Direct Observation of Sophorolipid Micelle Docking in Model Membranes and Cells by Single Particle Studies Reveals Optimal Fusion Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
Reagents for Tissue Culture
2.2. Preparation of Compounds
2.2.1. Production of Sophorolipids
2.2.2. Micelle Preparation and Purification
2.2.3. Liposomes Preparation
2.3. Data Acquisition
2.3.1. Data Acquisition for Single Liposome Experiments
2.3.2. Data Acquisition of CLSM Images
2.4. Live Cell Experiments
2.4.1. Cell Culture
2.4.2. Microscopy Cell Viability
2.4.3. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Cell Viability
2.5. Data Analysis
2.5.1. Data Analysis for Single Liposome Experiments
2.5.2. Exponential Fitting to Docking Rates
2.5.3. Quantification of CLSM Images
3. Results
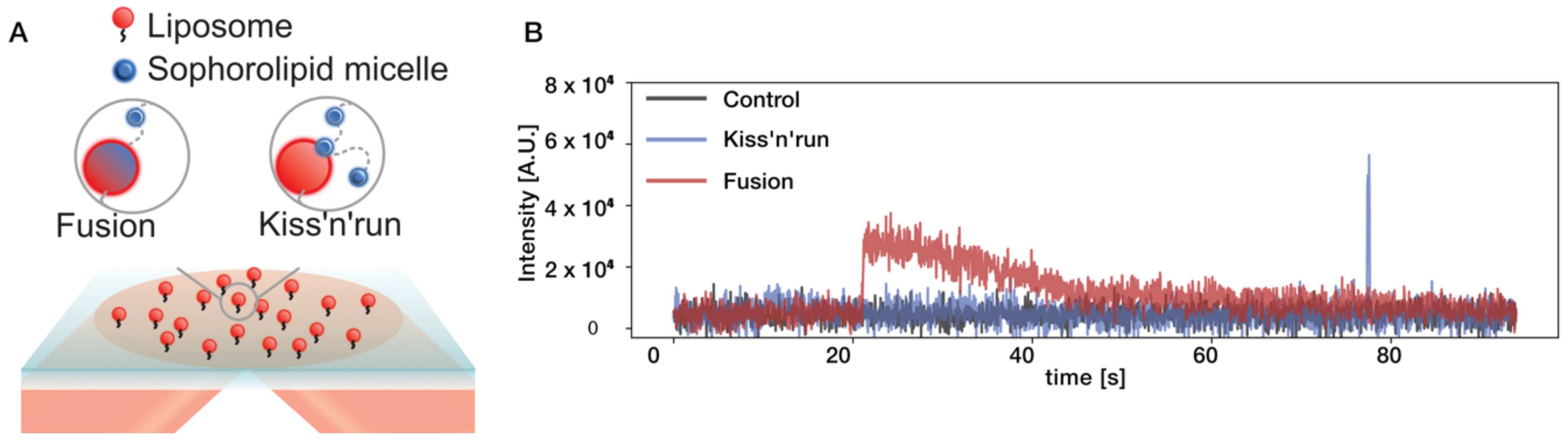
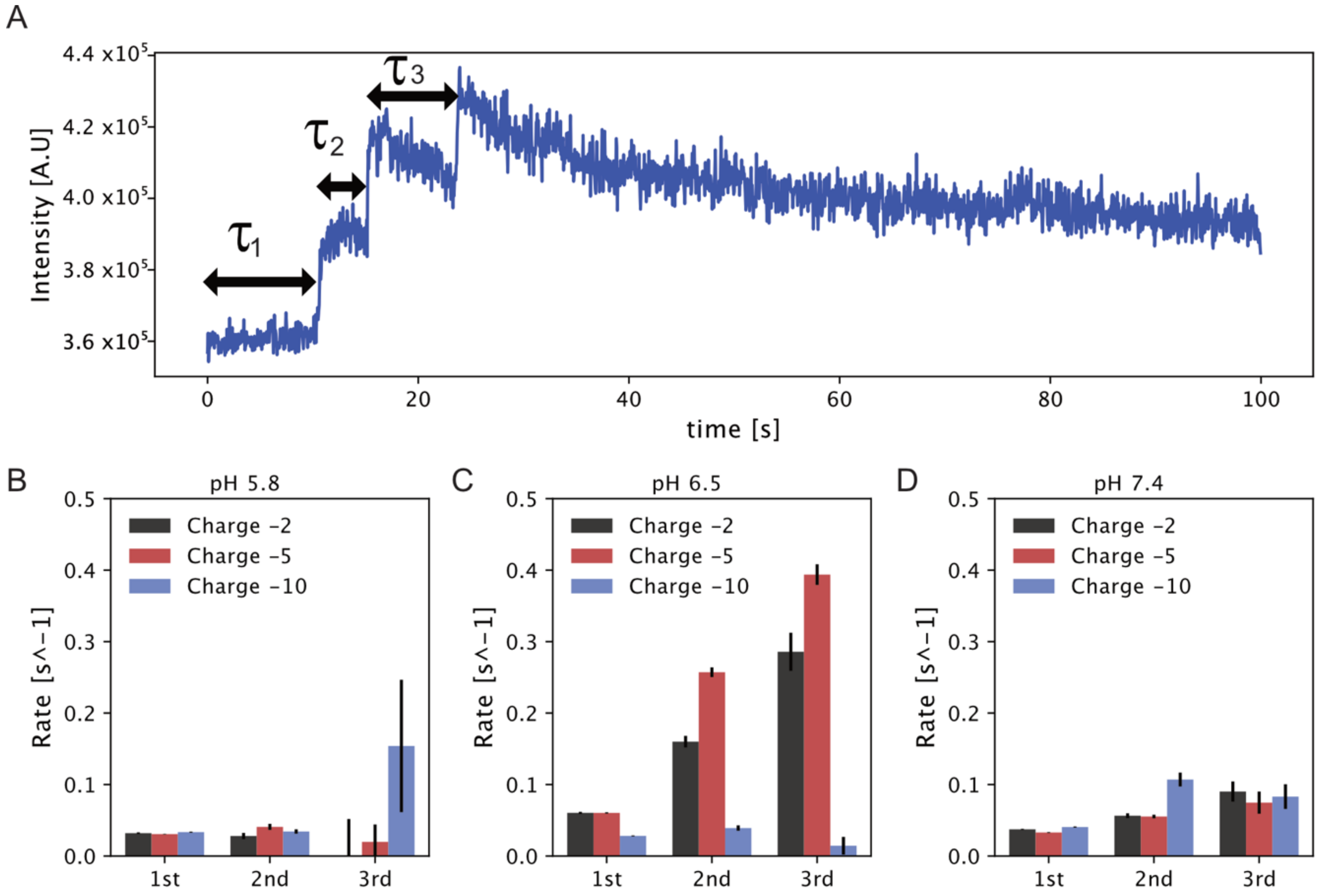
3.1. Sophorolipid Micelle Docking on Model Membranes
3.2. Sophorolipid Micelle Docking on Mammalian Cells
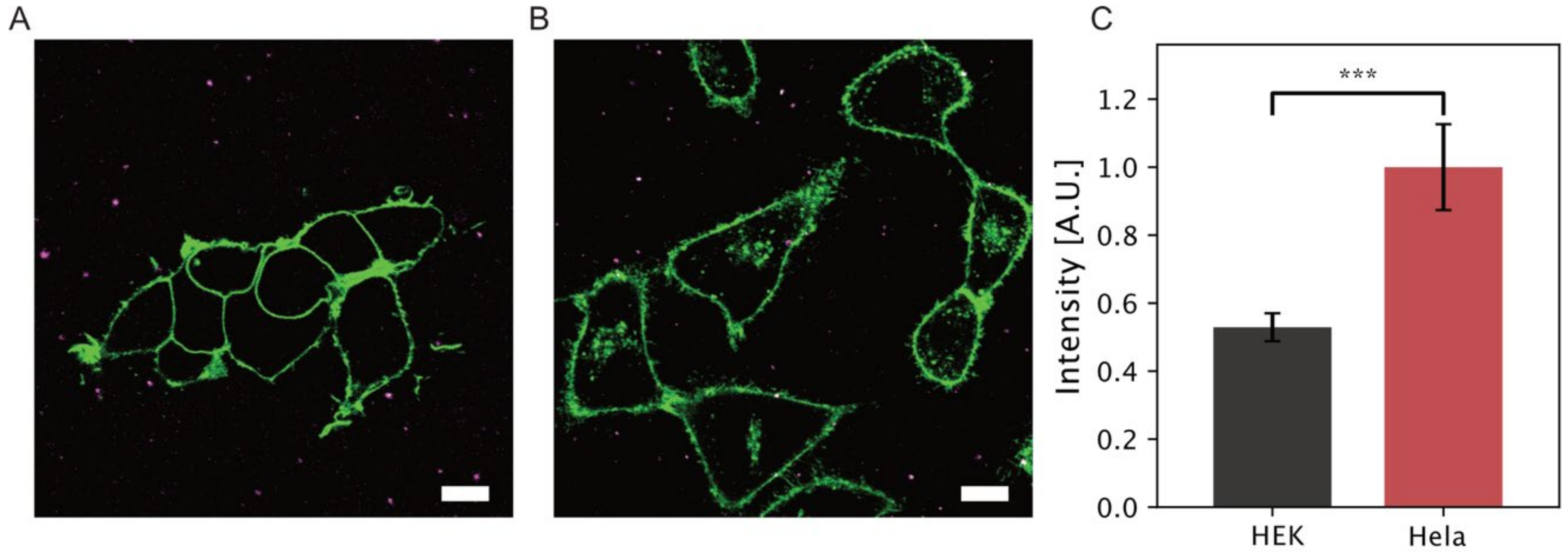
3.2.1. Sophorolipid uptake on Hela and HEK-293 Cells
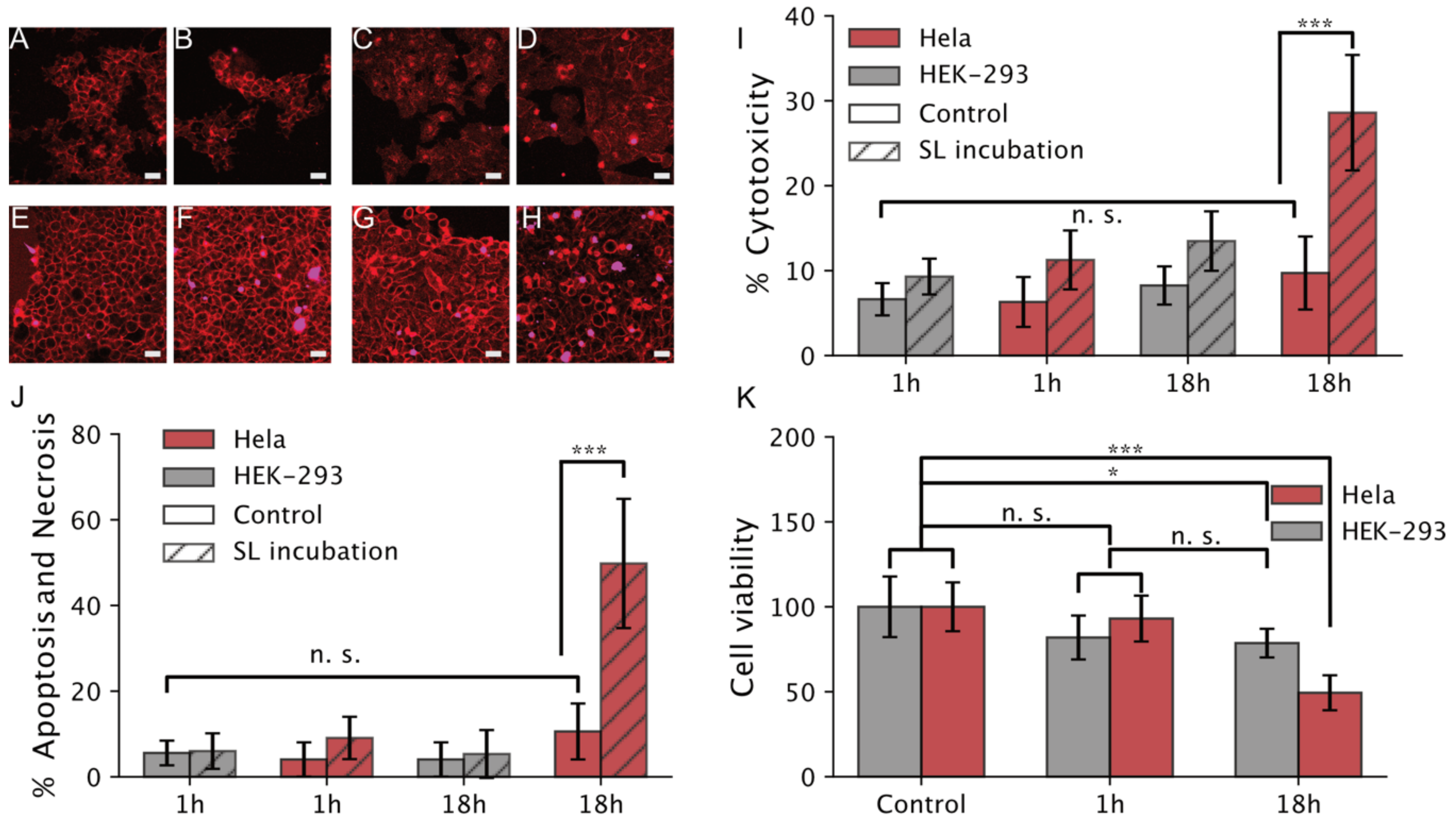
3.2.2. Cell Viability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Bogaert, I.N.A.; Zhang, J.; Soetaert, W. Microbial synthesis of sophorolipids. Process Biochem. 2011, 46, 821–833. [Google Scholar] [CrossRef]
- Zhang, L.; Somasundaran, P.; Singh, S.K.; Felse, A.P.; Gross, R. Synthesis and interfacial properties of sophorolipid derivatives. Colloids Surf. A Physicochem. Eng. Asp. 2004, 240, 75–82. [Google Scholar] [CrossRef]
- Van Bogaert, I.N.A.; Saerens, K.; De Muynck, C.; Develter, D.; Soetaert, W.; Vandamme, E.J. Microbial production and application of sophorolipids. Appl. Microbiol. Biotechnol. 