The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Grown Plant Tissues
2.2. Metabolomics Analyses
2.2.1. FTMS Analysis
2.2.2. FTMS Data Processing
2.2.3. Data Processing and Multivariate Statistics
2.2.4. Detection of Significant Ions and Linear Trends
2.2.5. Synthetic Biotransformations
2.2.6. Pathway Analysis
2.2.7. Hormonomics Analysis
2.2.8. Putative Compound ID
3. Results
3.1. Database Compilation and Multivariate Analysis
3.2. Logical Algorithms
3.3. Synthetic Biotransformations
3.4. Pairwise Analysis
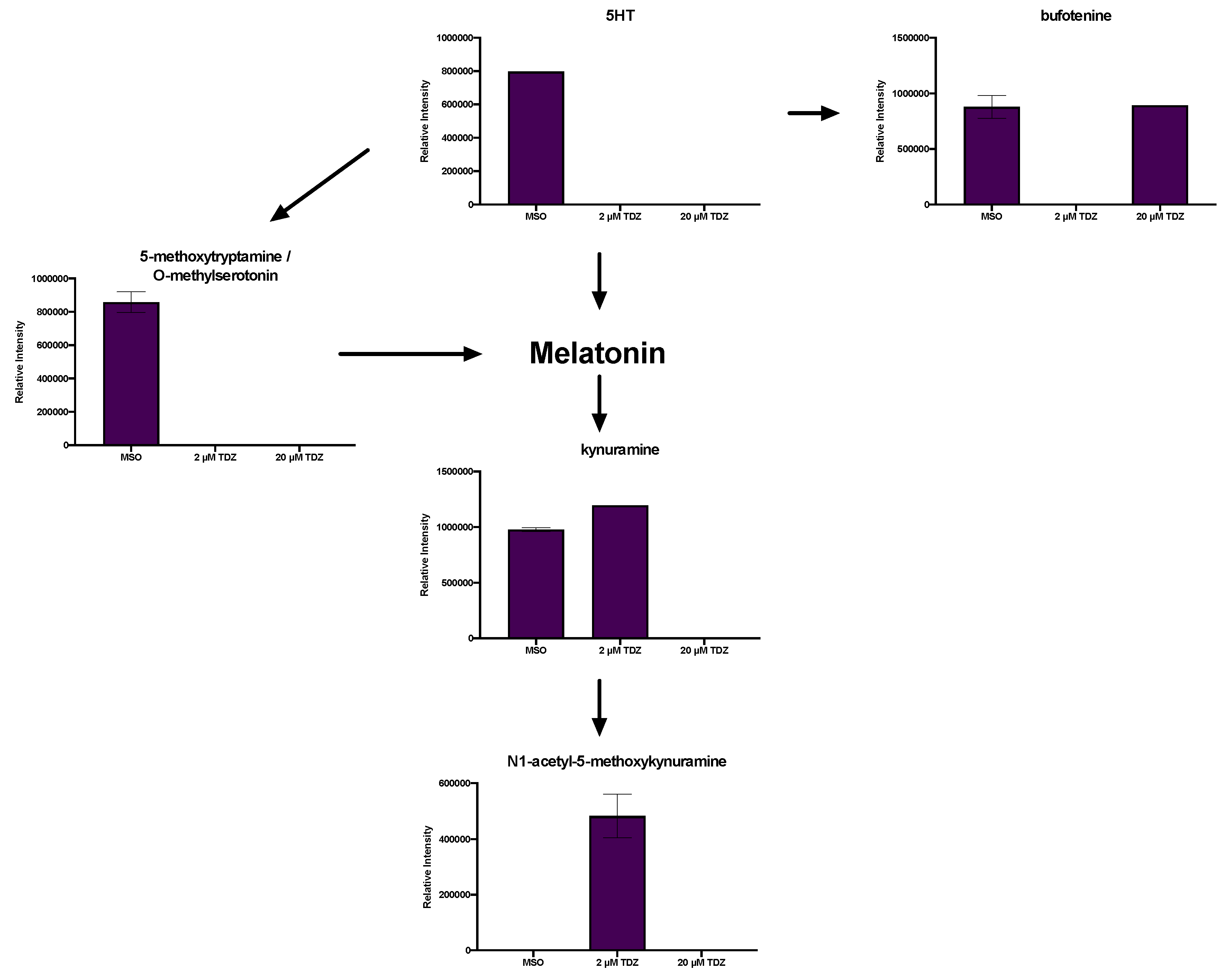
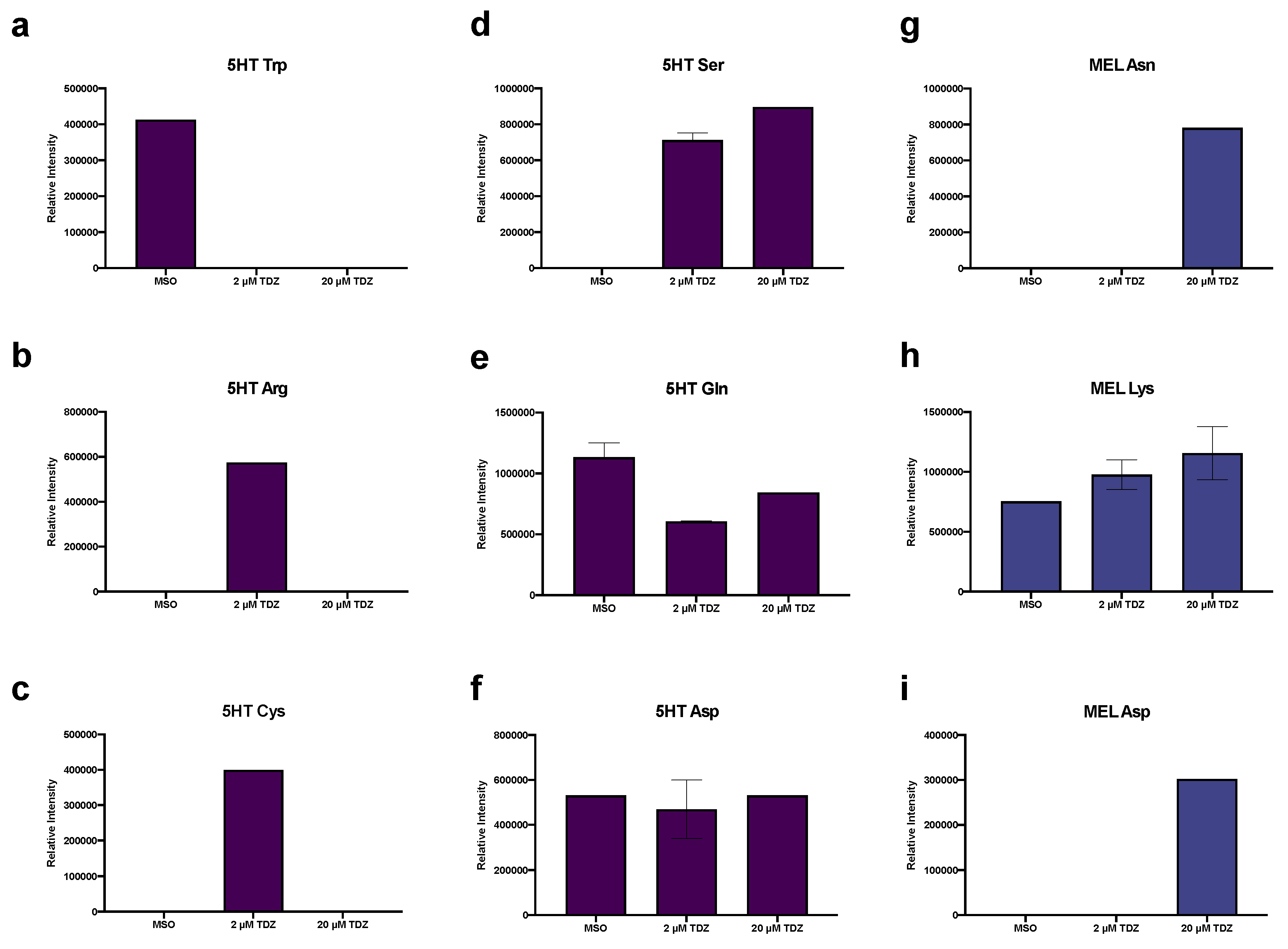
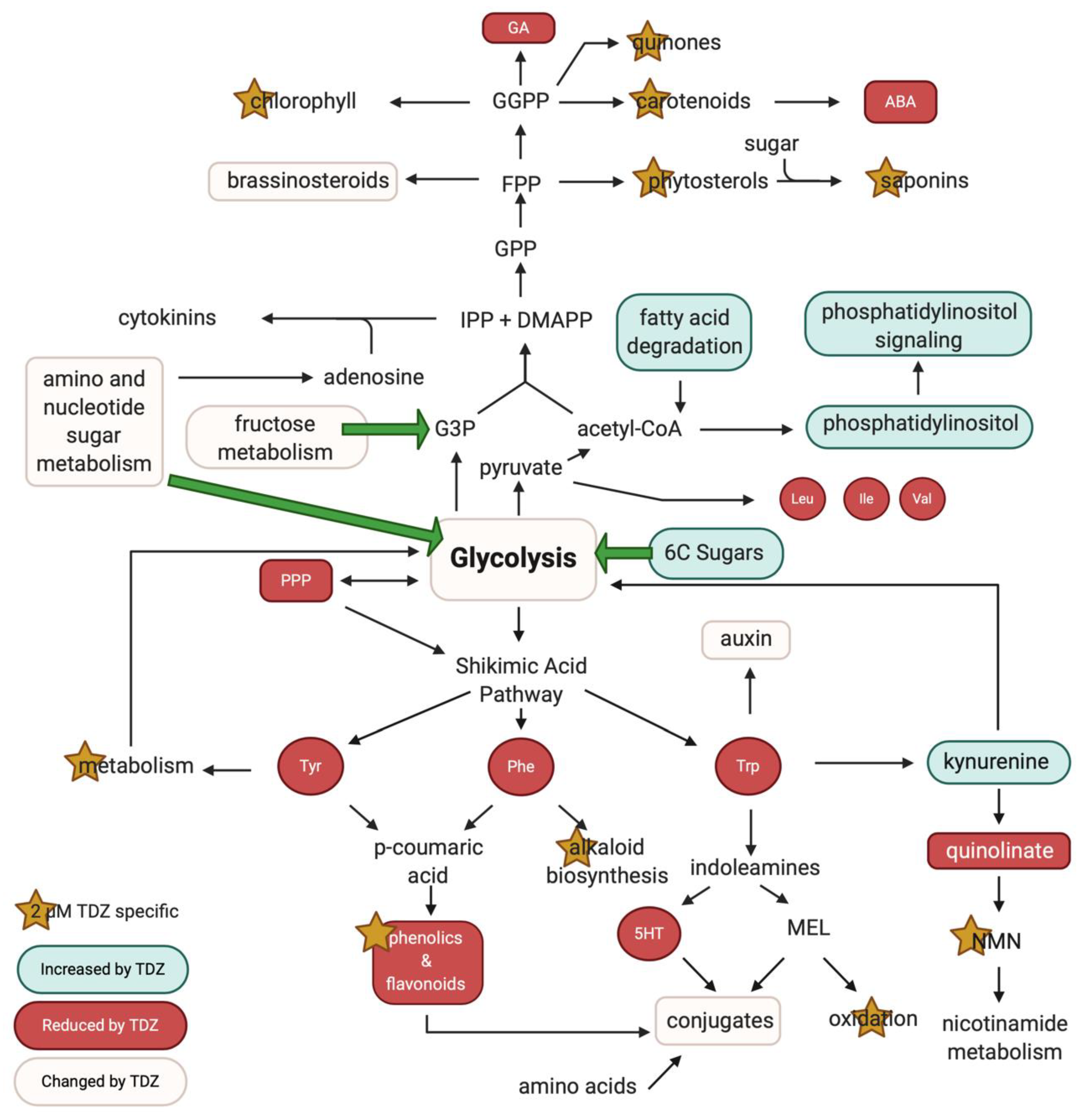
3.5. Pathway Analysis
3.6. Hormonomics Analysis
4. Discussion
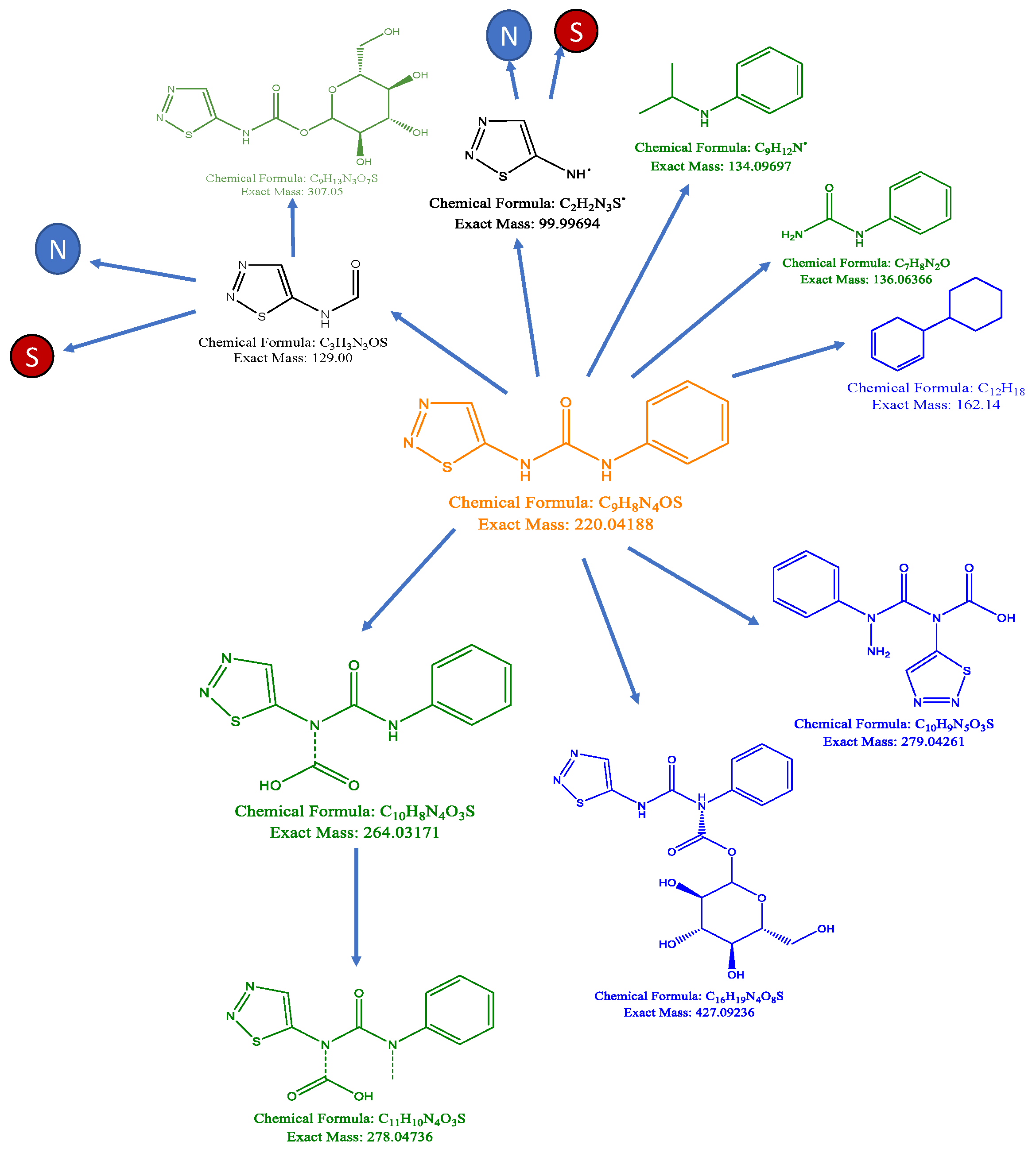
4.1. Metabolic Fate of TDZ in Plant Tissues
4.1.1. Catabolism of TDZ
4.1.2. Formation of Oligomers
4.1.3. TDZ-Conjugates
4.2. TDZ-Induced Metabolism
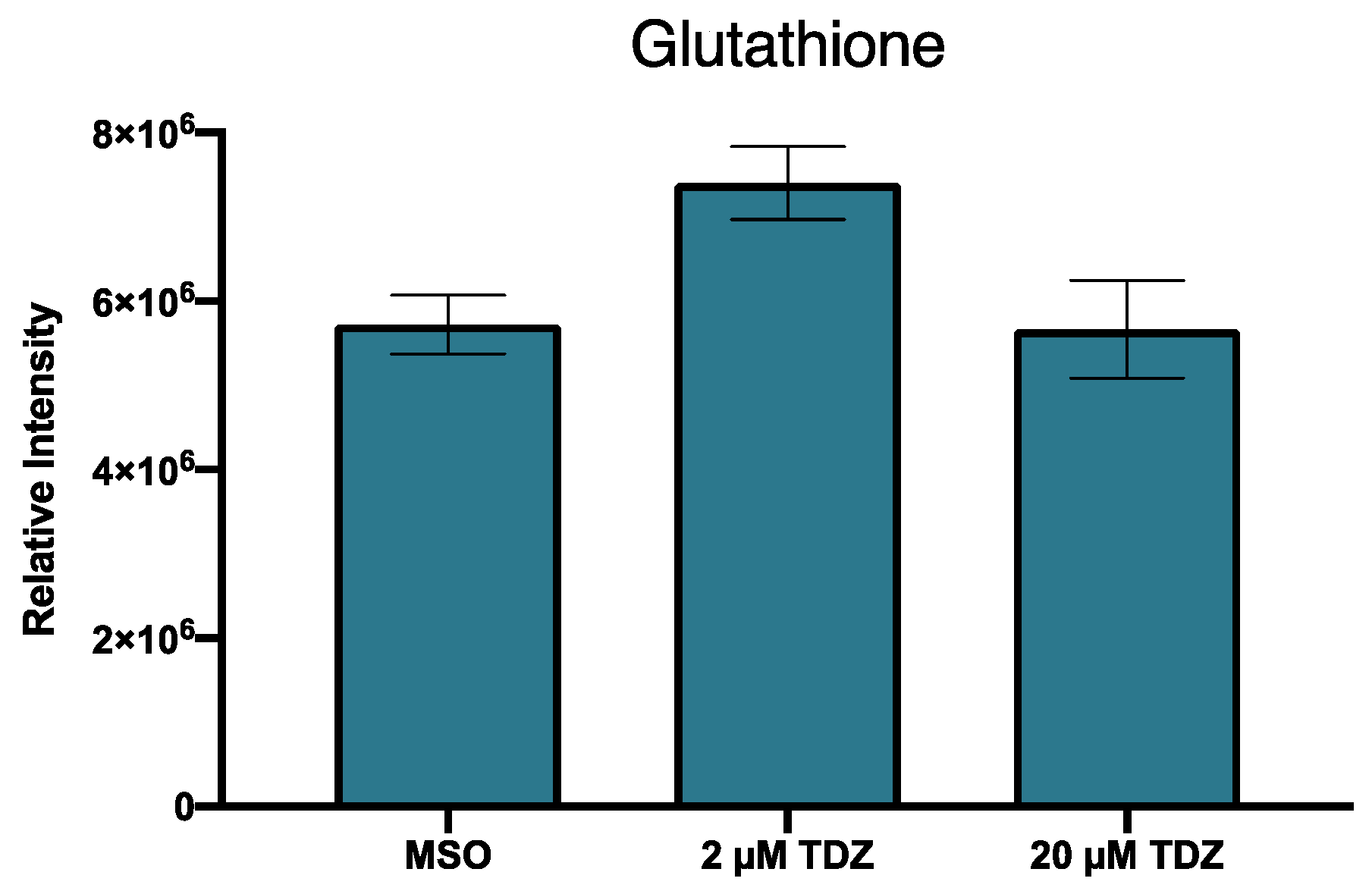
4.2.1. Primary Metabolism
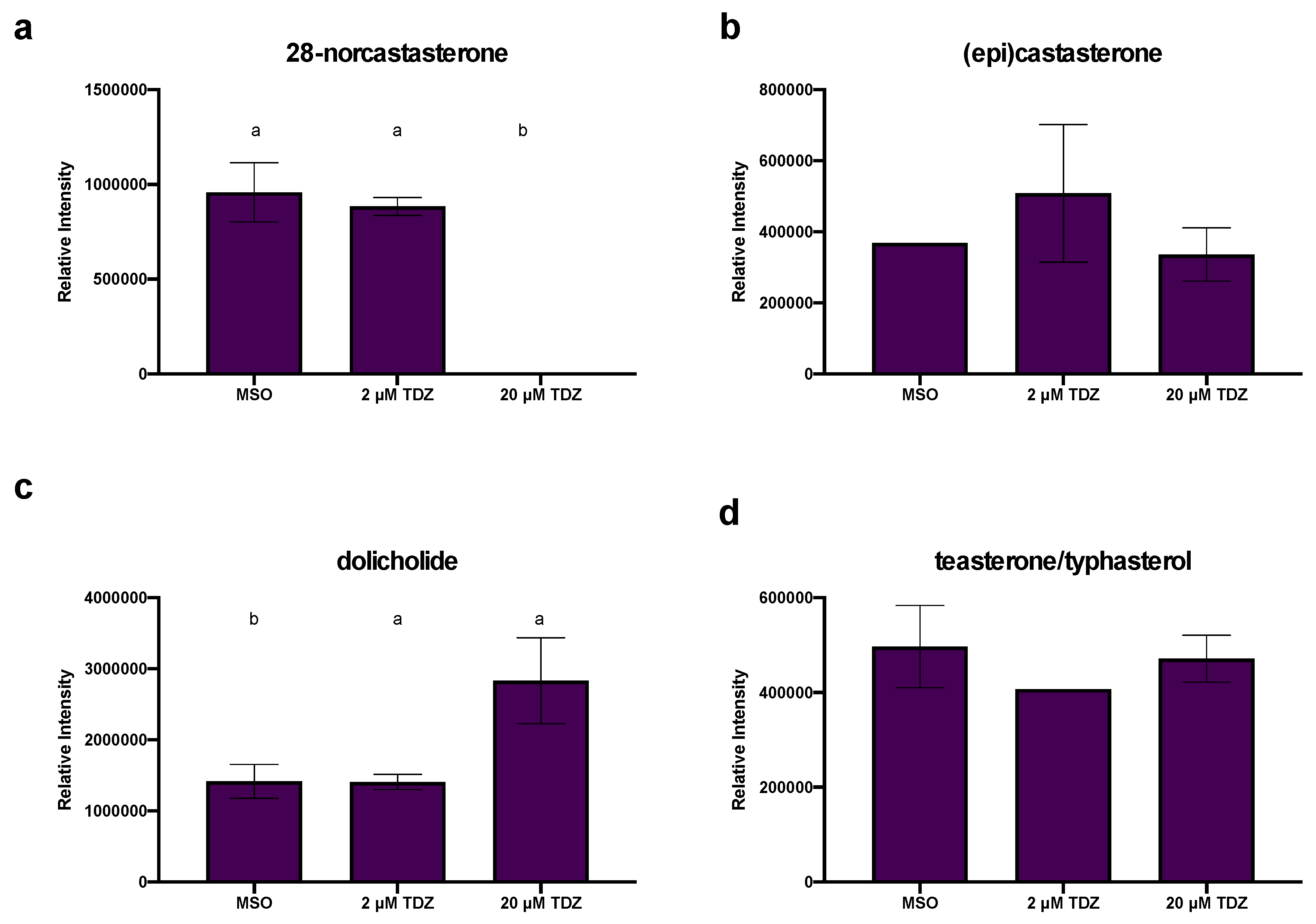
4.2.2. TDZ-Induced Terpene Metabolism
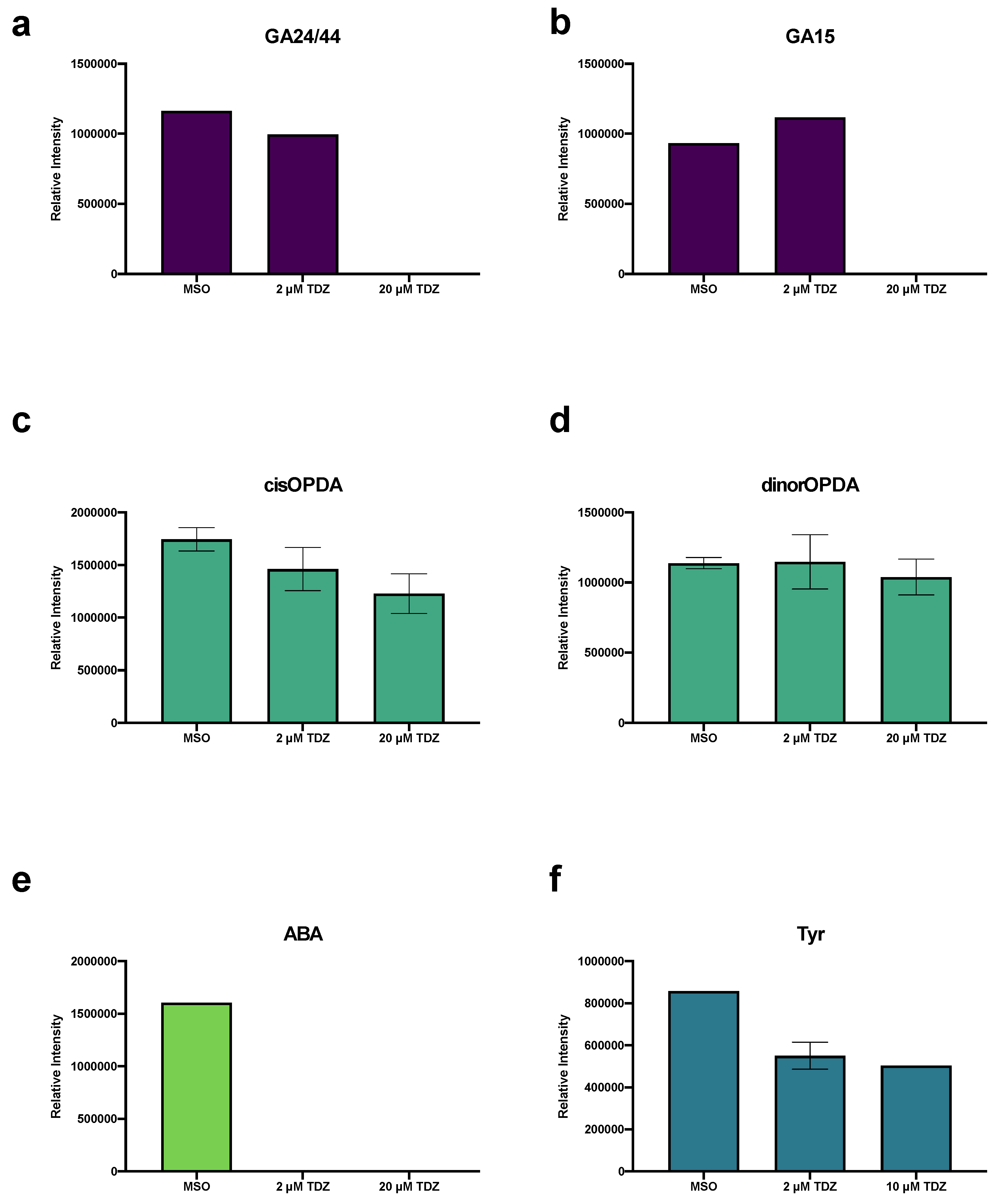
4.2.3. TDZ-Induced Plant Growth Regulators
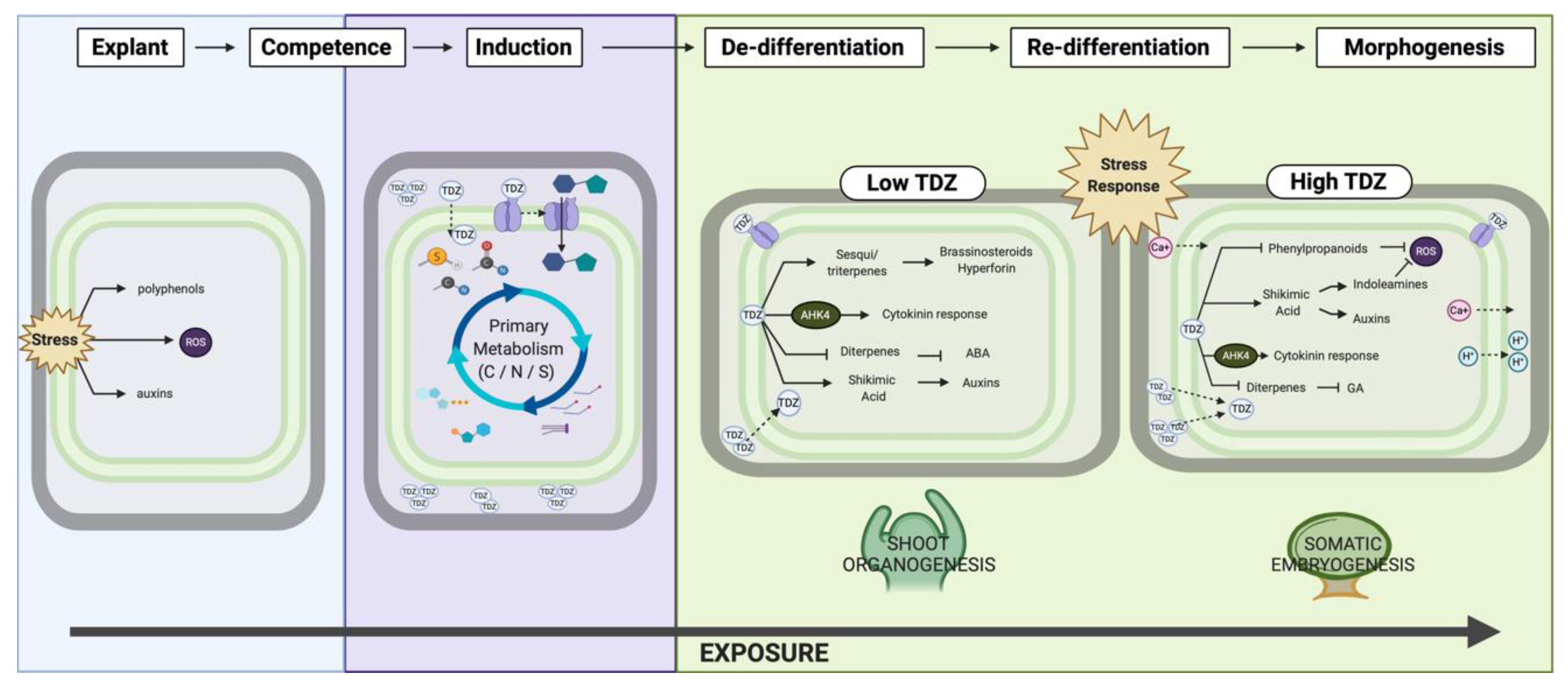
4.3. TDZ-Induced Morphogenesis
The Role of Stress
5. Conclusions
- TDZ is metabolized by plant cells to release bioavailable sulfur and nitrogen.
- TDZ forms oligomers in solutions and plant tissues.
- TDZ forms conjugates as inactive or storage forms of TDZ.
- TDZ increases uptake and catabolism of 5C and 6C sugars from the culture medium.
- TDZ inhibits biosynthesis of diterpene-derived metabolites and enhances synthesis of sesquiterpene and triterpene derivatives.
- TDZ mediates the rate of flux of metabolites through the shikimic acid pathway producing plant growth regulators and secondary metabolites.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mok, M.C.; Mok, D.W.S.; Armstrong, D.J.; Shudo, K.; Isogai, Y.; Okamoto, T. Cytokinin activity of N-phenyl-N′-1, 2,3-thiadiazol-5-ylurea (thidiazuron). Phytochemistry 1982, 21, 1509–1511. [Google Scholar] [CrossRef]
- Murthy, B.N.S.; Murch, S.J.; Saxena, P.K. Thidiazuron: A potent regulator of in vitro plant morphogenesis. Vitro Cell. Dev. Biol. Plant 1998, 34, 267–275. [Google Scholar] [CrossRef]
- Xu, J.; Chen, L.; Sun, H.; Wusiman, N.; Sun, W.; Li, B.; Gao, Y.; Kong, J.; Zhang, D.; Zhang, X.; et al. Crosstalk between cytokinin and ethylene signaling pathways regulates leaf abscission in cotton in response to chemical defoliants. J. Exp. Bot. 2019, 70, 1525–1538. [Google Scholar] [CrossRef]
- Xin, F.; Zhao, J.; Zhou, Y.; Wang, G.; Han, X.; Fu, W.; Deng, J.; Lan, Y. Effects of Dosage and Spraying Volume on Cotton Defoliants Efficacy: A Case Study Based on Application of Unmanned Aerial Vehicles. Agronomy 2018, 8, 85. [Google Scholar] [CrossRef]
- Xu, J.H.; Li, C.P.; Liu, Z.S.; Liu, S.J.; Zhang, D.W.; Ning, X.M.; Xie, D.J. Breeding and planting techniques of the high-quality and high-yield new variety Xinluzao 50. China Cotton 2011, 38, 35–36. [Google Scholar]
- Gormus, O.; Kurt, F.; Sabagh, A.E. Impact of defoliation timings and leaf pubescence on yield and fiber quality of cotton. J. Agric. Sci. Technol. 2017, 19, 903–915. [Google Scholar]
- Nisler, J.; Zatloukal, M.; Sobotka, R.; Pilný, J.; Zdvihalová, B.; Novák, O.; Strnad, M.; Spíchal, L. New Urea derivatives are effective anti-senescence Compounds acting most likely via a cytokinin-Independent mechanism. Front. Plant Sci. 2018, 9, 1225. [Google Scholar] [CrossRef]
- Toscano, S.; Trivellini, A.; Ferrante, A.; Romano, D. Physiological mechanisms for delaying the leaf yellowing of potted geranium plants. Sci. Hortic. 2018, 242, 146–154. [Google Scholar] [CrossRef]
- Famiani, F.; Proietti, P.; Pilli, M.; Battistelli, A.; Moscatello, S. Effects of application of thidiazuron (TDZ), gibberellic acid (GA 3), and 2,4-dichlorophenoxyacetic acid (2,4-D) on fruit size and quality of Actinidia deliciosa ‘Hayward’. N. Z. J. Crop Hortic. Sci. 2007, 35, 341–347. [Google Scholar] [CrossRef]
- Stern, R.; Shargal, A.; Flaishman, M. Thidiazuron increases fruit size of ‘Spadona’ and ‘Coscia’ pear (Pyrus communis L.). J. Hortic. Sci. Biotechnol. 2003, 78, 51–55. [Google Scholar] [CrossRef]
- Reynolds, A.; Wardle, D.; Zurowski, C.; Looney, N. Phenylureas CPPU and Thidiazuron Affect YieldComponents, Fruit Composition, and Storage Potential of Four Seedless Grape Selections. J. Am. Soc. Hortic. Sci. 1992, 1, 85–89. [Google Scholar] [CrossRef]
