Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms
Abstract
1. Introduction
2. Research Design and Methods
2.1. Animal Model Experiment
2.2. Biochemical Plasma Profile
2.3. Liver Triglyceride Determination
2.4. Standard Histological Analysis
2.5. RNA Preparation and Quantitative Real-Time Reverse Transcription–Polymerase Chain Reaction (RT-PCR) Analysis
2.6. Lipidomics
2.7. Tissue Molecular Imaging
2.8. Statistical Analysis
3. Results
3.1. Characteristics of the HFD Mice Model
3.1.1. HFD Mice Developed Obesity and Metabolic Disturbances
3.1.2. HFD-Induced Hepatic Steatosis in Mice
3.1.3. Expression of Genes Involved in Fatty Acid Intracellular Lipid Metabolism and Inflammation Pathways
3.2. Liver Triglyceride Composition in HFD-Fed Mice Revealed by Lipidomics
3.3. Correlations between Plasma Adipokines and Liver Triglycerides
3.4. Determination of Plasma FABP4 Origin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bonora, E.; Targher, G. Increased risk of cardiovascular disease and chronic kidney disease in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 372–381. [Google Scholar] [CrossRef]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Kanwal, F.; Kramer, J.R.; Duan, Z.; Yu, X.; White, D.; El-Serag, H.B. Trends in the Burden of Nonalcoholic Fatty Liver Disease in a United States Cohort of Veterans. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2016, 14, 301–308.E2. [Google Scholar] [CrossRef]
- Lambert, J.E.; Ramos-Roman, M.A.; Browning, J.D.; Parks, E.J. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology 2014, 146, 726–735. [Google Scholar] [CrossRef]
- Mittal, S.; El-Serag, H.B.; Sada, Y.H.; Kanwal, F.; Duan, Z.; Temple, S.; May, S.B.; Kramer, J.R.; Richardson, P.A.; Davila, J.A. Hepatocellular Carcinoma in the Absence of Cirrhosis in United States Veterans is Associated with Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2016, 14, 124–131.E1. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Titchenell, P.M.; Lazar, M.A.; Birnbaum, M.J. Unraveling the Regulation of Hepatic Metabolism by Insulin. Trends Endocrinol. Metab. 2017, 28, 497–505. [Google Scholar] [CrossRef]
- Samuel, V.T.; Shulman, G.I. The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux. J. Clin. Investig. 2016, 126, 12–22. [Google Scholar] [CrossRef]
- Yki-Jarvinen, H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2014, 2, 901–910. [Google Scholar] [CrossRef]
- Deacon, E.M.; Pettitt, T.R.; Webb, P.; Cross, T.; Chahal, H.; Wakelam, M.; Lord, J.M. Generation of diacylglycerol molecular species through the cell cycle: A role for 1-stearoyl, 2-arachidonyl glycerol in the activation of nuclear protein kinase C-betaII at G2/M. J. Cell Sci. 2002, 115 Pt 5, 983–989. [Google Scholar]
