Mouse WIF1 Is Only Modified with O-Fucose in Its EGF-like Domain III Despite Two Evolutionarily Conserved Consensus Sites
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Reconstruction and Sequence Conservation Analyses
2.2. Plasmid Constructs
2.3. Protein Expression and Purification
2.4. Glycosyltransferase Reactions
2.5. Click Chemistry Reactions
2.6. SDS-PAGE and Blotting Techniques
2.7. Automatic Modeling and Superimposition with X-ray Structures
2.8. Protein Digestions, Mass Spectrometry Analyses and Data Processing
2.9. Statistical Analysis
3. Results
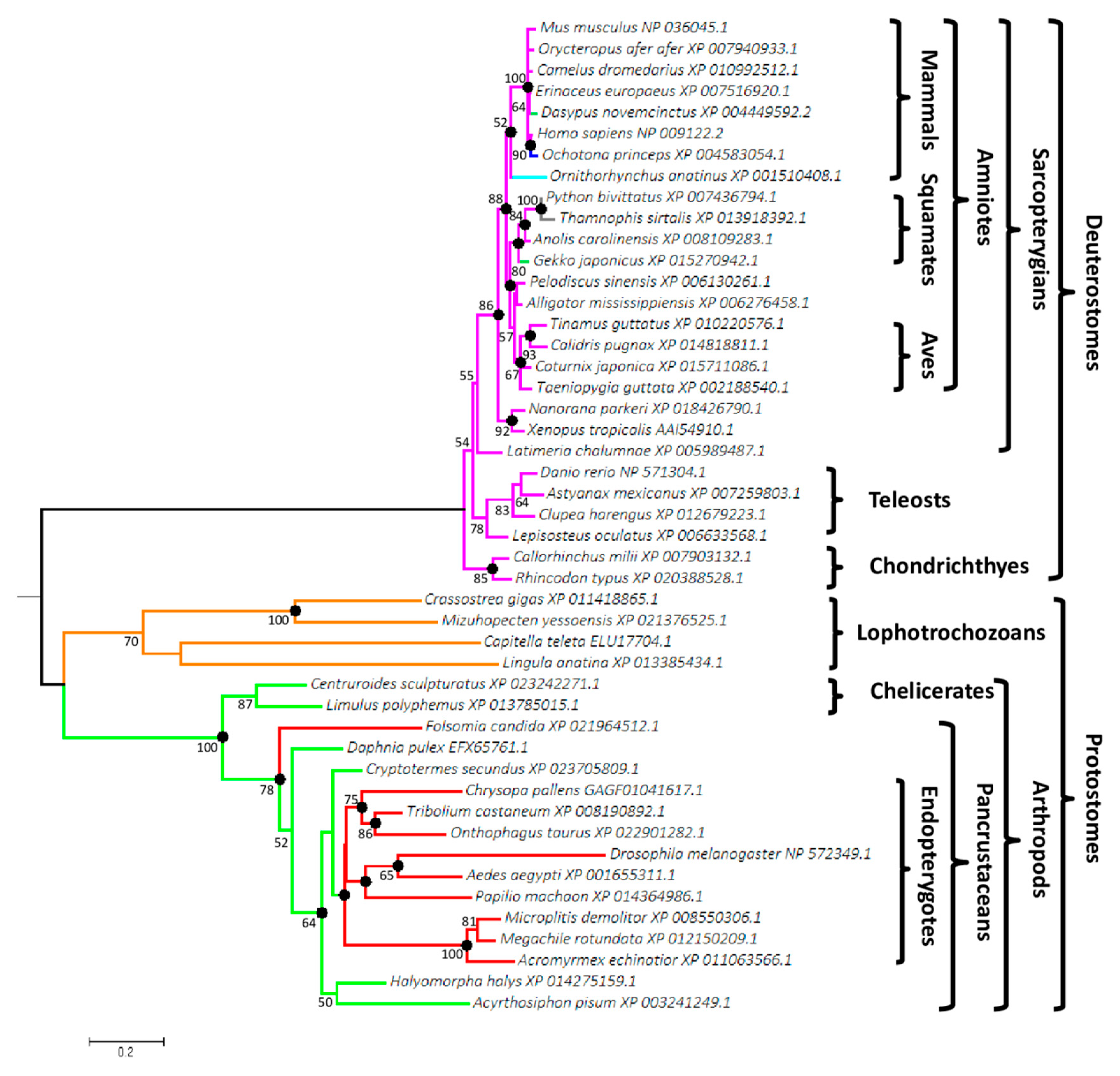
3.1. Wif1 Appeared in Bilaterian Ancestor
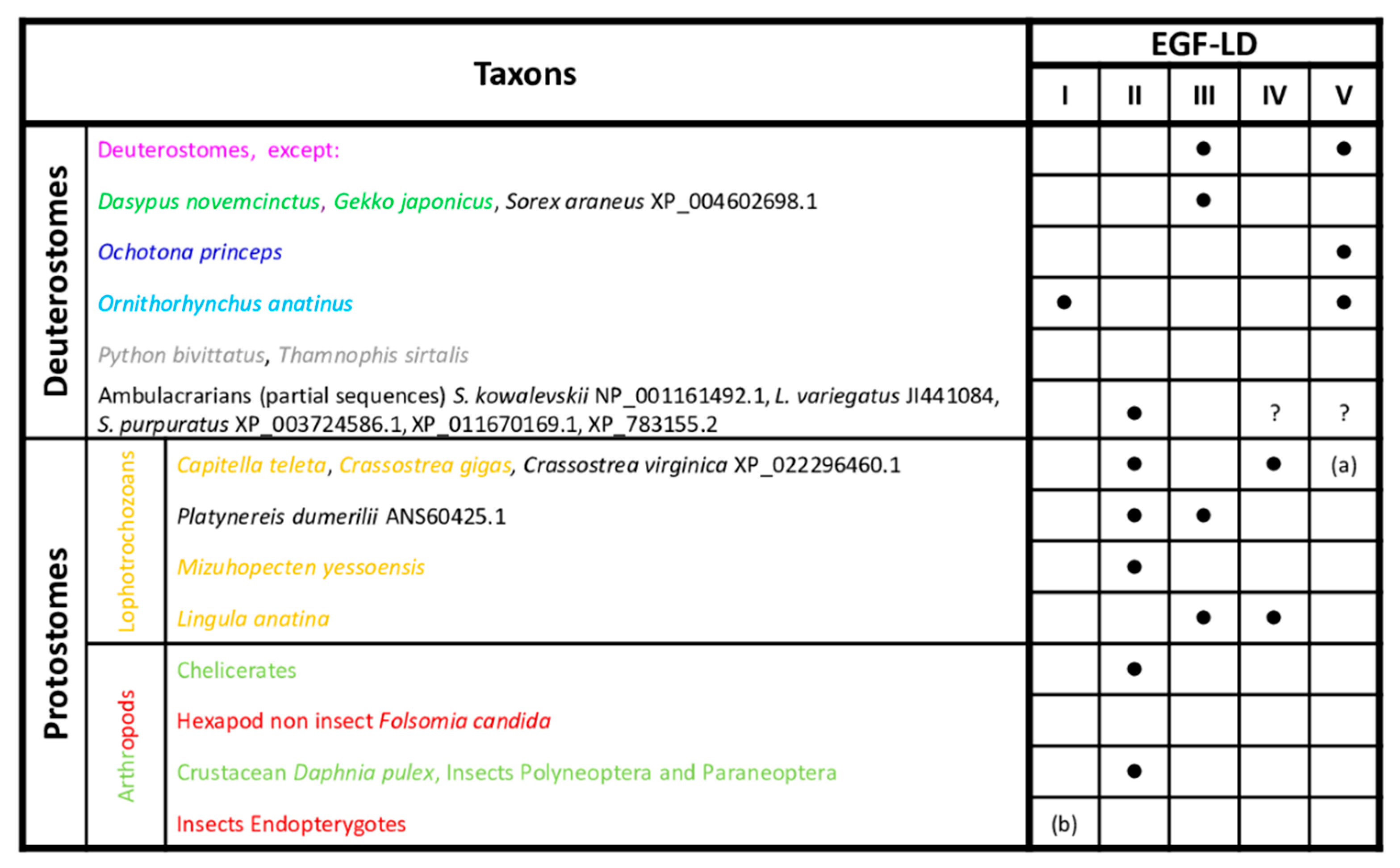
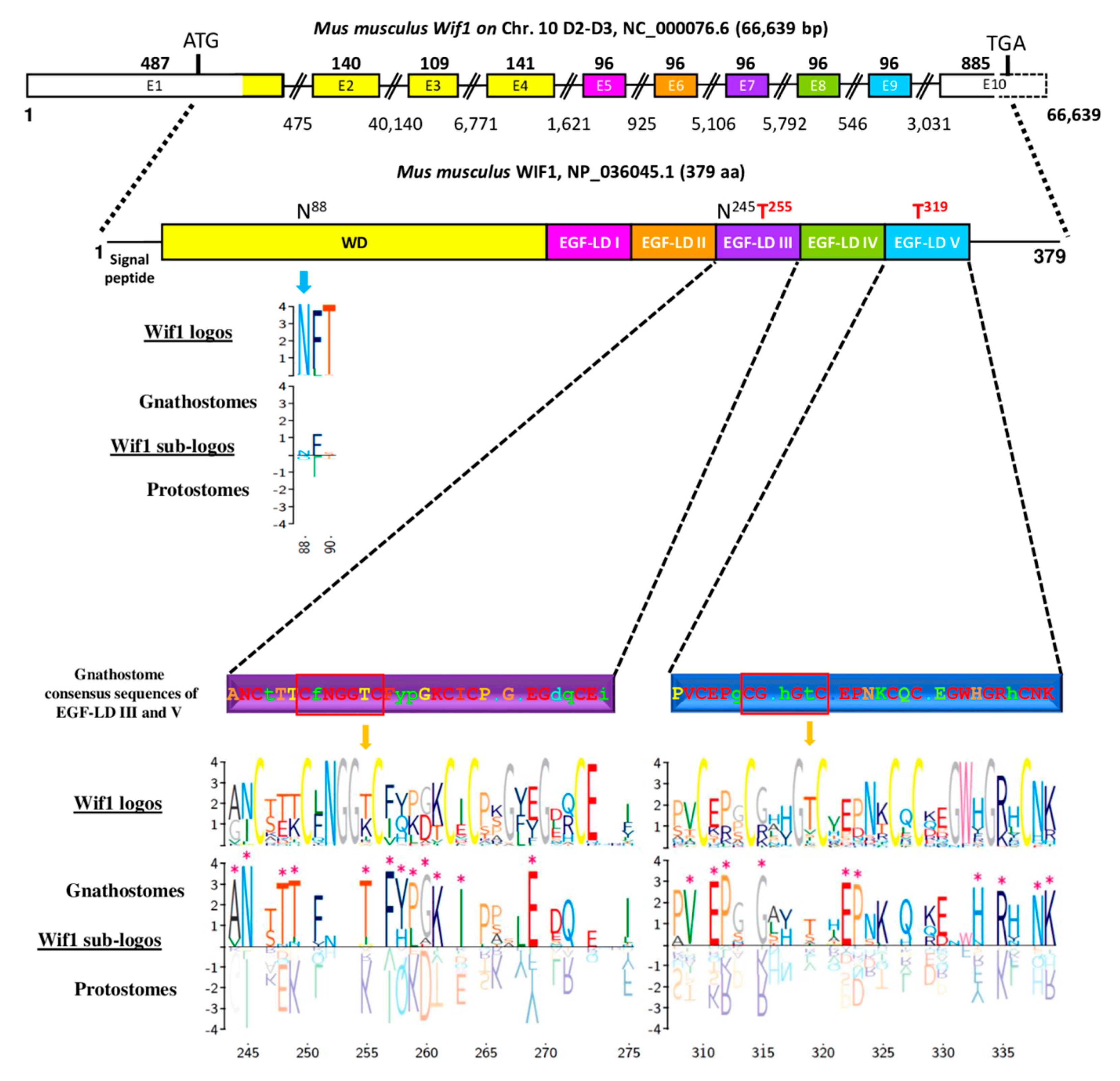
3.2. Predicted O-Fucosylation Sites Are Evolutionarily Conserved in Deuterostomes
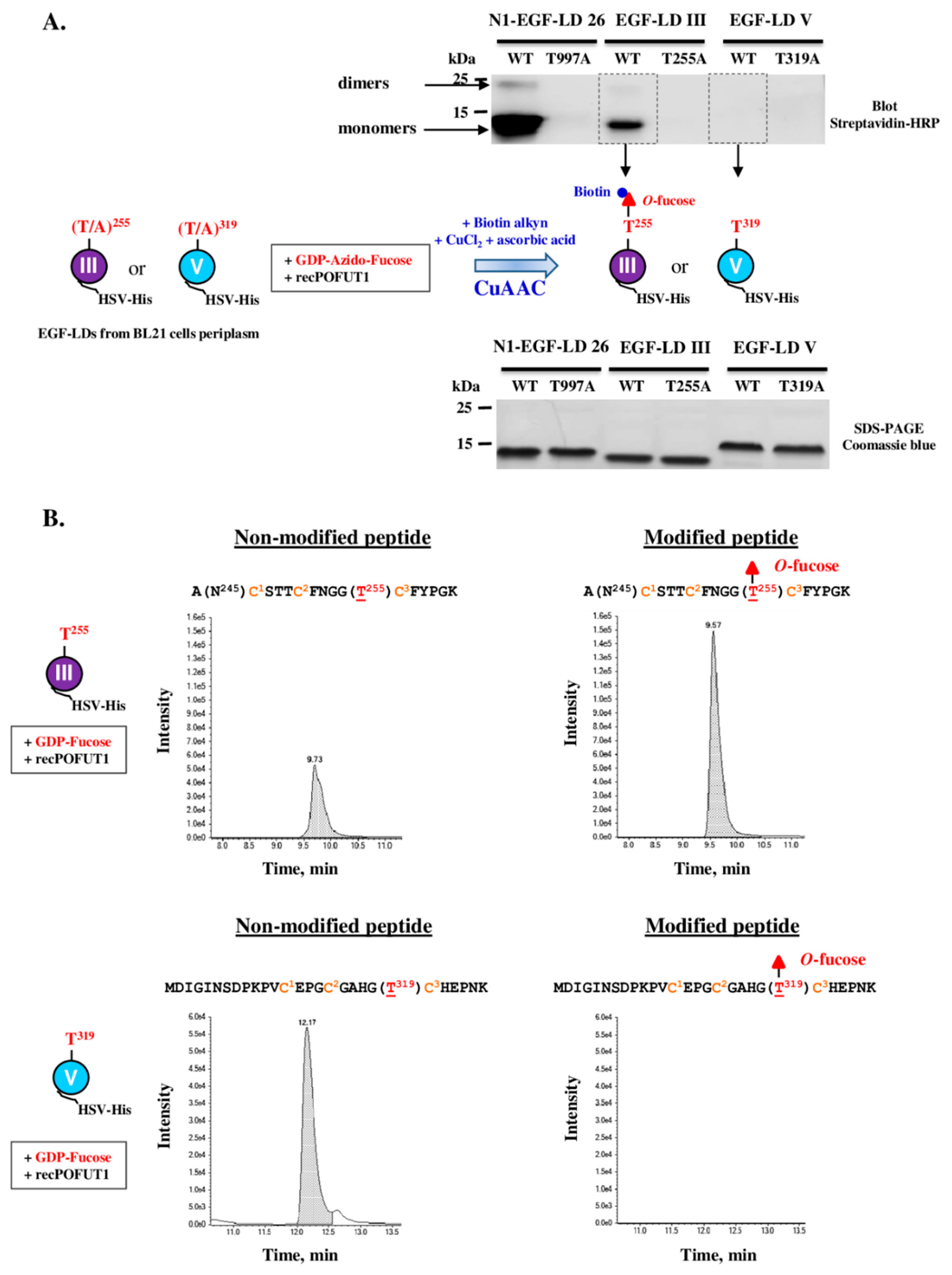
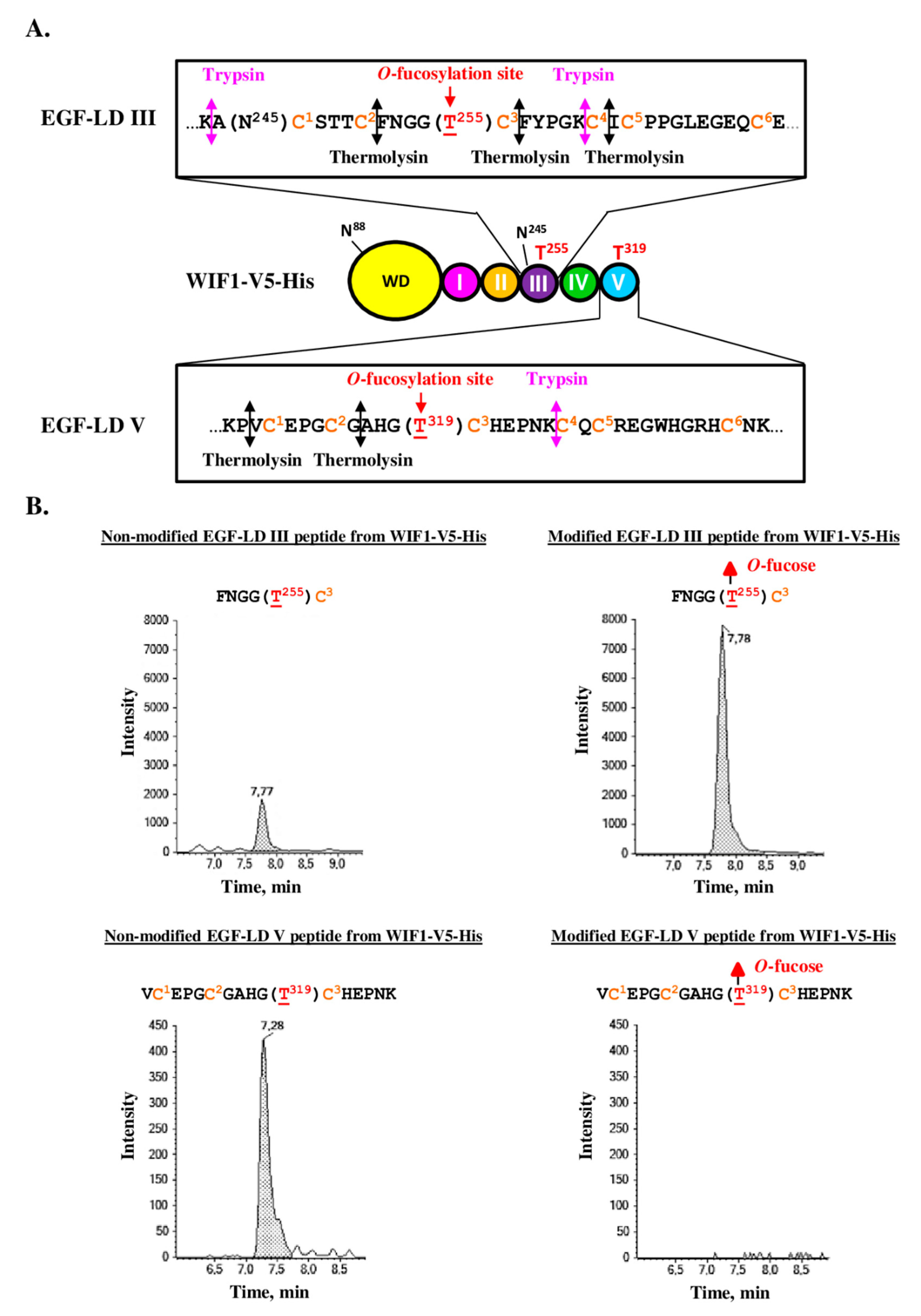
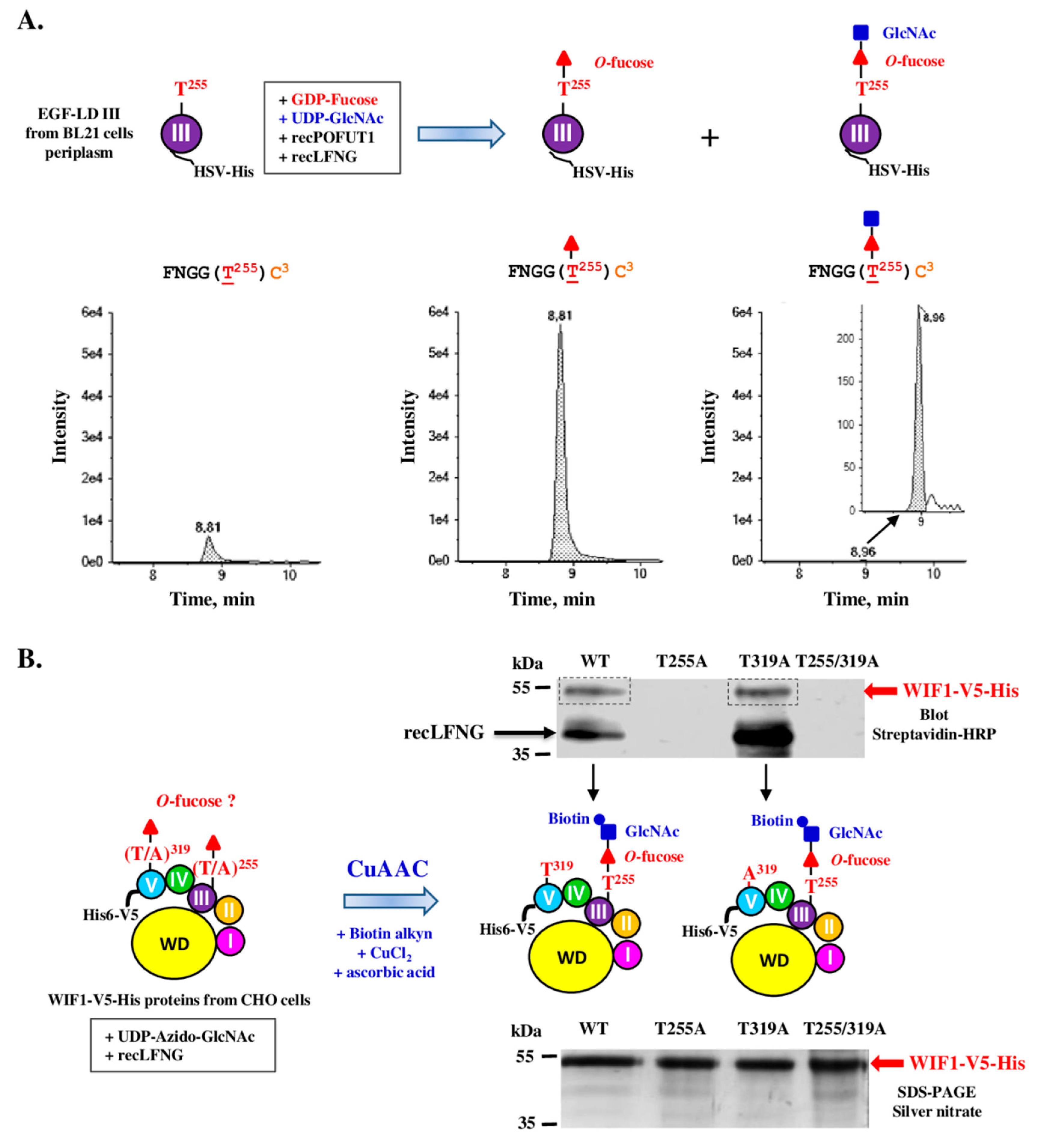
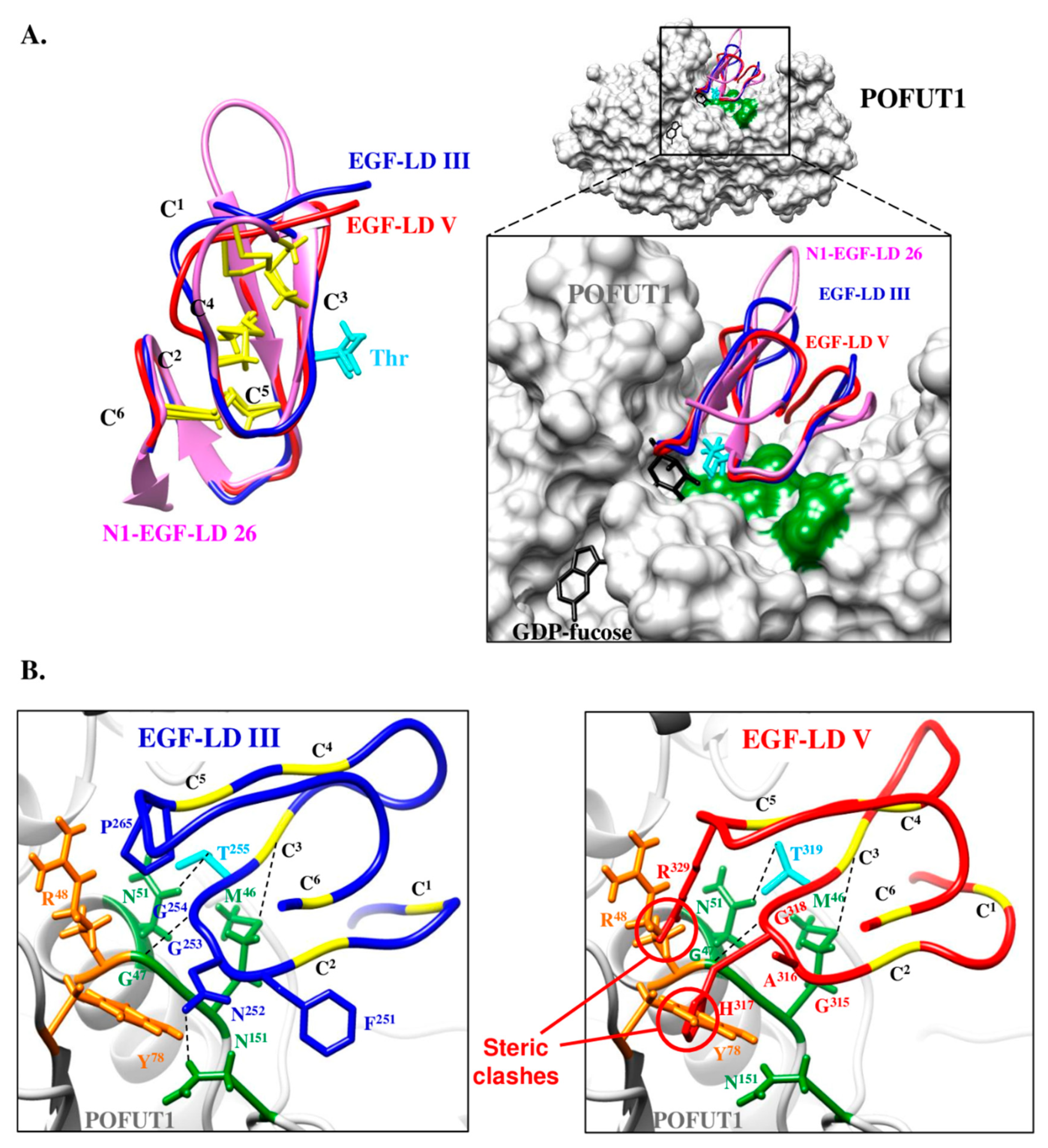
3.3. Only WIF1 EGF-LD III Can Be In Vitro Modified by O-Fucose
3.4. Full-Length Recombinant WIF1 Carried O-Fucose Only on Its EGF-LD III
3.5. Full-Length Recombinant WIF1 Carried a Non-Extended O-Fucose on Its EGF-LD III
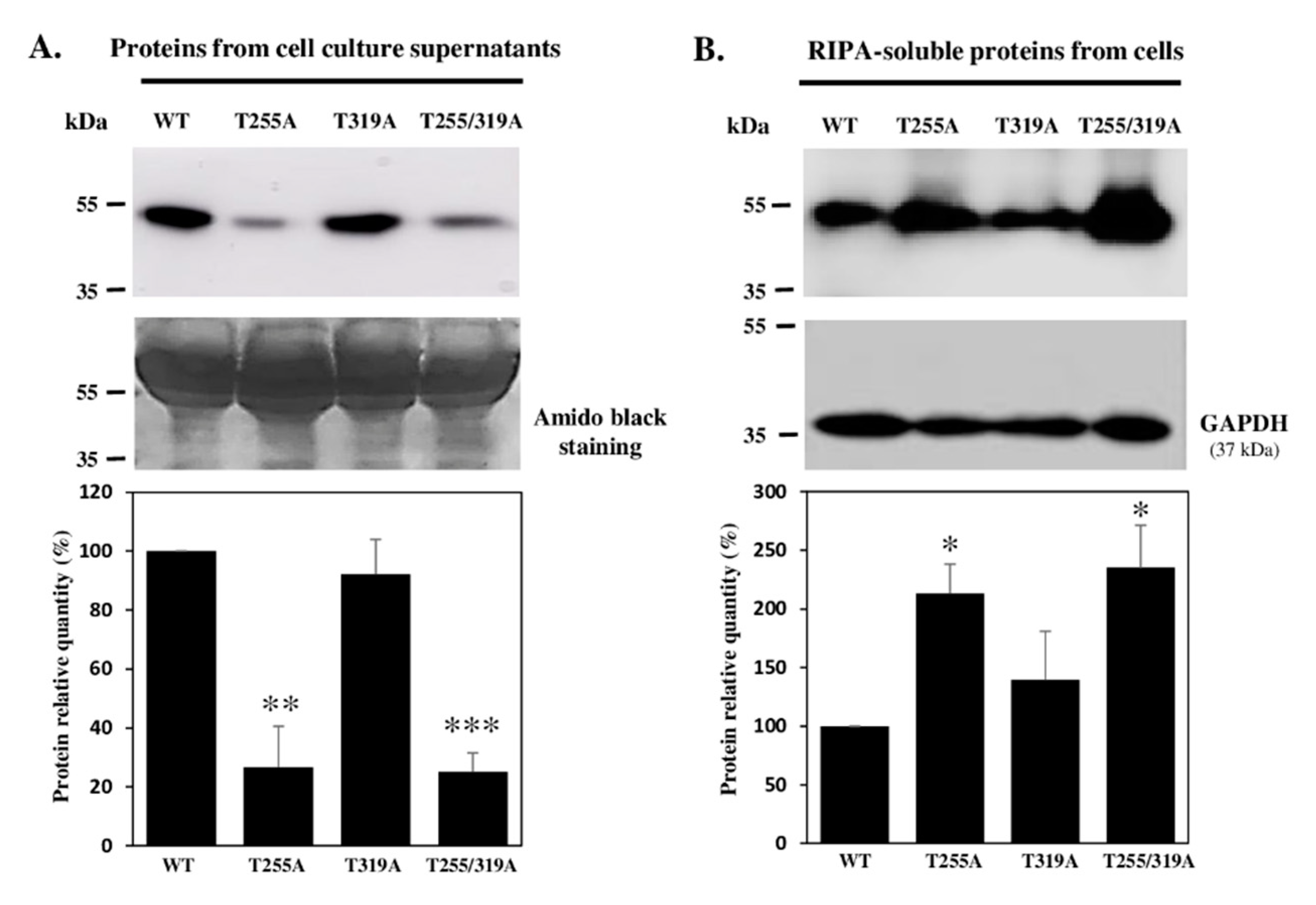
3.6. The O-Fucose Carried by T255 Was Required for Optimal Secretion of Recombinant WIF1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawano, Y.; Kypta, R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003, 116, 2627–2634. [Google Scholar] [CrossRef]