- Greene, D.W. Thidiazuron Effects on Fruit Set, Fruit Quality, and Return Bloom of Apples. Hortscience 1995, 30, 1238–1240. [Google Scholar] [CrossRef]
- Do Amarante, C.V.T.; Megguer, C.A.; Blum, L.E.B. Effect of preharvest spraying with thidiazuron on fruit quality and maturity of apples. Rev. Bras. Frutic. 2003, 25, 59–62. [Google Scholar] [CrossRef]
- Do Amarante, C.V.T.; Ernani, P.R.; Blum, L.E.B.; Megguer, C.A. Thidiazuron effects on shoot growth, return bloom, fruit set and nutrition of apples. Pesq. Agropec. Bras. 2002, 37, 1365–1372. [Google Scholar] [CrossRef]
- Pasa, M.S.; Silva, C.P.D.; Carra, B.; Brighenti, A.F.; Souza, A.L.K.D.; Petri, J.L. Thidiazuron (TDZ) increases fruit set and yield of ‘Hosui’ and ‘Packham’s Triumph’ pear trees. An. Acad. Bras. Ciênc. 2017, 89, 3103–3110. [Google Scholar] [CrossRef]
- Yang, Y.-Z.; Lin, D.-C.; Guo, Z.-Y. Promotion of fruit development in cucumber (Cucumis sativus) by thidiazuron. Sci. Hortic. 1992, 50, 47–51. [Google Scholar] [CrossRef]
- Lu, C.-Y. The use of thidiazuron in tissue culture. Vitro Cell. Dev. Biol. Plant 1993, 29, 92–96. [Google Scholar] [CrossRef]
- Guo, B.; Abbasi, B.H.; Zeb, A.; Xu, L.L.; Wei, Y.H. Thidiazuron: A multi-dimensional plant growth regulator. Afr. J. Biotechnol. 2011, 10, 8984–9000. [Google Scholar] [CrossRef]
- Debnath, S.C. Hidiazuron in Micropropagation of Small Fruits. In Thidiazuron: From Urea Derivative to Plant Growth Regulator; Ahmad, N., Faisal, M., Eds.; Springer: Singapore, 2018; pp. 139–158. [Google Scholar] [CrossRef]
- Huetteman, C.A.; Preece, J.E. Thidiazuron: A potent cytokinin for woody plant tissue culture. Plant Cell Tissue Organ Cult. 1993, 33, 105–119. [Google Scholar] [CrossRef]
- Hare, P.D.; Staden, J.V. Inhibitory Effect of Thidiazuron on the Activity of Cytokinin Oxidase Isolated from Soybean Callus. Plant Cell Physiol. 1994, 35, 1121–1125. [Google Scholar] [CrossRef]
- De Ferreira, W.M.; Kerbauy, G.B.; Kraus, J.E.; Pescador, R.; Suzuki, R.M. Thidiazuron influences the endogenous levels of cytokinins and IAA during the flowering of isolated shoots of Dendrobium. J. Plant Physiol. 2006, 163, 1126–1134. [Google Scholar] [CrossRef]
- Hothorn, M.; Dabi, T.; Chory, J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat. Chem. Biol. 2011, 7, 766–768. [Google Scholar] [CrossRef]
- Visser, C.; Qureshi, J.A.; Gill, R.; Saxena, P.K. Morphoregulatory role of thidiazuron substitution of auxin and cytokinin requirement for the induction of somatic embryogenesis in geranium hypocotyl cultures. Plant Physiol. 1992, 99, 1704–1707. [Google Scholar] [CrossRef]
- Hutchinson, M.J.; Saxena, P.K. Acetylsalicylic acid enhances and synchronizes thidiazuron-induced somatic embryogenesis in geranium (Pelargonium x hortorum Bailey) tissue cultures. Plant Cell Rep. 1996, 15, 512–515. [Google Scholar] [CrossRef]
- Naseem, A.; Mohammad, F. Thidiazuron: From Urea Derivative to Plant Growth Regulator; Springer: Singapore, 2018; pp. 1–36. [Google Scholar] [CrossRef]
- Murch, S.J.; Saxena, P.K. Molecular fate of thidiazuron and its effects on auxin transport in hypocotyls tissues of Pelargonium × hortorum Bailey. Plant Growth Regul. 2001, 35, 269–275. [Google Scholar] [CrossRef]
- Jones, M.P.A.; Yi, Z.; Murch, S.J.; Saxena, P.K. Thidiazuron-induced regeneration of Echinacea purpurea L.: Micropropagation in solid and liquid culture systems. Plant Cell Rep. 2007, 26, 13–19. [Google Scholar] [CrossRef]
- Murch, S.J.; Victor, J.M.R.; Saxena, P.K. Auxin, Calcium and Sodium in Somatic Embryogenesis of African Violet. Acta Hortic. 2003, 625, 201–209. [Google Scholar] [CrossRef]
- Hutchinson, M.J.; Senaratna, T.; Sahi, S.V.; Saxena, P.K. Light Mediates Endogenous Plant Growth Substances in Thidiazuron-induced Somatic Embryogenesis in Geranium Hypocotyl Cultures. J. Plant Biochem. Biotechnol. 2000, 9, 1–6. [Google Scholar] [CrossRef]
- Murch, S.J.; Krishnaraj, S.; Saxena, P.K. Thidiazuron-induced morphogenesis of Regal geranium (Pelargonium domesticum): A potential stress response. Physiol. Plant 1997. [Google Scholar] [CrossRef]