- Lambertucci, F.; Arboatti, A.; Sedlmeier, M.G.; Motiño, O.; Alvarez, M.D.L.; Ceballos, M.P.; Villar, S.R.; Roggero, E.; Monti, J.A.; Pisani, G.; et al. Disruption of tumor necrosis factor alpha receptor 1 signaling accelerates NAFLD progression in mice upon a high-fat diet. J. Nutr. Biochem. 2018, 58, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Macrae, K.; Stretton, C.; Lipina, C.; Blachnio-Zabielska, A.; Baranowski, M.; Gorski, J.; Marley, A.; Hundal, H.S. Defining the role of DAG, mitochondrial function, and lipid deposition in palmitate-induced proinflammatory signaling and its counter-modulation by palmitoleate. J. Lipid Res. 2013, 54, 2366–2378. [Google Scholar] [CrossRef] [PubMed]
- Boslem, E.; MacIntosh, G.; Preston, A.M.; Bartley, C.; Busch, A.K.; Fuller, M.; Laybutt, D.R.; Meikle, P.J.; Biden, T.J. A lipidomic screen of palmitate-treated MIN6 beta-cells links sphingolipid metabolites with endoplasmic reticulum (ER) stress and impaired protein trafficking. Biochem. J. 2011, 435, 267–276. [Google Scholar] [CrossRef]
- Arguello, G.; Balboa, E.; Arrese, M.; Zanlungo, S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim. Biophys. Acta 2015, 1852, 1765–1778. [Google Scholar] [CrossRef]
- Tamura, S.; Shimomura, I. Contribution of adipose tissue and de novo lipogenesis to nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Boutari, C.; Perakakis, N.; Mantzoros, C.S. Association of Adipokines with Development and Progression of Nonalcoholic Fatty Liver Disease. Endocrinol. Metab. 2018, 33, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Divella, R.; Mazzocca, A.; Daniele, A.; Sabba, C.; Paradiso, A. Obesity, Nonalcoholic Fatty Liver Disease and Adipocytokines Network in Promotion of Cancer. Int. J. Biol. Sci. 2019, 15, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Mantzoros, C.S. Leptin in health and disease: Facts and expectations at its twentieth anniversary. Metab. Clin. Exp. 2015, 64, 5–12. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kountouras, J.; Zavos, C. Nonalcoholic fatty liver disease: The pathogenetic roles of insulin resistance and adipocytokines. Curr. Mol. Med. 2009, 9, 299–314. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kountouras, J.; Zavos, C.; Tsiaousi, E. The role of adiponectin in the pathogenesis and treatment of non-alcoholic fatty liver disease. Diabetes Obes. Metab. 2010, 12, 365–383. [Google Scholar] [CrossRef]
- Hui, J.M.; Hodge, A.; Farrell, G.C.; Kench, J.G.; Kriketos, A.; George, J. Beyond insulin resistance in NASH: TNF-alpha or adiponectin? Hepatology 2004, 40, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Aronis, K.N.; Kountouras, J.; Raptis, D.D.; Vasiloglou, M.F.; Mantzoros, C.S. Circulating leptin in non-alcoholic fatty liver disease: A systematic review and meta-analysis. Diabetologia 2016, 59, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Zelber-Sagi, S.; Lotan, R.; Shlomai, A.; Webb, M.; Harrari, G.; Buch, A.; Kaluski, D.N.; Halpern, Z.; Oren, R. Predictors for incidence and remission of NAFLD in the general population during a seven-year prospective follow-up. J. Hepatol. 2012, 56, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Argentou, M.; Tiniakos, D.G.; Karanikolas, M.; Melachrinou, M.; Makri, M.G.; Kittas, C.; Kalfarentzos, F. Adipokine serum levels are related to liver histology in severely obese patients undergoing bariatric surgery. Obes. Surg. 2009, 19, 1313–1323. [Google Scholar] [CrossRef]