- Malinauskas, T.; Jones, E.Y. Extracellular modulators of Wnt signalling. Curr. Opin. Struct. Biol. 2014, 29, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Dequeant, M.L.; Pourquie, O. Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 2008, 9, 370–382. [Google Scholar] [CrossRef]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Ilyas, M. Wnt signalling and the mechanistic basis of tumour development. J. Pathol. 2005, 205, 130–144. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Hsieh, J.C.; Kodjabachian, L.; Rebbert, M.L.; Rattner, A.; Smallwood, P.M.; Samos, C.H.; Nusse, R.; Dawid, I.B.; Nathans, J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 1999, 398, 431–436. [Google Scholar] [CrossRef]
- Liepinsh, E.; Banyai, L.; Patthy, L.; Otting, G. NMR structure of the WIF domain of the human Wnt-inhibitory factor-1. J. Mol. Biol. 2006, 357, 942–950. [Google Scholar] [CrossRef]
- Malinauskas, T.; Aricescu, A.R.; Lu, W.; Siebold, C.; Jones, E.Y. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat. Struct. Mol. Biol. 2011, 18, 886–893. [Google Scholar] [CrossRef]
- Pennarubia, F.; Germot, A.; Pinault, E.; Maftah, A.; Legardinier, S. The single EGF-like domain of mouse PAMR1 is modified by O-Glucose, O-Fucose and O-GlcNAc. Glycobiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Moloney, D.J.; Haltiwanger, R. Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the Abruptex region. J. Biol. Chem. 2003, 278, 7775–7782. [Google Scholar] [CrossRef] [PubMed]