- Mithila, J.; Hall, J.; Victor, J.M.R.; Saxena, P. Thidiazuron induces shoot organogenesis at low concentrations and somatic embryogenesis at high concentrations on leaf and petiole explants of African violet (Saintpaulia ionantha Wendl.). Plant Cell Rep. 2003, 21, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, P.; Murch, S.J.; Sullivan, J.A.; Saxena, P.K. Micropropagation of Primulina dryas (Dunn) Mich. Möller & A. Webber: High frequency regeneration from leaf explants. Sci. Hortic. 2015, 192, 250–255. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Sullivan, J.A.; Murch, S.J.; Saxena, P. Development of an efficient protocol for high frequency in vitro regeneration of a horticultural plant Primulina tamiana (B.L. Burtt) Mich. Möller & A. Webber. Can. J. Plant Sci. 2014, 94, 1281–1287. [Google Scholar] [CrossRef][Green Version]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Turi, C.E.; Finley, J.; Shipley, P.R.; Murch, S.J.; Brown, P.N. Metabolomics for Phytochemical Discovery: Development of Statistical Approaches Using a Cranberry Model System. J. Nat. Prod. 2015, 78, 953–966. [Google Scholar] [CrossRef]
- Murthy, B.N.; Singh, R.P.; Saxena, P.K. Induction of high-frequency somatic embryogenesis in geranium (Pelargonium x hortorum Bailey cv Ringo Rose) cotyledonary cultures. Plant Cell Rep. 1996, 15, 423–426. [Google Scholar] [CrossRef]
- Shukla, M.; Sullivan, J.A.; Jain, S.M.; Murch, S.J.; Saxena, P.K. Micropropagation of African violet (Saintpaulia ionantha Wendl.). Methods Mol. Biol. 2013, 11013, 279–289. [Google Scholar] [CrossRef]
- Aharoni, A.; de Vos, C.H.R.; Verhoeven, H.A.; Maliepaard, C.A.; Kruppa, G.; Bino, R.; Goodenowe, D.B. Nontargeted Metabolome Analysis by Use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS J. Integr. Biol. 2002, 6, 217–234. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. From the Cover: Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef]
- Murch, S.J.; Rupasinghe, H.P.V.; Goodenowe, D.; Saxena, P.K. A metabolomic analysis of medicinal diversity in Huang-qin (Scutellaria baicalensis Georgi) genotypes: Discovery of novel compounds. Plant Cell Rep. 2004, 23, 419–425. [Google Scholar] [CrossRef]
- Murch, S.J.; Saxena, P.K. A melatonin-rich germplasm line of St John’s wort (Hypericum perforatum L.). J. Pineal Res. 2006, 41, 284–287. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Erland, L.; Turi, C.E.; Saxena, P.K.; Murch, S.J. Metabolomics and hormonomics to crack the code of filbert growth. Metabolomics 2020, 16, 62. [Google Scholar] [CrossRef]
- Brown, P.; Turi, C.; Shipley, P.; Murch, S.J. Comparisons of Large (Vaccinium macrocarponAit.) and Small (Vaccinium oxycoccos L., Vaccinium vitis-idaea L.) Cranberry in British Columbia by Phytochemical Determination, Antioxidant Potential, and Metabolomic Profiling with Chemometric Analysis. Planta Med. 2012, 78, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Turi, C.E.; Murch, S.J. Targeted and untargeted phytochemistry of Ligusticum canbyi: Indoleamines, phthalides, antioxidant potential, and use of metabolomics as a hypothesis-generating technique for compound discovery. Planta Med. 2013, 79, 1370–1379. [Google Scholar] [CrossRef]
- Lieb, H.E.; Still, C.C. Herbicide Metabolism in Plants: Specificity of peroxidases for aniline substrates. Plant Physiol. 2020, 44, 1672–1673. [Google Scholar] [CrossRef][Green Version]
- Mott, R.L.; Steward, F.C. Solute Accumulation in Plant Cells. Ann. Bot. 1972, 36, 621–639. [Google Scholar] [CrossRef]
- Brunkard, J.O.; Xu, M.; Scarpin, M.R.; Chatterjee, S.; Shemyakina, E.A.; Goodman, H.M.; Zambryski, P. TOR dynamically regulates plant cell–cell transport. Proc. Natl. Acad. Sci. USA 2020, 117, 5049–5058. [Google Scholar] [CrossRef]
- Bush, D.R. Identifying the pathways that control resource allocation in higher plants. Proc. Natl. Acad. Sci. USA 2020, 117, 8669–8671. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, S.; Ma, Y.; Song, M.; Song, Y.; Mu, S.; Li, Y.; Liu, X.; Ren, Y.; Gao, C.; et al. Carbon export from leaves is controlled via ubiquitination and phosphorylation of sucrose transporter SUC2. Proc. Natl. Acad. Sci. USA 2020, 117, 6223–6230. [Google Scholar] [CrossRef] [PubMed]
- Chupeau, M.-C.; Lemoine, M.; Chupeau, Y. Requirement of thidiazuron for healthy protoplast development to efficient tree regeneration of a hybrid poplar (Populus tremula ß p. alba). J. Plant Physiol. 1993, 141, 601–609. [Google Scholar] [CrossRef]
- Li, Z.; Traore, A.; Maximova, S.; Guiltinan, M.J. Somatic embryogenesis and plant regeneration from floral explants of cacao (Theobroma cacao L.) using thidiazuron. Vitro Cell. Dev. Biol. Plant 1998, 34, 293–299. [Google Scholar] [CrossRef]