- Senates, E.; Colak, Y.; Yesil, A.; Coşkunpinar, E.; Sahin, O.; Kahraman, O.T.; Şenateş, B.E.; Tuncer, I. Circulating resistin is elevated in patients with non-alcoholic fatty liver disease and is associated with steatosis, portal inflammation, insulin resistance and nonalcoholic steatohepatitis scores. Minerva Med. 2012, 103, 369–376. [Google Scholar] [PubMed]
- Jamali, R.; Razavizade, M.; Arj, A.; Aarabi, M.H. Serum adipokines might predict liver histology findings in non-alcoholic fatty liver disease. World J. Gastroenterol. 2016, 22, 5096–5103. [Google Scholar] [CrossRef]
- Rodriguez-Calvo, R.; Girona, J.; Alegret, J.M.; Bosquet, A.; Ibarretxe, D.; Masana, L. Role of the fatty acid-binding protein 4 in heart failure and cardiovascular disease. J. Endocrinol. 2017, 233, R173–R184. [Google Scholar] [CrossRef]
- Bosquet, A.; Guaita-Esteruelas, S.; Saavedra, P.; Rodríguez-Calvo, R.; Heras, M.; Girona, J.; Masana, L. Exogenous FABP4 induces endoplasmic reticulum stress in HepG2 liver cells. Atherosclerosis 2016, 249, 191–199. [Google Scholar] [CrossRef]
- Koh, J.H.; Shin, Y.G.; Nam, S.M.; Lee, M.Y.; Chung, C.H.; Shin, J.Y. Serum adipocyte fatty acid-binding protein levels are associated with nonalcoholic fatty liver disease in type 2 diabetic patients. Diabetes Care 2009, 32, 147–152. [Google Scholar]
- Kim, Y.C.; Cho, Y.K.; Lee, W.Y.; Kim, H.J.; Park, J.-H.; Park, D.I.; Sohn, C.I.; Jeon, W.K.; Kim, B.-I.; Park, S.-E. Serum adipocyte-specific fatty acid-binding protein is associated with nonalcoholic fatty liver disease in apparently healthy subjects. J. Nutr. Biochem. 2011, 22, 289–292. [Google Scholar] [CrossRef]
- Bosquet, A.; Girona, J.; Guaita-Esteruelas, S.; Heras, M.; Saavedra-Garcia, P.; Martínez-Micaelo, N.; Masana, L.; Rodríguez-Calvo, R. FABP4 inhibitor BMS309403 decreases saturated-fatty-acid-induced endoplasmic reticulum stress-associated inflammation in skeletal muscle by reducing p38 MAPK activation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Calvo, R.; Girona, J.; Rodriguez, M.; Samino, S.; Barroso, E.; De Gonzalo-Calvo, D.; Guaita-Esteruelas, S.; Heras, M.; Van Der Meer, R.W.; Lamb, H.J.; et al. Fatty acid binding protein 4 (FABP4) as a potential biomarker reflecting myocardial lipid storage in type 2 diabetes. Metab. Clin. Exp. 2019, 96, 12–21. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Liu, K.H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A Guideline to Univariate Statistical Analysis for LC/MS-Based Untargeted Metabolomics-Derived Data. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef]