- Moloney, D.J.; Shair, L.H.; Lu, F.M.; Xia, J.; Locke, R.; Matta, K.L.; Haltiwanger, R.S. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 2000, 275, 9604–9611. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Rampal, R.; Fung, E.; Darland, D.C.; Liu, M.; Martinez, M.C.; Donahue, C.P.; Navarro-Gonzalez, M.F.; Libby, P.; D’Amore, P.A.; et al. CADASIL mutations impair Notch3 glycosylation by Fringe. Hum. Mol. Genet. 2005, 14, 1631–1639. [Google Scholar] [CrossRef]
- Panin, V.M.; Shao, L.; Lei, L.; Moloney, D.J.; Irvine, K.D.; Haltiwanger, R.S. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 2002, 277, 29945–29952. [Google Scholar] [CrossRef]
- Harris, R.J.; Leonard, C.K.; Guzzetta, A.W.; Spellman, M.W. Tissue plasminogen activator has an O-linked fucose attached to threonine-61 in the epidermal growth factor domain. Biochemistry 1991, 30, 2311–2314. [Google Scholar] [CrossRef]
- Kentzer, E.J.; Buko, A.; Menon, G.; Sarin, V.K. Carbohydrate composition and presence of a fucose-protein linkage in recombinant human pro-urokinase. Biochem. Biophys. Res. Commun. 1990, 171, 401–406. [Google Scholar] [CrossRef]
- Harris, R.J.; Ling, V.T.; Spellman, M.W. O-linked fucose is present in the first epidermal growth factor domain of factor XII but not protein C. J. Biol. Chem. 1992, 267, 5102–5107. [Google Scholar]
- Nishimura, H.; Takao, T.; Hase, S.; Shimonishi, Y.; Iwanaga, S. Human factor IX has a tetrasaccharide O-glycosidically linked to serine 61 through the fucose residue. J. Biol. Chem. 1992, 267, 17520–17525. [Google Scholar]