- Wang, S.Y.; Ji, Z.L.; Sun, T.; Faust, M. Effect of thidiazuron on abscisic acid content in apple bud relative to dormancy. Physiol. Plant. 1987. [Google Scholar] [CrossRef]
- Wang, S.Y.; Ji, Z.L.; Faust, M. Metabolic changes associated with bud break induced by thidiazuron. J. Plant Growth Regul. 1987, 6, 85–95. [Google Scholar] [CrossRef]
- Kiba, T.; Takebayashi, Y.; Kojima, M.; Sakakibara, H. Sugar-induced de novo cytokinin biosynthesis contributes to Arabidopsis growth under elevated CO2. Sci Rep. 2019, 9, 7765. [Google Scholar] [CrossRef]
- EI-D, A.M.S.A.; Salama, A.; Wareing, P.F. Effects of Mineral Nutrition on Endogenous Cytokinins in Plants of Sunflower (Helianthus annuus L.). J. Exp. Bot. 1979, 30, 971–981. [Google Scholar] [CrossRef]
- Kamada-Nobusada, T.; Makita, N.; Kojima, M.; Sakakibara, H. Nitrogen-Dependent Regulation of De Novo Cytokinin Biosynthesis in Rice: The Role of Glutamine Metabolism as an Additional Signal. Plant. Cell Physiol. 2013, 54, 1881–1893. [Google Scholar] [CrossRef]
- Murch, S.J.; Saxena, P.K. Modulation of mineral and free fatty acid profiles during thidiazuron mediated somatic embryogenesis in peanuts (Arachis hypogeae L.). J. Plant Physiol. 1997, 151, 358–361. [Google Scholar] [CrossRef]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef]
- Mahmoud, S.S.; Croteau, R.B. Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci. 2002, 7, 366–373. [Google Scholar] [CrossRef]
- Stermer, B.A.; Bianchini, G.M.; Korth, K.L. Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 1994, 35, 1133–1140. [Google Scholar] [PubMed]
- Dewir, Y.H.; Nurmansyah; Naidoo, Y.; da Silva, J.A.T. Thidiazuron-induced abnormalities in plant tissue cultures. Plant Cell Rep. 2018, 37, 1451–1470. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, P.; Hu, K.; Xiao, Q.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Shen, J. Transcriptome profiling reveals cytokinin promoted callus regeneration in Brassica juncea. Plant Cell Tissue Organ Cult. 2020, 141, 191–206. [Google Scholar] [CrossRef]
- Mok, M.C.; Martin, R.C.; Dobrev, P.I.; Vanková, R.; Ho, P.S.; Yonekura-Sakakibara, K.; Sakakibara, H.; Mok, D.W.S. Topolins and Hydroxylated Thidiazuron Derivatives Are Substrates of Cytokinin O-Glucosyltransferase with Position Specificity Related to Receptor Recognition. Plant Physiol. 2005, 137, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Louie, G.V.; Bowman, M.E.; Moffitt, M.C.; Baiga, T.J.; Moore, B.S.; Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 2006, 13, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Erland, L.A.E.; Chattopadhyay, A.; Jones, A.M.P.; Saxena, P.K. Melatonin in plants and plant culture systems: Variability, stability and efficient quantification. Front. Plant Sci. 2016, 7, 108. [Google Scholar] [CrossRef]
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957, 11, 118–130. [Google Scholar]
- Charchoglyan, A.; Abrahamyan, A.; Fujii, I.; Boubakir, Z.; Gulder, T.A.M.; Kutchan, T.M.; Vardapetyan, H.; Bringmann, G.; Ebizuka, Y.; Beerhues, L. Differential accumulation of hyperforin and secohyperforin in Hypericum perforatum tissue cultures. Phytochemistry 2007, 68, 2670–2677. [Google Scholar] [CrossRef]
- Da Silva, J.A.T.; Dobránszki, J.; Ross, S. Phloroglucinol in plant tissue culture. Vitro Cell. Dev. Biol. Plant 2013, 49, 1–16. [Google Scholar] [CrossRef]
- Steward, F.C.; Bidwell, R.G.S.; Yemm, E.W. Nitrogen Metabolism, Respiration, and Growth of Cultured Plant Tissue: Part I. Experimental design, techniques, and recorded data: Part II. The interpretation of specific activity data in tracer experiments: Part III. Nitrogen metabolism and respiration of carrot tissue explants as revealed by experiments with C 14-Labelled Substrates. J. Exp. Bot. 1958, 9, 11–49. [Google Scholar]
- Steward, F.C.; Kent, A.E.; Mapes, M.O. Growth and organization in cultured cells: Sequential and synergistic effects of growth-regulating substances. Ann. N. Y. Acad. Sci. 1967, 144, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.; Saxena, P.K. Direct somatic embryogenesis and regeneration of plants from seedling explants of peanut (Arachis hypogaea): Promotive role of thidiazuron. Can. J. Bot. 1992, 70, 1186–1192. [Google Scholar] [CrossRef]