- Rafols, P.; Vilalta, D.; Torres, S.; Calavia, R.; Heijs, B.; McDonnell, L.A.; Brezmes, J.; Del Castillo, E.; Yanes, O.; Ramírez, N.; et al. Assessing the potential of sputtered gold nanolayers in mass spectrometry imaging for metabolomics applications. PLoS ONE 2018, 13, e0208908. [Google Scholar] [CrossRef]
- Rafols, P.; Castillo, E.D.; Yanes, O.; Brezmes, J.; Correig, X. Novel automated workflow for spectral alignment and mass calibration in MS imaging using a sputtered Ag nanolayer. Anal. Chim. Acta 2018, 1022, 61–69. [Google Scholar] [CrossRef]
- Rafols, P.; Torres, S.; Ramirez, N.; Del Castillo, E.; Yanes, O.; Brezmes, J.; Correig, X. rMSI: An R package for MS imaging data handling and visualization. Bioinformatics 2017, 33, 2427–2428. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Greco, D.; Kotronen, A.; Westerbacka, J.; Puig, O.; Arkkila, P.; Kiviluoto, T.; Laitinen, S.; Kolak, M.; Fisher, R.M.; Hamsten, A.; et al. Gene expression in human NAFLD. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G1281–G1287. [Google Scholar] [CrossRef]
- Coilly, A.; Desterke, C.; Guettier, C.; Samuel, D.; Chiappini, F. FABP4 and MMP9 levels identified as predictive factors for poor prognosis in patients with nonalcoholic fatty liver using data mining approaches and gene expression analysis. Sci. Rep. 2019, 9, 19785. [Google Scholar] [CrossRef] [PubMed]
- Kanuri, G.; Bergheim, I. In vitro and in vivo models of non-alcoholic fatty liver disease (NAFLD). Int. J. Mol. Sci. 2013, 14, 11963–11980. [Google Scholar] [CrossRef]
- Ferramosca, A.; Zara, V. Modulation of hepatic steatosis by dietary fatty acids. World J. Gastroenterol. 2014, 20, 1746–1755. [Google Scholar] [CrossRef]
- Simoes, I.C.M.; Janikiewicz, J.; Bauer, J.; Karkucińska-Więckowska, A.; Kalinowski, P.; Dobrzyn, A.; Wolski, A.; Pronicki, M.; Zieniewicz, K.; Dobrzyń, P.; et al. Fat and Sugar-A Dangerous Duet. A Comparative Review on Metabolic Remodeling in Rodent Models of Nonalcoholic Fatty Liver Disease. Nutrients 2019, 11, 2871. [Google Scholar] [CrossRef]
- Perry, R.J.; Samuel, V.T.; Petersen, K.F.; Shulman, G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 2014, 510, 84–91. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Zhou, Y.; Sadevirta, S.; Leivonen, M.; Arola, J.; Orešič, M.; Hyötyläinen, T.; Yki-Järvinen, H. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1167–1175. [Google Scholar] [CrossRef]
- Mauer, A.S.; Hirsova, P.; Maiers, J.L.; Shah, V.H.; Malhi, H. Inhibition of sphingosine 1-phosphate signaling ameliorates murine nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G300–G313. [Google Scholar] [CrossRef]
- Hirsova, P.; Ibrahim, S.H.; Gores, G.J.; Malhi, H. Lipotoxic lethal and sublethal stress signaling in hepatocytes: Relevance to NASH pathogenesis. J. Lipid Res. 2016, 57, 1758–1770. [Google Scholar] [CrossRef] [PubMed]
- Han, M.S.; Park, S.Y.; Shinzawa, K.; Kim, S.; Chung, K.W.; Lee, J.-H.; Kwon, C.H.; Lee, K.-W.; Lee, J.-H.; Park, C.K.; et al. Lysophosphatidylcholine as a death effector in the lipoapoptosis of hepatocytes. J. Lipid Res. 2008, 49, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, G.N. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol. Metab. 2016, 27, 84–95. [Google Scholar] [CrossRef]