2007, 76, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Borah, S.N.; Bora, A.; Deka, S. Production, characterization, and antifungal activity of a biosurfactant produced by Rhodotorula babjevae YS3. Microb. Cell Fact. 2017, 16, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Katsiwela, E.; Wagner, F. Antimicrobial effects of biosurfactants. Fett Wiss. Technol. 1989, 91, 363–366. [Google Scholar] [CrossRef]
- Vasudevan, S.; Prabhune, A.A. Photophysical studies on curcumin-sophorolipid nanostructures: Applications in quorum quenching and imaging. R. Soc. Open Sci. 2018, 5, 170865–170879. [Google Scholar] [CrossRef]
- Shah, V.; Doncel, G.F.; Seyoum, T.; Eaton, K.M.; Zalenskaya, I.; Hagver, R.; Azim, A.; Gross, R. Sophorolipids, microbial glycolipids with anti-human immunodeficiency virus and sperm-immobilizing activities. Antimicrob. Agents Chemother. 2005, 49, 4093–4100. [Google Scholar] [CrossRef]
- Rodrigues, L.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef]
- Kithur Mohamed, S.; Asif, M.; Nazari, M.V.; Baharetha, H.M.; Mahmood, S.; Yatim, A.R.M.; Abdul Majid, A.S.; Abdul Majid, A.M.S. Antiangiogenic activity of sophorolipids extracted from refined bleached deodorized palm olein. Indian J. Pharmacol. 2019, 51, 45–54. [Google Scholar] [CrossRef]
- Singh, P.K.; Wani, K.; Kaul-Ghanekar, R.; Prabhune, A.; Ogale, S. From micron to nano-curcumin by sophorolipid co-processing: Highly enhanced bioavailability, fluorescence, and anti-cancer efficacy. RSC Adv. 2014, 4, 60334–60341. [Google Scholar] [CrossRef]
- Akiyode, O.; George, D.; Getti, G.; Boateng, J. Systematic comparison of the functional physico-chemical characteristics and biocidal activity of microbial derived biosurfactants on blood-derived and breast cancer cells. J. Colloid Interface Sci. 2016, 479, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, I.A.C.; Faustino, C.M.C.; Guerreiro, P.S.; Frade, R.F.M.; Bronze, M.R.; Castro, M.F.; Ribeiro, M.H.L. Development of novel sophorolipids with improved cytotoxic activity toward MDA-MB-231 breast cancer cells. J. Mol. Recognit. 2015, 28, 155–165. [Google Scholar] [CrossRef]
