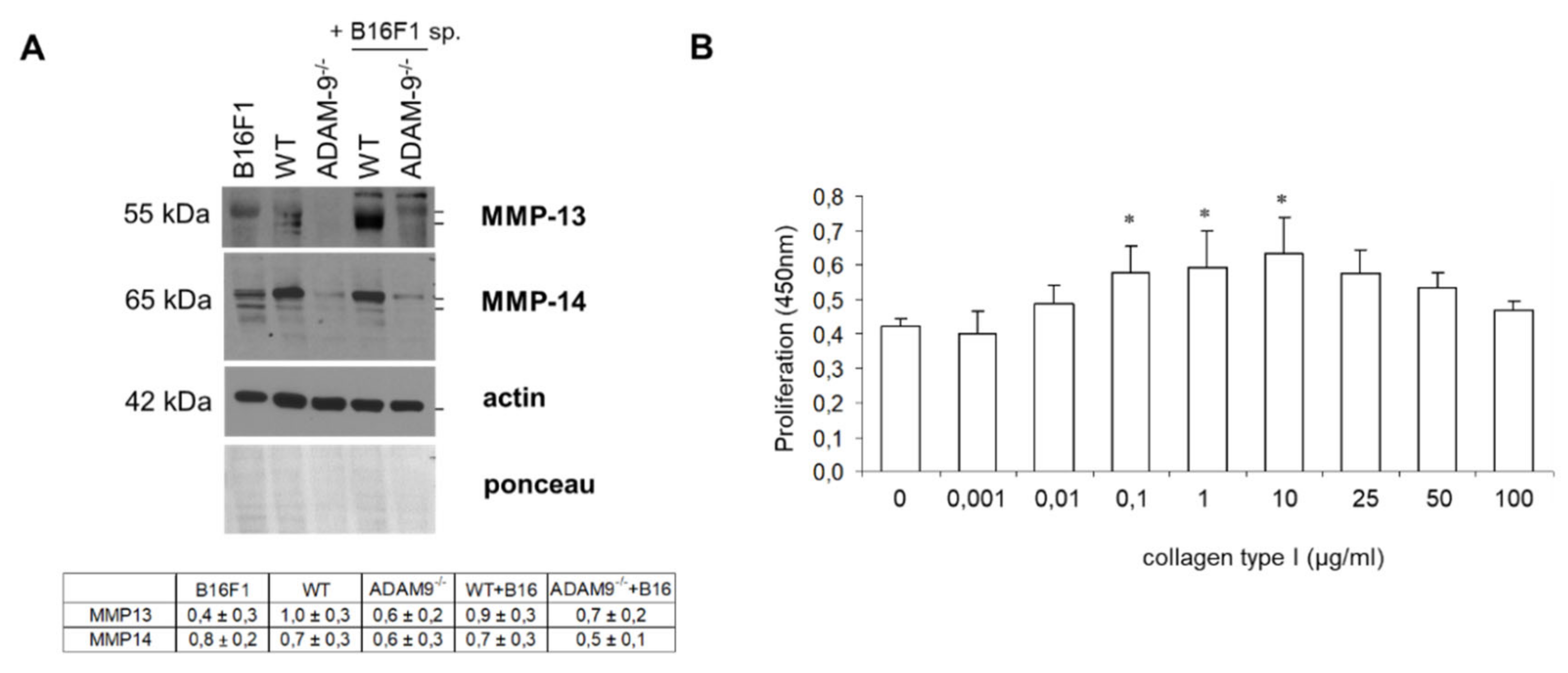
3.2. Increased Collagen Type I by Fibroblasts in the Absence of ADAM9
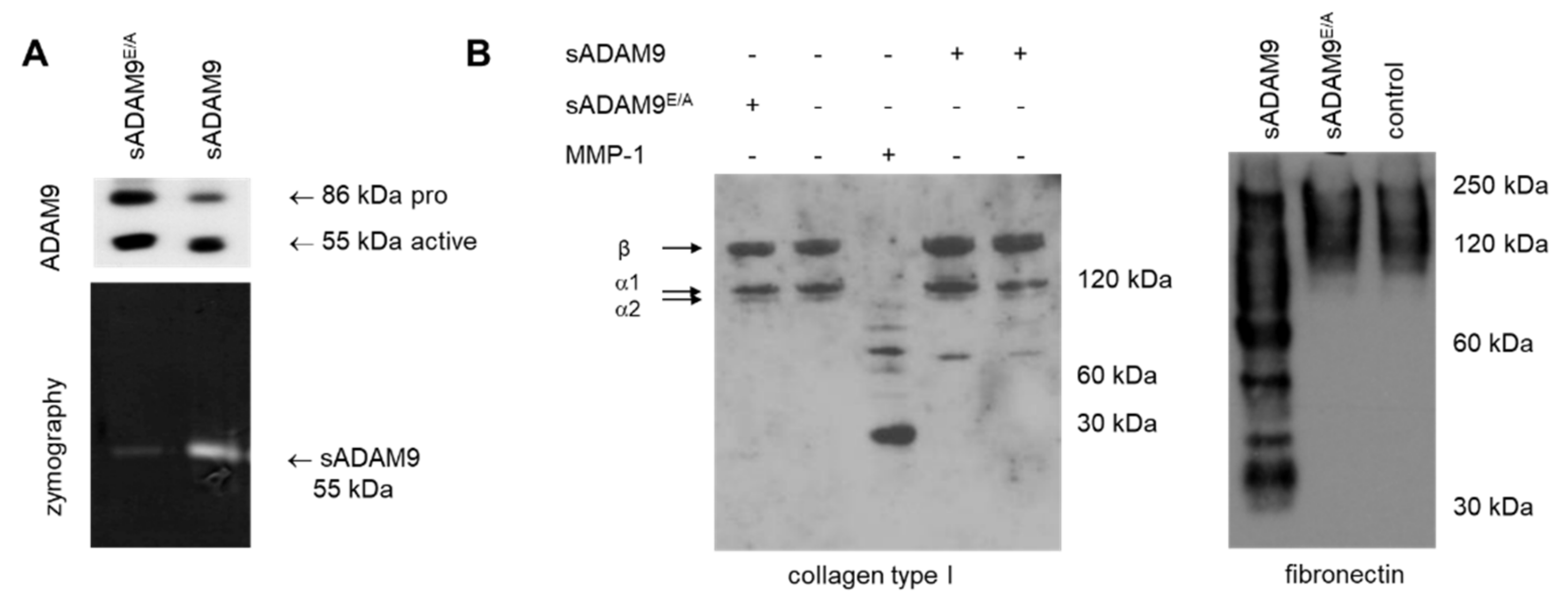
3.3. Impact of Collagen Type I Cleavage by ADAM9
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liotta, L.A.; Kohn, E.C. The microenvironment of the tumour-host interface. Nature 2001, 411, 375–379. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Giebeler, N.; Zigrino, P. A Disintegrin and Metalloprotease (ADAM): Historical Overview of Their Functions. Toxins 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, C.; McKie, N.; Buggy, Y.; Duggan, C.; Hill, A.D.; McDermott, E.; O’Higgins, N.; Duffy, M.J. Expression of ADAM-9 mRNA and protein in human breast cancer. Int. J. Cancer 2003, 105, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Peduto, L. ADAM9 as a potential target molecule in cancer. Curr. Pharm. Des. 2009, 15, 2282–2287. [Google Scholar] [CrossRef]
- Peduto, L.; Reuter, V.E.; Shaffer, D.R.; Scher, H.I.; Blobel, C.P. Critical function for ADAM9 in mouse prostate cancer. Cancer Res. 2005, 65, 9312–9319. [Google Scholar] [CrossRef]
- Zigrino, P.; Mauch, C.; Fox, J.W.; Nischt, R. Adam-9 expression and regulation in human skin melanoma and melanoma cell lines. Int. J. Cancer 2005, 116, 853–859. [Google Scholar] [CrossRef]
- Franzke, C.W.; Bruckner-Tuderman, L.; Blobel, C.P. Shedding of collagen XVII/BP180 in skin depends on both ADAM10 and ADAM9. J. Biol. Chem. 2009, 284, 23386–23396. [Google Scholar] [CrossRef]
- Schwettmann, L.; Tschesche, H. Cloning and expression in Pichia pastoris of metalloprotease domain of ADAM 9 catalytically active against fibronectin. Protein Expr. Purif. 2001, 21, 65–70. [Google Scholar] [CrossRef]
- Nath, D.; Slocombe, P.M.; Webster, A.; Stephens, P.E.; Docherty, A.J.; Murphy, G. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1 )integrin, leading to a marked induction of fibroblast cell motility. J. Cell Sci. 2000, 113, 2319–2328. [Google Scholar] [PubMed]
- Zigrino, P.; Nischt, R.; Mauch, C. The disintegrin-like and cysteine-rich domains of ADAM-9 mediate interactions between melanoma cells and fibroblasts. J. Biol. Chem. 2011, 286, 6801–6807. [Google Scholar] [CrossRef] [PubMed]