- Sa, R.; Zhang, W.; Ge, J.; Wei, X.; Zhou, Y.; Landzberg, D.R.; Wang, Z.; Han, X.; Chen, L.; Yin, H.; et al. Discovering a critical transition state from nonalcoholic hepatosteatosis to nonalcoholic steatohepatitis by lipidomics and dynamical network biomarkers. J. Mol. Cell Biol. 2016, 8, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.N.; Showalter, M.R.; Cajka, T.; Fan, S.; Pillai, V.V.; Fiehn, O.; Selvaraj, V. Metabolomic characteristics of cholesterol-induced non-obese nonalcoholic fatty liver disease in mice. Sci. Rep. 2017, 7, 6120. [Google Scholar] [CrossRef]
- Chitraju, C.; Trotzmuller, M.; Hartler, J.; Wolinski, H.; Thallinger, G.G.; Lass, A.; Zechner, R.; Zimmermann, R.; Koefeler, H.; Spener, F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 2012, 53, 2141–2152. [Google Scholar] [CrossRef]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Musso, G.; Cassader, M.; Paschetta, E.; Gambino, R. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology 2018, 155, 282–302.e288. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Jin, Y.; Chen, H.; Cao, M.; Li, W.; Luo, H.; Wu, Z. Huang-Qi San improves glucose and lipid metabolism and exerts protective effects against hepatic steatosis in high fat diet-fed rats. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 126, 109734. [Google Scholar] [CrossRef]
- Chen, J.W.; Kong, Z.L.; Tsai, M.L.; Lo, C.Y.; Ho, C.T.; Lai, C.S. Tetrahydrocurcumin ameliorates free fatty acid-induced hepatic steatosis and improves insulin resistance in HepG2 cells. J. Food Drug Anal. 2018, 26, 1075–1085. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, J.W.; Yang, M.S.; Park, C.; Kim, J.H.; Lim, C.W.; Kim, B. Beneficial Effects of Korean Red Ginseng in the Progression of Non-Alcoholic Steatohepatitis via FABP4 Modulation. Am. J. Chin. Med. 2018, 46, 1581–1607. [Google Scholar] [CrossRef] [PubMed]
- Hoo, R.L.; Lee, I.P.; Zhou, M.; Wong, J.Y.; Hui, X.; Xu, A.; Lam, K.S.L. Pharmacological inhibition of adipocyte fatty acid binding protein alleviates both acute liver injury and non-alcoholic steatohepatitis in mice. J. Hepatol. 2013, 58, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Wang, Y.; Xu, J.Y.; Stejskal, D.; Tam, S.; Zhang, J.-L.; Wat, N.M.; Wong, W.K.; Lam, K.S.L. Adipocyte fatty acid-binding protein is a plasma biomarker closely associated with obesity and metabolic syndrome. Clin. Chem. 2006, 52, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Sekiya, M.; Ertunc, M.E.; Burak, M.F.; Mayers, J.R.; White, A.; Inouye, K.; Rickey, L.M.; Ercal, B.C.; Furuhashi, M.; et al. Adipocyte lipid chaperone AP2 is a secreted adipokine regulating hepatic glucose production. Cell Metab. 2013, 17, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Kralisch, S.; Ebert, T.; Lossner, U.; Jessnitzer, B.; Stumvoll, M.; Fasshauer, M. Adipocyte fatty acid-binding protein is released from adipocytes by a non-conventional mechanism. Int. J. Obes. 2014, 38, 1251–1254. [Google Scholar] [CrossRef]