- Bjoern, S.; Foster, D.C.; Thim, L.; Wiberg, F.C.; Christensen, M.; Komiyama, Y.; Pedersen, A.H.; Kisiel, W. Human plasma and recombinant factor VII. Characterization of O-glycosylations at serine residues 52 and 60 and effects of site-directed mutagenesis of serine 52 to alanine. J. Biol. Chem. 1991, 266, 11051–11057. [Google Scholar]
- Kim, M.L.; Chandrasekharan, K.; Glass, M.; Shi, S.; Stahl, M.C.; Kaspar, B.; Stanley, P.; Martin, P.T. O-fucosylation of muscle agrin determines its ability to cluster acetylcholine receptors. Mol. Cell Neurosci. 2008, 39, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, J.M.; Muller, S.; Hanisch, F.G.; Paulsson, M.; Wagener, R. O-glucosylation and O-fucosylation occur together in close proximity on the first epidermal growth factor repeat of AMACO (VWA2 protein). J. Biol. Chem. 2008, 283, 17846–17854. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, S.G.; Foley, S.; Kaffashan, A.; Hronowski, X.; Zichittella, A.E.; Yeo, C.Y.; Miatkowski, K.; Adkins, H.B.; Damon, B.; Whitman, M.; et al. Fucosylation of Cripto is required for its ability to facilitate nodal signaling. J. Biol. Chem. 2001, 276, 37769–37778. [Google Scholar] [CrossRef]
- Alfaro, J.F.; Gong, C.X.; Monroe, M.E.; Aldrich, J.T.; Clauss, T.R.; Purvine, S.O.; Wang, Z.; Camp II, D.G.; Shabanowitz, J.; Stanley, P.; et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. USA 2012, 109, 7280–7285. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Nara, N.; Kawakita, Y.; Takeshima, Y.; Arakawa, M.; Katoh, M.; Morita, S.; Iwatsuki, K.; Tanaka, K.; Okamoto, S.; et al. Cloning of cDNA encoding a regeneration-associated muscle protease whose expression is attenuated in cell lines derived from Duchenne muscular dystrophy patients. Am. J. Pathol. 2004, 164, 1773–1782. [Google Scholar] [CrossRef]
- Wang, Y.; Shao, L.; Shi, S.; Harris, R.J.; Spellman, M.W.; Stanley, P.; Haltiwanger, R.S. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 2001, 276, 40338–40345. [Google Scholar] [CrossRef]
- Loriol, C.; Dupuy, F.; Rampal, R.; Dlugosz, M.A.; Haltiwanger, R.S.; Maftah, A.; Germot, A. Molecular evolution of protein O-fucosyltransferase genes and splice variants. Glycobiology 2006, 16, 736–747. [Google Scholar] [CrossRef]
- Wouters, M.A.; Rigoutsos, I.; Chu, C.K.; Feng, L.L.; Sparrow, D.B.; Dunwoodie, S.L. Evolution of distinct EGF domains with specific functions. Protein Sci. 2005, 14, 1091–1103. [Google Scholar] [CrossRef]
- Li, Z.; Han, K.; Pak, J.E.; Satkunarajah, M.; Zhou, D.; Rini, J.M. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 2017, 13, 757–763. [Google Scholar] [CrossRef]
- Lira-Navarrete, E.; Valero-Gonzalez, J.; Villanueva, R.; Martinez-Julvez, M.; Tejero, T.; Merino, P.; Panjikar, S.; Hurtado-Guerrero, R. Structural insights into the mechanism of protein O-fucosylation. PLoS ONE 2011, 6, e25365. [Google Scholar] [CrossRef]