- Zigrino, P.; Steiger, J.; Fox, J.W.; Loffek, S.; Schild, A.; Nischt, R.; Mauch, C. Role of ADAM-9 disintegrin-cysteine-rich domains in human keratinocyte migration. J. Biol. Chem. 2007, 282, 30785–30793. [Google Scholar] [CrossRef] [PubMed]
- Giebeler, N.; Schonefuss, A.; Landsberg, J.; Tuting, T.; Mauch, C.; Zigrino, P. Deletion of ADAM-9 in HGF/CDK4 mice impairs melanoma development and metastasis. Oncogene 2017, 36, 5058–5067. [Google Scholar] [CrossRef]
- Abety, A.N.; Fox, J.W.; Schonefuss, A.; Zamek, J.; Landsberg, J.; Krieg, T.; Blobel, C.; Mauch, C.; Zigrino, P. Stromal fibroblast-specific expression of ADAM-9 modulates proliferation and apoptosis in melanoma cells in vitro and in vivo. J. Investig. Dermatol. 2012, 132, 2451–2458. [Google Scholar] [CrossRef]
- Florin, L.; Knebel, J.; Zigrino, P.; Vonderstrass, B.; Mauch, C.; Schorpp-Kistner, M.; Szabowski, A.; Angel, P. Delayed wound healing and epidermal hyperproliferation in mice lacking JunB in the skin. J. Investig. Dermatol. 2006, 126, 902–911. [Google Scholar] [CrossRef]
- Roghani, M.; Becherer, J.D.; Moss, M.L.; Atherton, R.E.; Erdjument-Bromage, H.; Arribas, J.; Blackburn, R.K.; Weskamp, G.; Tempst, P.; Blobel, C.P. Metalloprotease-disintegrin MDC9: Intracellular maturation and catalytic activity. J. Biol. Chem. 1999, 274, 3531–3540. [Google Scholar] [CrossRef]
- Zigrino, P.; Ayachi, O.; Schild, A.; Kaltenberg, J.; Zamek, J.; Nischt, R.; Koch, M.; Mauch, C. Loss of epidermal MMP-14 expression interferes with angiogenesis but not with re-epithelialization. Eur. J. Cell. Biol. 2012, 91, 748–756. [Google Scholar] [CrossRef]
- Zigrino, P.; Kuhn, I.; Bauerle, T.; Zamek, J.; Fox, J.W.; Neumann, S.; Licht, A.; Schorpp-Kistner, M.; Angel, P.; Mauch, C. Stromal expression of MMP-13 is required for melanoma invasion and metastasis. J. Investig. Dermatol. 2009, 129, 2686–2693. [Google Scholar] [CrossRef]
- Mikesh, L.M.; Aramadhaka, L.R.; Moskaluk, C.; Zigrino, P.; Mauch, C.; Fox, J.W. Proteomic anatomy of human skin. J. Proteomics 2013, 84, 190–200. [Google Scholar] [CrossRef]