- Schlottmann, I.; Ehrhart-Bornstein, M.; Wabitsch, M.; Bornstein, S.R.; Lamounier-Zepter, V. Calcium-dependent release of adipocyte fatty acid binding protein from human adipocytes. Int. J. Obes. 2014, 38, 1221–1227. [Google Scholar] [CrossRef]
- Mita, T.; Furuhashi, M.; Hiramitsu, S.; Ishii, J.; Hoshina, K.; Ishimura, S.; Fuseya, T.; Watanabe, Y.; Tanaka, M.; Ohno, K.; et al. FABP4 is secreted from adipocytes by adenyl cyclase-PKA- and guanylyl cyclase-PKG-dependent lipolytic mechanisms. Obesity 2015, 23, 359–367. [Google Scholar] [CrossRef]
- Fu, S.; Watkins, S.M.; Hotamisligil, G.S. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012, 15, 623–634. [Google Scholar] [CrossRef]
- Drosatos, K.; Drosatos-Tampakaki, Z.; Khan, R.; Homma, S.; Schulze, P.C.; Zannis, V.I.; Goldberg, I.J. Inhibition of c-Jun-N-terminal kinase increases cardiac peroxisome proliferator-activated receptor alpha expression and fatty acid oxidation and prevents lipopolysaccharide-induced heart dysfunction. J. Biol. Chem. 2011, 286, 36331–36339. [Google Scholar] [CrossRef]
- Shen, T.; Chen, X.; Li, Y.; Tang, X.; Jiang, X.; Yu, C.; Zheng, Y.; Guo, H.; Ling, W. Interleukin-17A exacerbates high-fat diet-induced hepatic steatosis by inhibiting fatty acid beta-oxidation. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1510–1518. [Google Scholar] [CrossRef]
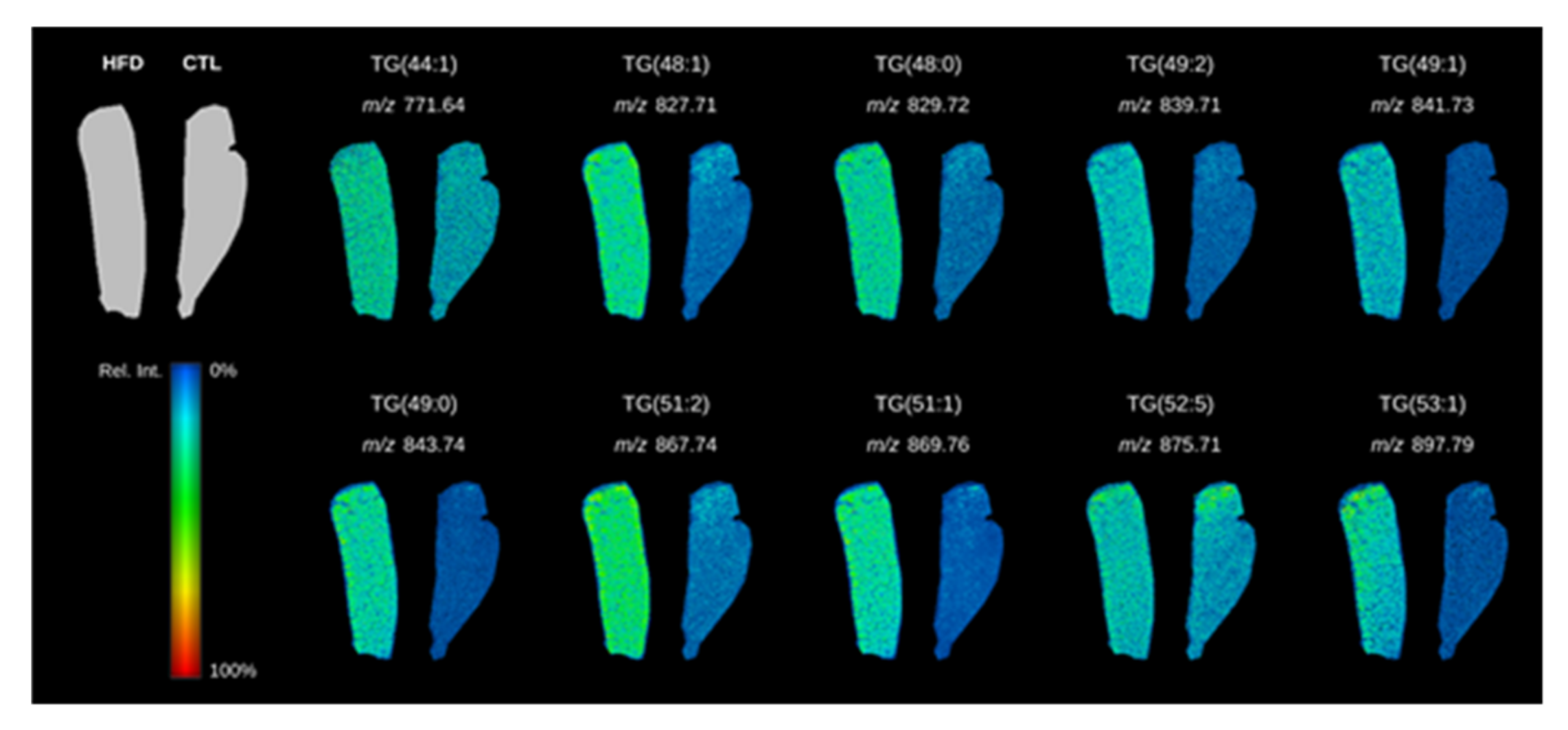
| HFD vs. STD | |||||
|---|---|---|---|---|---|
| m/z | Triglycerides | Adduct Ion | Formula | Fold Change | p-Value |
| 862.7867 | TG(51:2) | NH4+ | C54H100O6 | 7.82 | 0.004 |
| 864.8020 | TG(51:1) | NH4+ | C54H102O6 | 5.40 | 0.008 |
| 836.7710 | TG(49:1) | NH4+ | C52H98O6 | 5.16 | 0.009 |
| 892.8326 | TG(53:1) | NH4+ | C56H106O6 | 4.70 | 0.006 |
| 796.7388 | TG(46:0) | NH4+ | C49H94O6 | 4.14 | <0.001 |
| 839.7103 | TG(49:2) | Na+ | C52H96O6 | 3.83 | 0.001 |
| 838.7856 | TG(49:0) | NH4+ | C52H100O6 | 3.59 | 0.004 |
| 771.6474 | TG(44:1) | Na+ | C47H88O6 | 3.21 | 0.001 |
| 810.7535 | TG(47:0) | NH4+ | C50H96O6 | 3.21 | 0.007 |
| 768.7076 | TG(44:0) | NH4+ | C47H90O6 | 3.04 | 0.001 |
| 752.6759 | TG(43:1) | NH4+ | C46H86O6 | 3.01 | 0.010 |
| 822.7558 | TG(48:1) | NH4+ | C51H96O6 | 2.31 | 0.003 |
| 870.7509 | TG(52:5) | NH4+ | C55H96O6 | 1.98 | 0.008 |
| 824.7706 | TG(48:0) | NH4+ | C51H98O6 | 1.53 | 0.005 |
| Leptin | Resistin | Adiponectin | FABP4 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STD | HFD | STD | HFD | STD | HFD | STD | HFD | |||||||||
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| TG(51:2) | −0.083 | 0.894 | 0.071 | 0.929 | 0.348 | 0.566 | 0.227 | 0.773 | 0.100 | 0.850 | −0.669 | 0.217 | 0.013 | 0.980 | 0.977 | 0.003 |
| TG(51:1) | −0.255 | 0.679 | 0.000 | 1.000 | −0.273 | 0.656 | 0.216 | 0.784 | 0.039 | 0.942 | −0.695 | 0.193 | −0.169 | 0.749 | 0.989 | 0.011 |