- Rampal, R.; Arboleda-Velasquez, J.F.; Nita-Lazar, A.; Kosik, K.S.; Haltiwanger, R.S. Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 2005, 280, 32133–32140. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, L.M.; Dlugosz, M.; Luther, K.B.; Haltiwanger, R.S.; Majerus, E.M. O-fucosylation is required for ADAMTS13 secretion. J. Biol. Chem. 2007, 282, 17014–17023. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Suzuki, T.; Dohmae, N.; Simizu, S. O-Fucosylation of CCN1 is required for its secretion. FEBS Lett. 2015, 589, 3287–3293. [Google Scholar] [CrossRef]
- Panin, V.M.; Papayannopoulos, V.; Wilson, R.; Irvine, K.D. Fringe modulates Notch-ligand interactions. Nature 1997, 387, 908–912. [Google Scholar] [CrossRef]
- Rana, N.A.; Haltiwanger, R.S. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 2011, 21, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kakuda, S.; Haltiwanger, R.S. Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Dev. Cell 2017, 40, 193–201. [Google Scholar] [CrossRef]
- Pennarubia, F.; Pinault, E.; Maftah, A.; Legardinier, S. In vitro acellular method to reveal O-fucosylation on EGF-like domains. Glycobiology 2018. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar] [CrossRef]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol. Direct. 2008, 3, 20. [Google Scholar] [CrossRef]
- Beitz, E. TEXshade: Shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef]