- Mazzocca, A.; Coppari, R.; De Franco, R.; Cho, J.Y.; Libermann, T.A.; Pinzani, M.; Toker, A. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res. 2005, 65, 4728–4738. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H. Stromal reaction in cancer tissue: Pathophysiologic significance of the expression of matrix-degrading enzymes in relation to matrix turnover and immune/inflammatory reactions. Pathol. Int. 1998, 48, 1–9. [Google Scholar] [CrossRef] [PubMed]
- van Kempen, L.C.; Rijntjes, J.; Mamor-Cornelissen, I.; Vincent-Naulleau, S.; Gerritsen, M.J.; Ruiter, D.J.; van Dijk, M.C.; Geffrotin, C.; van Muijen, G.N. Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int. J. Cancer 2008, 122, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Geho, D.H.; Bandle, R.W.; Clair, T.; Liotta, L.A. Physiological mechanisms of tumor-cell invasion and migration. Physiology 2005, 20, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, S.; Gibertini, S.; Mora, M. Altered production of extra-cellular matrix components by muscle-derived Duchenne muscular dystrophy fibroblasts before and after TGF-beta1 treatment. Cell Tissue Res. 2010, 339, 397–410. [Google Scholar] [CrossRef]
- Mauch, C.; Zamek, J.; Abety, A.N.; Grimberg, G.; Fox, J.W.; Zigrino, P. Accelerated wound repair in ADAM-9 knockout animals. J. Investig. Dermatol. 2010, 130, 2120–2130. [Google Scholar] [CrossRef]
- Visse, R.; Nagase, H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: Structure, function, and biochemistry. Circ. Res. 2003, 92, 827–839. [Google Scholar] [CrossRef]
- Lukashev, M.E.; Werb, Z. ECM signalling: Orchestrating cell behaviour and misbehaviour. Trends Cell. Biol 1998, 8, 437–441. [Google Scholar] [CrossRef]
- Airola, K.; Karonen, T.; Vaalamo, M.; Lehti, K.; Lohi, J.; Kariniemi, A.L.; Keski-Oja, J.; Saarialho-Kere, U.K. Expression of collagenases-1 and -3 and their inhibitors TIMP-1 and -3 correlates with the level of invasion in malignant melanomas. Br. J. Cancer 1999, 80, 733–743. [Google Scholar] [CrossRef]
- Duffy, M.J.; Mullooly, M.; O’Donovan, N.; Sukor, S.; Crown, J.; Pierce, A.; McGowan, P.M. The ADAMs family of proteases: New biomarkers and therapeutic targets for cancer? Clin. Proteomics 2011, 8, 9. [Google Scholar] [CrossRef]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Perez-Gomez, B.; Palacios, J.; Pollan, M.; Linares, J.; Serrano, S.; Saez-Castillo, A.I.; Sanchez, L.; et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef] [PubMed]