- Li, H.; Guo, W.; Ma, X.-J.; Li, J.-S.; Song, X. In vitro and in vivo anticancer activity of sophorolipids to human cervical cancer. Appl. Biochem. Biotechnol. 2017, 181, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Dubey, P.; Raina, P.; Prabhune, A.; Ghanekar, R.K. Cetyl alcohol and oleic acid sophorolipids exhibit anticancer activity. Int. J. Pharm. Pharm. Sci. 2016, 8, 399–402. [Google Scholar]
- Shao, L.; Song, X.; Ma, X.; Li, H.; Qu, Y. Bioactivities of sophorolipid with different structures against human esophageal cancer cells. J. Surg. Res. 2012, 173, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Reddy, E.M.; Prabhune, A.; Pokharkar, V.; Shiras, A.; Prasad, B.L.V. Cytotoxicity of sophorolipid-gellan gum-gold nanoparticle conjugates and their doxorubicin loaded derivatives towards human glioma and human glioma stem cell lines. Nanoscale 2011, 3, 575–580. [Google Scholar] [CrossRef]
- Callaghan, B.; Lydon, H.; Roelants, S.L.K.W.; Van Bogaert, I.N.A.; Marchant, R.; Banat, I.M.; Mitchell, C.A. Lactonic Sophorolipids Increase Tumor Burden in Apcmin+/- Mice. PLoS ONE 2016, 11, 1–16. [Google Scholar] [CrossRef]
- Chen, J.; Song, X.; Zhang, H.; Qu, Y. Production, structure elucidation and anticancer properties of sophorolipid from Wickerhamiella domercqiae. Enzyme Microb. Technol. 2006, 39, 501–506. [Google Scholar] [CrossRef]
- Chen, J.; Song, X.; Zhang, H.; Qu, Y.-B.; Miao, J.-Y. Sophorolipid produced from the new yeast strain Wickerhamiella domercqiae induces apoptosis in H7402 human liver cancer cells. Appl. Microbiol. Biotechnol. 2006, 72, 52–59. [Google Scholar] [CrossRef]