- Beitz, E. Subfamily logos: Visualization of sequence deviations at alignment positions with high information content. BMC Bioinform. 2006, 7, 313. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Telford, M.J.; Budd, G.E.; Philippe, H. Phylogenomic Insights into Animal Evolution. Curr. Biol. 2015, 25, R876–R887. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Brinkmann, H.; Philippe, H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007, 7, S4. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Hernandez, D.; Sierra, J.; Ortigao-Farias, J.R.; Guerrero, I. The WIF domain of the human and Drosophila Wif-1 secreted factors confers specificity for Wnt or Hedgehog. Development 2012, 139, 3849–3858. [Google Scholar] [CrossRef] [PubMed]
- Deschuyter, M.; Pennarubia, F.; Pinault, E.; Legardinier, S.; Maftah, A. Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer. Cancers 2020, 12, 1430. [Google Scholar] [CrossRef]
- Moloney, D.J.; Panin, V.M.; Johnston, S.H.; Chen, J.; Shao, L.; Wilson, R.; Wang, Y.; Stanley, P.; Irvine, K.D.; Haltiwanger, R.S.; et al. Fringe is a glycosyltransferase that modifies Notch. Nature 2000, 406, 369–375. [Google Scholar] [CrossRef]
- Wu, Z.L.; Tatge, T.J.; Grill, A.E.; Zou, Y. Detecting and Imaging O-GlcNAc Sites Using Glycosyltransferases: A Systematic Approach to Study O-GlcNAc. Cell Chem. Biol. 2018, 25, 1428–1435.e3. [Google Scholar] [CrossRef]
- Haltiwanger, R.S. Regulation of signal transduction pathways in development by glycosylation. Curr. Opin. Struct. Biol. 2002, 12, 593–598. [Google Scholar] [CrossRef]