- Wenstrup, R.J.; Florer, J.B.; Brunskill, E.W.; Bell, S.M.; Chervoneva, I.; Birk, D.E. Type V collagen controls the initiation of collagen fibril assembly. J. Biol. Chem. 2004, 279, 53331–53337. [Google Scholar] [CrossRef] [PubMed]
- Kalamajski, S.; Aspberg, A.; Oldberg, A. The decorin sequence SYIRIADTNIT binds collagen type I. J. Biol Chem. 2007, 282, 16062–16067. [Google Scholar] [CrossRef] [PubMed]
- Labrousse, A.L.; Ntayi, C.; Hornebeck, W.; Bernard, P. Stromal reaction in cutaneous melanoma. Crit Rev. Oncol. Hematol. 2004, 49, 269–275. [Google Scholar] [CrossRef]
- van Kempen, L.C.; van Muijen, G.N.; Ruiter, D.J. Stromal responses in human primary melanoma of the skin. Front. Biosci 2005, 10, 2922–2931. [Google Scholar] [CrossRef][Green Version]
- van Kempen, L.C.; Rijntjes, J.; Claes, A.; Blokx, W.A.; Gerritsen, M.J.; Ruiter, D.J.; van Muijen, G.N. Type I collagen synthesis parallels the conversion of keratinocytic intraepidermal neoplasia to cutaneous squamous cell carcinoma. J. Pathol. 2004, 204, 333–339. [Google Scholar] [CrossRef]
- Aoudjit, F.; Vuori, K. Integrin signaling in cancer cell survival and chemoresistance. Chemother Res. Pract. 2012, 2012, 283181. [Google Scholar] [CrossRef]
- Henriet, P.; Zhong, Z.D.; Brooks, P.C.; Weinberg, K.I.; DeClerck, Y.A. Contact with fibrillar collagen inhibits melanoma cell proliferation by up-regulating p27KIP1. Proc. Natl. Acad. Sci. USA 2000, 97, 10026–10031. [Google Scholar] [CrossRef]
- Huh, S.J.; Chen, Y.L.; Friedman, S.L.; Liao, J.; Huang, H.J.; Cavenee, W.K.; Robertson, G.P. KLF6 Gene and early melanoma development in a collagen I-rich extracellular environment. J. Natl. Cancer Inst. 2010, 102, 1131–1147. [Google Scholar] [CrossRef]
- Kielty, C.M.; Shuttleworth, C.A. Synthesis and assembly of fibrillin by fibroblasts and smooth muscle cells. J. Cell Sci. 1993, 106, 167–173. [Google Scholar]
- Zigrino, P.; Brinckmann, J.; Niehoff, A.; Lu, Y.; Giebeler, N.; Eckes, B.; Kadler, K.E.; Mauch, C. Fibroblast-Derived MMP-14 Regulates Collagen Homeostasis in Adult Skin. J. Investig. Dermatol. 2016, 136, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Jarvelainen, H.T.; Iruela-Arispe, M.L.; Kinsella, M.G.; Sandell, L.J.; Sage, E.H.; Wight, T.N. Expression of decorin by sprouting bovine aortic endothelial cells exhibiting angiogenesis in vitro. Exp. Cell Res. 1992, 203, 395–401. [Google Scholar] [CrossRef][Green Version]