- Malik, K.A.; Saxena, P.K. Thidiazuron Induces High-Frequency Shoot Regeneration in Intact Seedlings of Pea (Pisum-Sativum), Chickpea (Cicer-Arietinum) and Lentil (Lens-Culinaris). Aust. J. Plant Physiol. 1992, 19, 731–740. [Google Scholar] [CrossRef]
- Saxena, P.K.; Malik, K.A.; Gill, R. Induction by thidiazuron of somatic embryogenesis in intact seedlings of peanut. Planta 1992, 187, 421–424. [Google Scholar] [CrossRef]
- Murch, S.J.; Campbell, S.S.B.; Saxena, P.K. The role of serotonin and melatonin in plant morphogenesis: Regulation of auxin-induced root organogenesis in in vitro-cultured explants of st. John’s Wort (Hypericum perforatum L.). Vitro Cell. Dev. Biol. Plant 2001, 37, 786–793. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Murch, S.J.; Reiter, R.J.; Saxena, P.K. A new balancing act: The many roles of melatonin and serotonin in plant growth and development. Plant Signal. Behav. 2015, 10, e1096469. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Shukla, M.R.; Singh, A.S.; Murch, S.J.; Saxena, P.K. Melatonin and serotonin: Mediators in the symphony of plant morphogenesis. J. Pineal Res. 2018, 64, e12452. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Erland, L.A.E.; Jones, A.M.P.; Saxena, P.K. Indoleamines and phenylpropanoids modify development in the bryophyte Plagiomnium cuspidatum. Vitro Cell. Dev. Biol. Plant 2018, 54, 454–464. [Google Scholar] [CrossRef]
- Street, H. Embryogenesis and chemically induced organogenesis. In Plant and Cell Tissue Culture: Principles and Application; Sharp, W.R., Larsen, R.P.O., Paddock, E.F., Raghavan, V., Eds.; Ohio State University Press: Columbus, OH, USA, 1979; pp. 123–153. [Google Scholar]


| Metabolite Mass | Putative ID * | Class | Associated KEGG Metabolic Pathway(s) | M | 0 | 2 | 20 |
|---|---|---|---|---|---|---|---|
| 254.05791 | (Z)-4′,6-Dihydroxyaurone | Aurone flavonoid | Flavonoid biosynthesis Isoflavonoid biosynthesis | 254.0579 | 62336170 | 35136500 | 18638300 |
| Chrysophanol | Anthraquinone | ||||||
| Chrysin | Flavone | ||||||
| 7,4′-Dihydroxyflavone | Flavone | ||||||
| Anhydroglycinol | Pterocarpan | ||||||
| Daidzein | Isoflavone | ||||||
| formononetin methylated | Isoflavone | ||||||
| 5,7-Dihydroxyisoflavone | Isoflavone | ||||||
| 254.058126 | (Z)-4′,6-Dihydroxyaurone | Aurone flavonoid | Flavonoid biosynthesis Isoflavonoid biosynthesis | 254.0579 | 62336170 | 35136500 | 18638300 |
| Chrysophanol | Anthraquinone | ||||||
| Chrysin | Flavone | ||||||
| 7,4′-Dihydroxyflavone | Flavone | ||||||
| Anhydroglycinol | Pterocarpan | ||||||
| Daidzein | Isoflavone | ||||||
| formononetin methylated | Isoflavone | ||||||
| 5,7-Dihydroxyisoflavone | Isoflavone | ||||||
| 297.266779 | Palmitoleoyl ethanolamide | N-acylethanolamines | Glycerophosphate metabolism | 297.2668 | 29559440 | 19908290 | 10659625 |
| 202.045236 | Ethyl aconitate | Tricarboxylic acid derivative | Citrate Cycle | 202.0477 | 83643600 | 54038350 | 64287550 |
| 364.09741 | Gerberinol | 4-Hydroxycoumarin | Phenylpropanoid biosynthesis/Biosynthesis of phenylpropanoids Phenylpropanoid biosynthesis | 364.0947 | 64478035 | 54169390 | 78307450 |
| 230.076354 | Aspartyl-Proline | Dipeptide | Arginine and proline metabolism Glutathione metabolism | 230.0903 | 17390700 | 22240300 | 30965200 |
| (2S,3′S)-alpha-Amino-2-carboxy-5-oxo-1-pyrrolidinebutanoic acid | Proline derivative | ||||||
| Prolyl-Aspartate | Dipeptide | ||||||
| 1-(gamma-Glutamylamino)cyclopropanecarboxylic acid | Dipeptide | ||||||
| Aspartyl-L-proline | Dipeptide | ||||||
| 230.076536 | Aspartyl-Proline | Dipeptide | Arginine and proline metabolism Glutathione metabolism | 230.0903 | 17390700 | 22240300 | 30965200 |
| (2S,3′S)-alpha-Amino-2-carboxy-5-oxo-1-pyrrolidinebutanoic acid | Proline derivative | ||||||
| Prolyl-Aspartate | Dipeptide | ||||||
| 1-(gamma-Glutamylamino)cyclopropanecarboxylic acid | Dipeptide | ||||||
| Aspartyl-L-proline | Dipeptide | ||||||
| 278.094295 | Methionyl-Glutamate | Dipeptide | Glutathione metabolism | 278.0936 | 24758300 | 26486100 | 13594000 |
| Glutamylmethionine | Dipeptide | ||||||
| N-gamma-L-Glutamyl-L-methionine | Dipeptide | ||||||
| 278.094511 | Methionyl-Glutamate | Dipeptide | Glutathione metabolism | 278.0936 | 24758300 | 26486100 | 13594000 |
| Glutamylmethionine | Dipeptide | ||||||
| N-gamma-L-Glutamyl-L-methionine | Dipeptide | ||||||
| 278.09498 | Methionyl-Glutamate | Dipeptide | Glutathione metabolism | 278.0936 | 24758300 | 25678000 | 13594000 |