- Taylor, P.; Takeuchi, H.; Sheppard, D.; Chillakuri, C.; Lea, S.M.; Haltiwanger, R.S.; Handford, P.A. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc. Natl. Acad. Sci. USA 2014, 111, 7290–7295. [Google Scholar] [CrossRef]
- Bruckner, K.; Perez, L.; Clausen, H.; Cohen, S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 2000, 406, 411–415. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pennarubia, F.; Pinault, E.; Al Jaam, B.; Brun, C.E.; Maftah, A.; Germot, A.; Legardinier, S. Mouse WIF1 Is Only Modified with O-Fucose in Its EGF-like Domain III Despite Two Evolutionarily Conserved Consensus Sites. Biomolecules 2020, 10, 1250. https://doi.org/10.3390/biom10091250
Pennarubia F, Pinault E, Al Jaam B, Brun CE, Maftah A, Germot A, Legardinier S. Mouse WIF1 Is Only Modified with O-Fucose in Its EGF-like Domain III Despite Two Evolutionarily Conserved Consensus Sites. Biomolecules. 2020; 10(9):1250. https://doi.org/10.3390/biom10091250
Chicago/Turabian StylePennarubia, Florian, Emilie Pinault, Bilal Al Jaam, Caroline E. Brun, Abderrahman Maftah, Agnès Germot, and Sébastien Legardinier. 2020. "Mouse WIF1 Is Only Modified with O-Fucose in Its EGF-like Domain III Despite Two Evolutionarily Conserved Consensus Sites" Biomolecules 10, no. 9: 1250. https://doi.org/10.3390/biom10091250
APA StylePennarubia, F., Pinault, E., Al Jaam, B., Brun, C. E., Maftah, A., Germot, A., & Legardinier, S. (2020). Mouse WIF1 Is Only Modified with O-Fucose in Its EGF-like Domain III Despite Two Evolutionarily Conserved Consensus Sites. Biomolecules, 10(9), 1250. https://doi.org/10.3390/biom10091250