- Iozzo, R.V. Matrix proteoglycans: From molecular design to cellular function. Annu Rev. Biochem. 1998, 67, 609–652. [Google Scholar] [CrossRef] [PubMed]
- Weber, I.T.; Harrison, R.W.; Iozzo, R.V. Model structure of decorin and implications for collagen fibrillogenesis. J. Biol. Chem. 1996, 271, 31767–31770. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H. Activation mechanisms of matrix metalloproteinases. Biol. Chem. 1997, 378, 151–160. [Google Scholar]
- Zigrino, P.; Mauch, C. Proteases in Melanoma. In Melanoma Development; Bosserhoff, A., Ed.; Springer: Vienna, Austria, 2011; pp. 165–179. [Google Scholar] [CrossRef]
- Lee, M.H.; Rapti, M.; Knauper, V.; Murphy, G. Threonine 98, the pivotal residue of tissue inhibitor of metalloproteinases (TIMP)-1 in metalloproteinase recognition. J. Biol. Chem. 2004, 279, 17562–17569. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abety, A.N.; Pach, E.; Giebeler, N.; Fromme, J.E.; Aramadhaka, L.R.; Mauch, C.; Fox, J.W.; Zigrino, P. Loss of ADAM9 Leads to Modifications of the Extracellular Matrix Modulating Tumor Growth. Biomolecules 2020, 10, 1290. https://doi.org/10.3390/biom10091290
Abety AN, Pach E, Giebeler N, Fromme JE, Aramadhaka LR, Mauch C, Fox JW, Zigrino P. Loss of ADAM9 Leads to Modifications of the Extracellular Matrix Modulating Tumor Growth. Biomolecules. 2020; 10(9):1290. https://doi.org/10.3390/biom10091290
Chicago/Turabian StyleAbety, Anna N., Elke Pach, Nives Giebeler, Julia E. Fromme, Lavakumar Reddy Aramadhaka, Cornelia Mauch, Jay W. Fox, and Paola Zigrino. 2020. "Loss of ADAM9 Leads to Modifications of the Extracellular Matrix Modulating Tumor Growth" Biomolecules 10, no. 9: 1290. https://doi.org/10.3390/biom10091290
APA StyleAbety, A. N., Pach, E., Giebeler, N., Fromme, J. E., Aramadhaka, L. R., Mauch, C., Fox, J. W., & Zigrino, P. (2020). Loss of ADAM9 Leads to Modifications of the Extracellular Matrix Modulating Tumor Growth. Biomolecules, 10(9), 1290. https://doi.org/10.3390/biom10091290