| Glutamylmethionine | Dipeptide | ||||||
| N-gamma-L-Glutamyl-L-methionine | Dipeptide | ||||||
| 612.475375 | DG | Diacylglycerol | >10 | 612.4754 | 26360600 | 22255850 | 17457400 |
| 364.097879 | Gerberinol | 4-Hydroxycoumarin | Phenylpropanoid biosynthesis Biosynthesis of phenylpropanoids Phenylpropanoid biosynthesis | 364.0947 | 49914200 | 42166150 | 58372250 |
| 364.098061 | Gerberinol | 4-Hydroxycoumarin | Phenylpropanoid biosynthesis/Biosynthesis of phenylpropanoids Phenylpropanoid biosynthesis | 364.0947 | 49824750 | 42156490 | 57558100 |
| 364.098277 | Gerberinol | 4-Hydroxycoumarin | Phenylpropanoid biosynthesis/Biosynthesis of phenylpropanoids Phenylpropanoid biosynthesis | 364.0947 | 49824750 | 42220140 | 57474850 |
| Metabolite Mass | Putative ID * | Class | Associated KEGG Metabolic Pathway(s) | Monoisotopic Mass | 0 | 2 | 20 | R2 (Linear) a |
|---|---|---|---|---|---|---|---|---|
| 162.14085 | Pregeijerene | Unsaturated Hydrocarbon | 162.1408506 | 0 | 846985 | 862490 | 0.347 | |
| 1,3-Diisopropylbenzene | Aromatic hydrocarbon | 162.1408506 | ||||||
| 192.027005 | dehydroascorbate (bicyclic form) | Ascorbate | Pentose and glucoronate interconversions Ascorbate and aldarate metabolism | 192.0270026 | 0 | 524910 | 502955 | 0.298 |
| Isocitric acid | Ascorbate | 192.0270026 | ||||||
| Citric acid | Ascorbate | 192.0270026 | ||||||
| 2,3-Diketo-L-gulonate | Ascorbate degradation | 192.0270026 | ||||||
| D-Glucaro-1,4-lactone | Monosaccharide | 192.0270026 | ||||||
| 196.109945 | Loliolide | carotenoid metabolite; benzofuran | Furfural degradation | 196.1099444 | 0 | 1036230 | 902080 | 0.226 |
| 3-Methylidene-4-oxopentyl angelate | Fatty acid ester | 196.1099444 | ||||||
| Isobutyl 2-furanpropionate | Fatty acid ester | 196.1099444 | ||||||
| Hexyl 2-furoate | Fuoric acid ester | 196.1099444 | ||||||
| 270.052825 | 4,6-dihydroxy-2-[(4-hydroxyphenyl)methylidene]-2,3-dihydro-1-benzofuran-3-one | Aurone flavonoid | Flavonoid biosynthesis Isoflavonoid biosynthesis Biosynthesis of phenylpropanoids | 270.0528234 | 0 | 1392765 | 1071285 | 0.146 |
| Rheinanthrone | Anthracenecarboxylic acid | 270.0528234 | ||||||
| Aloeemodin | Anthraquinone | 270.0528234 | ||||||
| Rhababerone | Anthraquinone | 270.0528234 | ||||||
| 1,2,8-Trihydroxy-3-methylanthraquinone | Anthraquinone | 270.0528234 | ||||||
| Emodin | Anthraquinone | 270.0528234 | ||||||
| Apigenin | Flavone | 270.0528234 | ||||||
| Norizalpinin | Hydroxyflavonol | 270.0528234 | ||||||
| 3,4′,7-Trihydroxyflavone | Hydroxyflavonol | 270.0528234 | ||||||
| 6-Hydroxydaidzein | Isoflavone | 270.0528234 | ||||||
| Genistein | Isoflavone | 270.0528234 | ||||||
| 330.122909 | Tyramine-betaxanthin | Betalain | Betalain biosynthesis | 330.1216 | 0 | 924825 | 892190 | 0.303 |
| 352.068037 | a-L-threo-4-Hex-4-enopyranuronosyl-D-galacturonic acid | Glucoronic acid derivative | Pentose and glucoronate interconversions | 352.064176 | 0 | 1220300 | 1574900 | 0.545 |
| 406.107975 | (R)-Apiumetin glucoside | Coumarin glycoside | Tyrosine metabolism Benzoate degradation Flavonoid biosynthesis | 406.1263823 | 0 | 578125 | 1001885 | 0.752 |
| Edulisin VI | Furanocoumarin | 406.1263823 | ||||||
| Benzyl 2,6-dihydroxybenzoate 2-glucoside | Phenolic glycoside | 406.1263823 | ||||||
| omega-Salicoyisalicin | Phenolic glycoside | 406.1263823 | ||||||
| Flacourtin | Phenolic glycoside | 406.1263823 | ||||||
| Dihydroresveratrol 3-glucuronide | Stilbene glycoside | 406.1263823 | ||||||
| Piceatannol 4′-glucoside | Stilbene glycoside | 406.1263823 | ||||||
| (E)-Oxyresveratrol 3′-O-b-D-glucoside | Stilbene glycoside | 406.1263823 | ||||||
| Astringin | Stilbene glycoside | 406.1263823 | ||||||
| 500.31379 | Theaflavin 3,3′-digallate | Polyphenol | Flavonoid biosynthesis Brassinosteroid biosynthesis Sesquiterpenoid and triterpenoid biosynthesis | 500.3148863 | 0 | 1350235 | 2631535 | 0.813 |
| Physalolactone B | Sterol | 500.3148863 | ||||||
| medicagenate | Triterpene | 500.3148863 | ||||||
| Ganolucidic acid A | Triterpene | 500.3148863 | ||||||
| Ganoderic acid beta | Triterpene derivative | 500.3148863 | ||||||
| 520.101648 | Melitric acid B | Polyphenol | Phenylpropanoid biosynthesis | 520.1005615 | 0 | 442050 | 511095 | 0.454 |
| 536.38656 | Hyperforin | Polycyclic Polyprenylated Acylphloroglucinol | Sesquiterpenoid and triterpenoid biosynthesis Aminobenzoate degradation | 536.3865392 | 0 | 1302600 | 6563250 | 0.990 |