- Nawale, L.; Dubey, P.; Chaudhari, B.; Sarkar, D.; Prabhune, A. Anti-proliferative effect of novel primary cetyl alcohol derived sophorolipids against human cervical cancer cells HeLa. PLoS ONE 2017, 12, 1–14. [Google Scholar] [CrossRef]
- Fu, S.L.; Wallner, S.R.; Bowne, W.B.; Hagler, M.D.; Zenilman, M.E.; Gross, R.; Bluth, M.H. Sophorolipids and their derivatives are lethal against human pancreatic cancer cells. J. Surg. Res. 2008, 148, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Kasture, M.; Singh, S.; Patel, P.; Joy, P.A.; Prabhune, A.A.; Ramana, C.V.; Prasad, B.L.V. Multiutility sophorolipids as nanoparticle capping agents: Synthesis of stable and water dispersible Co nanoparticles. Langmuir 2007, 23, 11409–11412. [Google Scholar] [CrossRef] [PubMed]
- Baccile, N.; Noiville, R.; Stievano, L.; Van Bogaert, I. Sophorolipids-functionalized iron oxide nanoparticles. Phys. Chem. Chem. Phys. 2013, 15, 1606–1620. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Mukherji, R.; Joshi-Navare, K.; Banerjee, A.; Gokhale, R.; Nagane, S.; Prabhune, A.; Ogale, S. Fluorescent sophorolipid molecular assembly and its magnetic nanoparticle loading: A pulsed laser process. Green Chem. 2013, 15, 943–953. [Google Scholar] [CrossRef]
- Rashad, M.M.; Nooman, M.U.; Ali, M.M.; Al-kashef, A.S.; Mahmoud, A.E. Production, characterization and anticancer activity of Candida bombicola sophorolipids by means of solid state fermentation of sunflower oil cake and soybean oil. Grasas y Aceites 2014, 65, 1–10. [Google Scholar] [CrossRef]
- Hatzakis, N.S.; Bhatia, V.K.; Larsen, J.; Madsen, K.L.; Bolinger, P.-Y.; Kunding, A.H.; Castillo, J.; Gether, U.; Hedegård, P.; Stamou, D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009, 5, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.M.; Bolinger, P.-Y.; Hatzakis, N.S.; Mortensen, M.W.; Stamou, D. Mixing subattolitre volumes in a quantitative and highly parallel manner with soft matter nanofluidics. Nat. Nanotechnol. 2011, 7, 51–55. [Google Scholar] [CrossRef]