| TG(49:1) | −0.206 | 0.739 | 0.049 | 0.951 | −0.148 | 0.813 | 0.108 | 0.892 | 0.164 | 0.756 | −0.632 | 0.253 | −0.412 | 0.418 | 0.984 | 0.016 |
| TG(53:1) | −0.006 | 0.992 | −0.075 | 0.925 | −0.500 | 0.391 | 0.253 | 0.747 | −0.352 | 0.493 | −0.711 | 0.178 | 0.121 | 0.820 | 0.974 | 0.026 |
| TG(49:2) | 0.377 | 0.532 | 0.268 | 0.732 | 0.097 | 0.877 | 0.166 | 0.834 | 0.047 | 0.930 | −0.493 | 0.399 | −0.543 | 0.266 | 0.990 | 0.010 |
| TG(49:0) | 0.815 | 0.093 | 0.015 | 0.985 | 0.308 | 0.614 | 0.277 | 0.723 | −0.141 | 0.791 | −0.747 | 0.147 | −0.403 | 0.428 | 0.993 | 0.007 |
| TG(44:1) | 0.393 | 0.513 | 0.322 | 0.678 | −0.405 | 0.499 | −0.263 | 0.737 | −0.143 | 0.787 | 0.112 | 0.857 | −0.296 | 0.568 | 0.892 | 0.108 |
| TG(48:1) | 0.434 | 0.465 | 0.251 | 0.749 | −0.745 | 0.148 | 0.013 | 0.987 | −0.579 | 0.228 | −0.181 | 0.770 | 0.594 | 0.214 | 0.994 | 0.006 |
| TG(52:5) | 0.188 | 0.762 | 0.228 | 0.772 | 0.247 | 0.689 | 0.264 | 0.736 | 0.072 | 0.893 | −0.585 | 0.301 | −0.496 | 0.317 | 0.973 | 0.027 |
| TG(48:0) | 0.938 | 0.018 | 0.030 | 0.970 | 0.257 | 0.677 | 0.467 | 0.533 | −0.086 | 0.871 | −0.745 | 0.149 | −0.075 | 0.888 | 0.925 | 0.075 |
| B (95% CI) | R2 | |
|---|---|---|
| TG(51:2) | 8.26 (6.41 to 10.11) | 0.997 |
| TG(51:1) | 84.64 (46.60 to 122.69) | 0.988 |
| TG(49:1) | 68.18 (30.73 to 105.62) | 0.985 |
| TG(53:1) | 21.52 (6.36 to 36.68) | 0.967 |
| TG(49:2) | 21.28 (11.86 to 30.70) | 0.991 |
| TG(49:0) | 6.52 (4.25 to 8.80) | 0.990 |
| TG(44:1) | 4.59 (−2.49 to 11.68) | 0.977 |
| TG(48:1) | 101.27 (68.00 to 134.53) | 0.996 |
| TG(52:5) | 1.72 (0.49 to 2.96) | 0.974 |
| TG(48:0) | 10.68 (−2.71 to 24.08) | 0.882 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Calvo, R.; Samino, S.; Girona, J.; Martínez-Micaelo, N.; Ràfols, P.; García-Altares, M.; Guaita-Esteruelas, S.; Junza, A.; Heras, M.; Yanes, O.; et al. Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms. Biomolecules 2020, 10, 1275. https://doi.org/10.3390/biom10091275
Rodríguez-Calvo R, Samino S, Girona J, Martínez-Micaelo N, Ràfols P, García-Altares M, Guaita-Esteruelas S, Junza A, Heras M, Yanes O, et al. Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms. Biomolecules. 2020; 10(9):1275. https://doi.org/10.3390/biom10091275
Chicago/Turabian StyleRodríguez-Calvo, Ricardo, Sara Samino, Josefa Girona, Neus Martínez-Micaelo, Pere Ràfols, María García-Altares, Sandra Guaita-Esteruelas, Alexandra Junza, Mercedes Heras, Oscar Yanes, and et al. 2020. "Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms" Biomolecules 10, no. 9: 1275. https://doi.org/10.3390/biom10091275
APA StyleRodríguez-Calvo, R., Samino, S., Girona, J., Martínez-Micaelo, N., Ràfols, P., García-Altares, M., Guaita-Esteruelas, S., Junza, A., Heras, M., Yanes, O., Correig, X., & Masana, L. (2020). Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms. Biomolecules, 10(9), 1275. https://doi.org/10.3390/biom10091275