| 568.119495 | Neobignonoside | Flavonoid glycoside | Flavonoid biosynthesis Polyphenol biosynthesis Biosynthesis of secondary metabolites | 568.1216909 | 0 | 403535 | 591625 | 0.643 |
| Chrysophanol 8-(6-galloylglucoside) | Anthraquinone | 568.1216909 | ||||||
| 570.155971 | Apiumoside | Psoralen | Biosynthesis of phenylpropanoids | 570.1737264 | 0 | 1204850 | 1155410 | 0.299 |
| 616.503989 | DG | Diacylglycerol | >10 | 616.5066753 | 0 | 461850 | 446055 | 0.304 |
| 636.473874 | DG | Diacylglycerol | >10 | 636.4753752 | 0 | 318400 | 380860 | 0.483 |
| 684.234252 | Maltulose | Glycerolipid | Glycerolipid metabolism | 684.2324231 | 0 | 600440 | 1097880 | 0.781 |
| Galabiose | Glycerolipid | 684.2324231 | ||||||
| 694.377879 | Capsoside A | Glycosyldiacylglycerol | Glycerolipid metabolism | 694.3775712 | 0 | 551420 | 567555 | 0.356 |
| 778.41126 | Periandrin V | Triterpene glycoside | Sesquiterpenoid and triterpenoid metabolism | 778.4139567 | 0 | 835800 | 810640 | 0.308 |
| 810.437975 | 9-Hentriacontanone | Ketone | Sesquiterpenoid and triterpenoid metabolism | 810.4401714 | 0 | 3421550 | 4660950 | 0.588 |
| Phytolaccoside D2 | Triterpene | 810.4401714 | ||||||
| Phytolaccoside D | Triterpene | 810.4401714 | ||||||
| Elatoside H | Triterpene saponin | 810.4401714 | ||||||
| Cynarasaponin E | Triterpene saponin | 810.4401714 | ||||||
| Lucyoside J | Triterpene saponin | 810.4401714 | ||||||
| Azukisaponin III | Triterpene saponin | 810.4401714 | ||||||
| Spinasaponin B | Triterpene saponin | 810.4401714 | ||||||
| 880.752675 | TG | Triacylglycerol | Glycerolipid metabolism | 880.7519909 | 0 | 2626300 | 2252500 | 0.215 |
| Metabolite Mass | M-TDZ | M-2TDZ | Putative ID * | Class | Monoisotopic Mass | Formula | 0 | 2 | 20 | R2 (Linear) |
|---|---|---|---|---|---|---|---|---|---|---|
| 432.072153 | 212.030271 | 4-Nitroso-2,6-dinitrotoluene | Dinitrotoluene | 211.0229 | C7H5N3O5 | 0 | 409980 | 585255 | 0.624 | |
| 2-Methyl-1,5-dinitro-3-nitrosobenzene | Dinitrotoluene | 211.0229 | C7H5N3O5 | |||||||
| 7.003782 | No match | |||||||||
| 456.0691 | 236.027218 | No match | 0 | 480545 | 712690 | 0.652 | ||||
| 16.993165 | Oxygen | Element | 15.999 | O | ||||||
| 465.049091 | 245.007209 | Coumaric acid sulfate | Phenylpropanoid | 244.0042 | C9H8O6S | 0 | 813650 | 1281750 | 0.693 | |
| 25.973156 | Magnesium | element | 24.305 | Mg | ||||||
| 676.200542 | 456.15866 | 5,10-Methenyltetrahydrofolate | Tetrahydrofolic acid | 455.1553 | C20H21N7O6 | 0 | 6572500 | 5052450 | 0.146 | |
| 237.124607 | Phenylalanylalanine | Dipeptide | 236.1161 | C12H16N2O3 | ||||||
| 706.21458 | 486.172698 | 485.594 | C29H27NO4S ** | 0 | 1335030 | 2554800 | 0.804 | |||
| 267.138645 | 3-Phenylpropyl cinnamate | Cinnamic acid ester | 266.1307 | C18H18O2 | ||||||
| 718.210422 | 498.16854 | 498.55474 | C23H26N6O5S ** | 0 | 1729150 | 1555300 | 0.250 | |||
| 279.134487 | Tyrosyl proline | Dipeptide | 278.1267 | C14H18N2O4 | ||||||
| L-Phenylalanylyl-L-hydroxyproline | Dipeptide | 278.1267 | C14H18N2O4 | |||||||
| 726.359148 | 506.317266 | LysoPE | Llysophospholipid | 505.3168 | C25H48NO7P | 0 | 1226900 | 864805 | 0.101 | |
| 287.283213 | Hexanal dihexyl acetal | Acetal | 286.2872 | C18H38O2 | ||||||
| 760.488734 | 540.446852 | 539.794 | C29H57N5O4 | 0 | 473410 | 578200 | 0.500 | |||
| 321.412799 | ||||||||||
| 826.411427 | 606.369545 | 605.8403 | C32H47N9OS ** | 0 | 1501580 | 1451550 | 0.305 | |||
| 387.335492 | 1-Phenyl-1,3-eicosandeione | Alkyl phenylketone | 386.3185 | C26H42O2 | ||||||
| 961.423907 | 741.382025 | C38H56N6O5S2 ** | 0 | 1241850 | 1256550 | 0.342 | ||||
| 522.347972 | LysoPC | Lysophospholipid | 521.3481 | C26H52NO7P | ||||||
| Lysolecithin | Phosphatidylcholine | 521.3481 | C26H52NO7P |
| Test | Comparison/Pattern | Pathways Significantly Modulated | Combined p-Values |
|---|---|---|---|
| t-test | 0 vs. 2 µM TDZ | Steroid biosynthesis Folate biosynthesis Histidine metabolism | |
| 0 vs. 20 µM TDZ | Steroid biosynthesis Sesquiterpene and triterpene biosynthesis | ||
| Volcano Analysis | 0 vs. 2 µM TDZ | Purine biosynthesis Starch and sucrose metabolism Steroid biosynthesis | |
| 0 vs. 20 µM TDZ | Glycolysis/gluconeogenesis Pentose phosphate pathway Fructose and mannose metabolism Galactose metabolism Fatty acid degradation | ||