- Clifton, L.A.; Campbell, R.A.; Sebastiani, F.; Campos-Terán, J.; Gonzalez-Martinez, J.F.; Björklund, S.; Sotres, J.; Cárdenas, M. Design and use of model membranes to study biomolecular interactions using complementary surface-sensitive techniques. Adv. Colloid Interface Sci. 2020, 277, 102118–102140. [Google Scholar] [CrossRef]
- Pomorski, T.G.; Nylander, T.; Cárdenas, M. Model cell membranes: Discerning lipid and protein contributions in shaping the cell. Adv. Colloid Interface Sci. 2014, 205, 207–220. [Google Scholar] [CrossRef]
- Rau, U.; Heckmann, R.; Wray, V.; Lang, S. Enzymatic conversion of a sophorolipid into a glucose lipid. Biotechnol. Lett. 1999, 21, 973–977. [Google Scholar] [CrossRef]
- Thomsen, R.P.; Malle, M.G.; Okholm, A.H.; Krishnan, S.; Bohr, S.S.-R.; Sørensen, R.S.; Ries, O.; Vogel, S.; Simmel, F.C.; Hatzakis, N.S.; et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 2019, 10, 5655–5665. [Google Scholar] [CrossRef] [PubMed]
- Bohr, S.S.-R.; Thorlaksen, C.; Kühnel, R.M.; Günther Pomorski, T.; Hatzakis, N.S. Label free fluorescence quantification of hydrolytic enzyme activity on native substrates reveal how lipase function depends on membrane curvature. Langmuir 2020, 36, 6473–6481. [Google Scholar] [CrossRef] [PubMed]
- Bech, E.M.; Kaiser, A.; Bellmann-Sickert, K.; Nielsen, S.S.-R.; Sørensen, K.K.; Elster, L.; Hatzakis, N.; Pedersen, S.L.; Beck-Sickinger, A.G.; Jensen, K.J. Half-Life Extending Modifications of Peptide YY3-36 Direct Receptor-Mediated Internalization. Mol. Pharm. 2019, 16, 3665–3677. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Mudaliar, P.; Jaleel, A.; Srikanth, J.; Sreekumar, E. High throughput proteomic analysis and a comparative review identify the nuclear chaperone, Nucleophosmin among the common set of proteins modulated in Chikungunya virus infection. J. Proteom. 2015, 120, 126–141. [Google Scholar] [CrossRef]
- Lin, P.-H.; Lin, H.-Y.; Kuo, C.-C.; Yang, L.-T. N-terminal functional domain of Gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting. J. Biomed. Sci. 2015, 22, 44–62. [Google Scholar] [CrossRef]
- Rodríguez Castaño, P.; Parween, S.; Pandey, A.V. Bioactivity of curcumin on the cytochrome P450 enzymes of the steroidogenic pathway. Int. J. Mol. Sci. 2019, 20, 4606. [Google Scholar] [CrossRef]
- Bohr, S.S.-R.; Lund, P.M.; Kallenbach, A.S.; Pinholt, H.; Thomsen, J.; Iversen, L.; Svendsen, A.; Christensen, S.M.; Hatzakis, N.S. Direct observation of Thermomyces lanuginosus lipase diffusional states by Single Particle Tracking and their remodeling by mutations and inhibition. Sci. Rep. 2019, 9, 16169–16180. [Google Scholar] [CrossRef]
- Wan, F.; Bohr, S.S.-R.; Kłodzińska, S.N.; Jumaa, H.; Huang, Z.; Nylander, T.; Thygesen, M.B.; Sørensen, K.K.; Jensen, K.J.; Sternberg, C.; et al. Ultrasmall TPGS-PLGA Hybrid Nanoparticles for Site-Specific Delivery of Antibiotics into Pseudomonas aeruginosa Biofilms in Lungs. ACS Appl. Mater. Interfaces 2020, 12, 380–389. [Google Scholar] [CrossRef]
- Joshi-Navare, K.; Prabhune, A. A biosurfactant-sophorolipid acts in synergy with antibiotics to enhance their efficiency. BioMed Res. Int. 2013, 2013, 512495–512503. [Google Scholar] [CrossRef]
- Veshaguri, S.; Christensen, S.M.; Kemmer, G.C.; Ghale, G.; Møller, M.P.; Lohr, C.; Christensen, A.L.; Justesen, B.H.; Jørgensen, I.L.; Schiller, J.; et al. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science 2016, 351, 1469–1473. [Google Scholar] [CrossRef]
- Stella, S.; Mesa, P.; Thomsen, J.; Paul, B.; Alcón, P.; Jensen, S.B.; Saligram, B.; Moses, M.E.; Hatzakis, N.S.; Montoya, G. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 2018, 175, 1856–1871. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.C.-H.; Gandhi, J.G.; Zia, R.N.; Paszek, M.J. Physical biology of the cancer cell glycocalyx. Nat. Phys. 2018, 14, 658–669. [Google Scholar] [CrossRef]
- Newell, K.J.; Tannock, I.F. Reduction of intracellular pH as a possible mechanism for killing cells in acidic regions of solid tumors: Effects of carbonylcyanide-3-chlorophenylhydrazone. Cancer Res. 1989, 49, 4477–4482. [Google Scholar] [PubMed]
- Persi, E.; Duran-Frigola, M.; Damaghi, M.; Roush, W.R.; Aloy, P.; Cleveland, J.L.; Gillies, R.J.; Ruppin, E. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat. Commun. 2018, 9, 2997–3008. [Google Scholar] [CrossRef]
- Bowen, M.E.; Weninger, K.; Brunger, A.T.; Chu, S. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J. 2004, 87, 3569–3584. [Google Scholar] [CrossRef]
- Kiessling, V.; Liang, B.; Kreutzberger, A.J.B.; Tamm, L.K. Planar Supported Membranes with Mobile SNARE Proteins and Quantitative Fluorescence Microscopy Assays to Study Synaptic Vesicle Fusion. Front. Mol. Neurosci. 2017, 10, 72–80. [Google Scholar] [CrossRef]
- Xie, Y.; Xianyu, Y.; Wang, N.; Yan, Z.; Liu, Y.; Zhu, K.; Hatzakis, N.S.; Jiang, X. Functionalized gold nanoclusters identify highly reactive oxygen species in living organisms. Adv. Funct. Mater. 2018, 28, 1702026–1702033. [Google Scholar] [CrossRef]
- Darne, P.A.; Mehta, M.R.; Agawane, S.B.; Prabhune, A.A. Bioavailability studies of curcumin–sophorolipid nano-conjugates in the aqueous phase: Role in the synthesis of uniform gold nanoparticles. RSC Adv. 2016, 6, 68504–68514. [Google Scholar] [CrossRef]