| Pattern Hunter | Linear Increase 0 < 2 < 20 µM TDZ | N-glycan biosynthesis | 0.01 |
| Fructose and mannose metabolism | 0.02 | ||
| Phosphatidylinositol signaling | 0.03 | ||
| Amino sugar and nucleotide sugar metabolism | 0.04 | ||
| Inositol phosphate metabolism | 0.04 | ||
| Arachidonic acid metabolism | 0.04 | ||
| Glycolysis/gluconeogenesis | 0.05 | ||
| Galactose metabolism | 0.05 | ||
| Linear Decrease 0 > 2 > 20 µM TDZ | Porphyrin and chlorophyll metabolism | 0 | |
| Phosphatidylinositol signaling | 0.01 | ||
| Starch and sucrose metabolism | 0.01 | ||
| Galactose metabolism | 0.01 | ||
| Inositol phosphate metabolism | 0.01 | ||
| Glycolysis/gluconeogenesis | 0.02 | ||
| Pentose phosphate pathway | 0.02 | ||
| Valine, leucine, isoleucine Biosynthesis | 0.02 | ||
| Fructose and mannose Metabolism | 0.02 | ||
| Ascorbate and aldarate Metabolism | 0.03 | ||
| Caffeine metabolism | 0.04 | ||
| Amino sugar and nucleotide Sugar metabolism | 0.04 | ||
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.05 | ||
| Flavonoid biosynthesis | 0.05 | ||
| Pentose and glucoronate interconversions | 0.05 | ||
| Peak at 2 µM TDZ 2 > 20 > 0 | Porphyrin and chlorophyll metabolism | 0 | |
| Tyrosine metabolism | 0.02 | ||
| Tropane, piperidine and pyridine alkaloid biosynthesis | 0.03 | ||
| Sesquiterpenoid and triterpenoid biosynthesis | 0.05 | ||
| Dip at 2 µM TDZ 2 < 20 < 0 | Carotenoid biosynthesis | 0.05 | |
| Galactose metabolism | 0.05 | ||
| Flavonoid biosynthesis | 0.05 |
| Test | Putative FooDB ID | Metabolite Class | Associated KEGG Pathway(s) |
|---|---|---|---|
| SAM | Triterpene saponin Lucyoside M Spinasaponin A Cynarasaponin C Calendulaglucoside E | Triterpene saponin | Sesquiterpenoid and triterpenoid biosynthesis |
| EBAM | Gerberinol | 4-Hydroxycoumarin | Phenylpropanoid biosynthesis/Biosynthesis of phenylpropanoids Phenylpropanoid biosynthesis |
| Xanthotoxol glucoside | Coumarin glucoside | Biosynthesis of secondary metabolites | |
| 1-Methyl-3-(2-thiazolyl)-1H-indole | Camalexin derivative | MAPK Signaling pathway-plant | |
| Triglyceride | Lipid | Triacylglycerol biosynthesis | |
| a-L-threo-4-Hex-4-enopyranuronosyl-D-galacturonic acid | Glucuronic acid derivative | Pentose and glucoronate interconversions Ascorbate and aldarate metabolism Amino sugar and nucleaotide sugar metabolism Inositol phosphate metabolism | |
| 4-Methylumbelliferone glucuronide | Glucoronic acid derivative/hydroxycoumarin | ||
| Chlorogenoquinone | Quinic acid derivative | Phenylalanine, tyrosine and tryptophan biosynthesis |
| Putative ID | Metabolite Mass | Monoisotopic Mass | 2 µM TDZ (% of Control) | 20 µM TDZ (% of Control) |
|---|---|---|---|---|
| Salicin phosphate | 366.0716 | 366.073955 | 150.16 | 200.33 |
| Disaccharide | 342.1162 | 342.115732 | 85.58 | 116.90 |
| Arbutin | 272.0896 | 272.087101 | 76.67 | 277.72 |
| Arbutin-phosphate | 352.0559 | 352.068037 | 74.68 | 93.34 |
| Salicin | 286.1053 | 286.09 | 42.09 | 51.85 |
| Glucose/fructose | 180.0634 | 180.06339 | 88.79 | 92.48 |
| 2-Dehydro-d-gluconate | 194.0427 | 194.05791 | 69.25 | 74.66 |
| 6C Disaccharide-P | 422.0825 | 422.082547 | 60.09 | 92.73 |
| 5C Monosaccharide-P | 230.0192 | 230.019157 | 72.19 | 50.21 |
| 6C Monosaccharide-P | 260.0297 | 260.029722 | 605.62 | 55.80 |
| 7C Monosaccharide-P | 290.0403 | 290.049016 | 113.01 | 49.21 |
| deoxy-ribose-P | 214.0242 | 214.008851 | 101.84 | 91.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erland, L.A.E.; Giebelhaus, R.T.; Victor, J.M.R.; Murch, S.J.; Saxena, P.K. The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity. Biomolecules 2020, 10, 1253. https://doi.org/10.3390/biom10091253
Erland LAE, Giebelhaus RT, Victor JMR, Murch SJ, Saxena PK. The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity. Biomolecules. 2020; 10(9):1253. https://doi.org/10.3390/biom10091253
Chicago/Turabian StyleErland, Lauren A. E., Ryland T. Giebelhaus, Jerrin M. R. Victor, Susan J. Murch, and Praveen K. Saxena. 2020. "The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity" Biomolecules 10, no. 9: 1253. https://doi.org/10.3390/biom10091253
APA StyleErland, L. A. E., Giebelhaus, R. T., Victor, J. M. R., Murch, S. J., & Saxena, P. K. (2020). The Morphoregulatory Role of Thidiazuron: Metabolomics-Guided Hypothesis Generation for Mechanisms of Activity. Biomolecules, 10(9), 1253. https://doi.org/10.3390/biom10091253