| pH | 5.8 | 6.5 | 7.4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SURFACE CHARGE | −2 | −5 | −10 | −2 | −5 | −10 | −2 | −5 | −10 |
| LIPOSOMES | 1242 | 1608 | 1817 | 2011 | 1214 | 1666 | 1399 | 1427 | 1711 |
| TOTAL EVENTS | 902 | 1164 | 1192 | 1388 | 1903 | 1315 | 1370 | 1495 | 1427 |
| LIPOSOMES WITH EVENTS | 845 | 1083 | 1093 | 1197 | 1244 | 1229 | 1195 | 1325 | 1309 |
| KISS-AND-RUNS | 537 | 1197 | 1192 | 1221 | 1040 | 1512 | 1891 | 2218 | 1911 |
| EVENTS PER LIPOSOME | 0.73 | 0.72 | 0.66 | 0.69 | 1.57 | 0.79 | 0.97 | 1.04 | 0.83 |
| LIKELIHOOD OF MULTIPLE EVENTS | 0.33 | 0.32 | 0.36 | 0.3 | 0.52 | 0.35 | 0.39 | 0.39 | 0.34 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.K.; Bohr, S.S.-R.; Hatzakis, N.S. Direct Observation of Sophorolipid Micelle Docking in Model Membranes and Cells by Single Particle Studies Reveals Optimal Fusion Conditions. Biomolecules 2020, 10, 1291. https://doi.org/10.3390/biom10091291
Singh PK, Bohr SS-R, Hatzakis NS. Direct Observation of Sophorolipid Micelle Docking in Model Membranes and Cells by Single Particle Studies Reveals Optimal Fusion Conditions. Biomolecules. 2020; 10(9):1291. https://doi.org/10.3390/biom10091291
Chicago/Turabian StyleSingh, Pradeep Kumar, Søren S.-R. Bohr, and Nikos S. Hatzakis. 2020. "Direct Observation of Sophorolipid Micelle Docking in Model Membranes and Cells by Single Particle Studies Reveals Optimal Fusion Conditions" Biomolecules 10, no. 9: 1291. https://doi.org/10.3390/biom10091291
APA StyleSingh, P. K., Bohr, S. S.-R., & Hatzakis, N. S. (2020). Direct Observation of Sophorolipid Micelle Docking in Model Membranes and Cells by Single Particle Studies Reveals Optimal Fusion Conditions. Biomolecules, 10(9), 1291. https://doi.org/10.3390/biom10091291







