Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator
Abstract
1. Introduction
2. Materials and Methods
2.1. Modelling, Parameterization and MD Simulation
2.2. Markov Chain Monte Carlo (MCMC) Simulations
2.3. Analysis and Visualization
2.4. MM-GBSA Analysis
3. Results
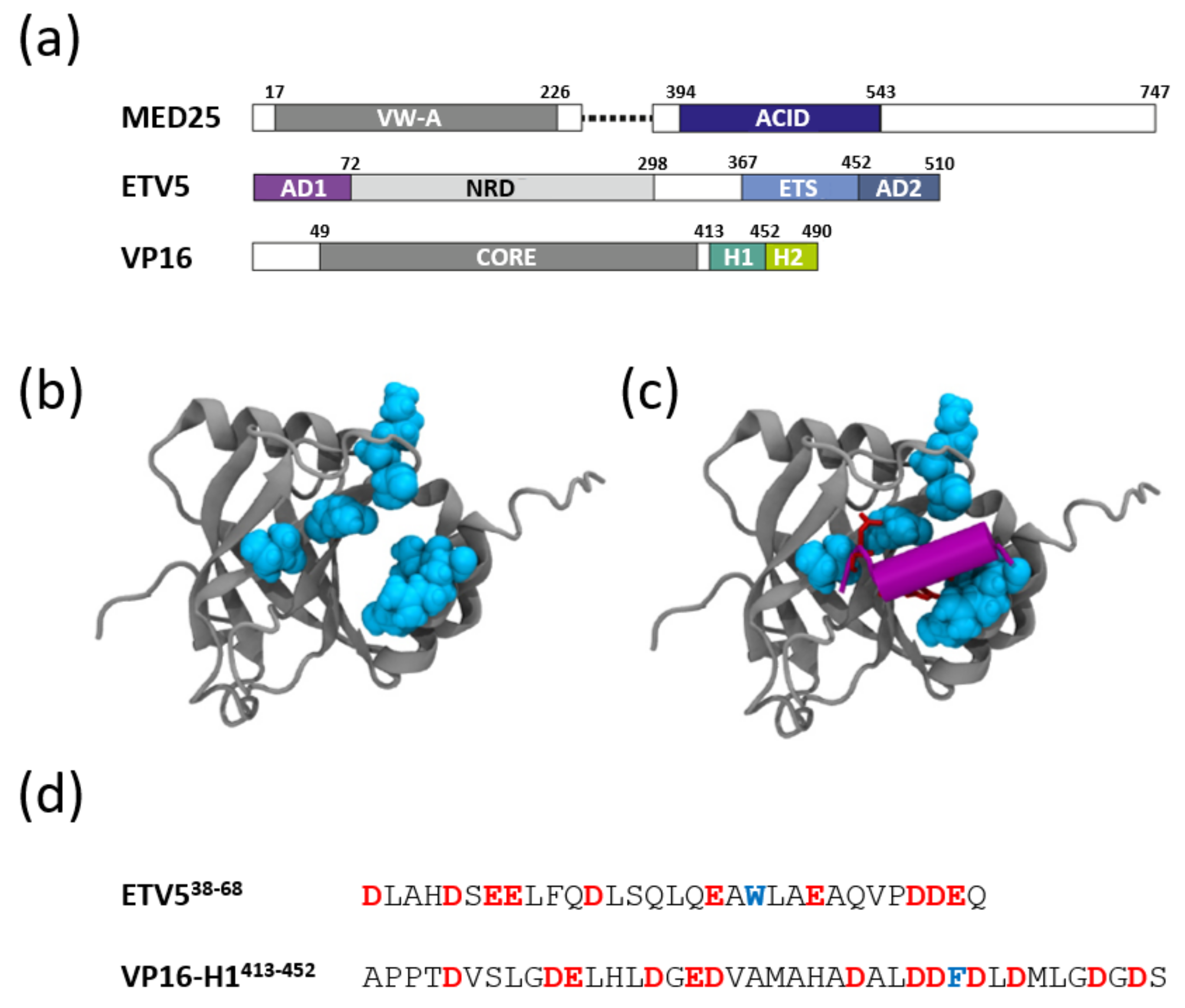
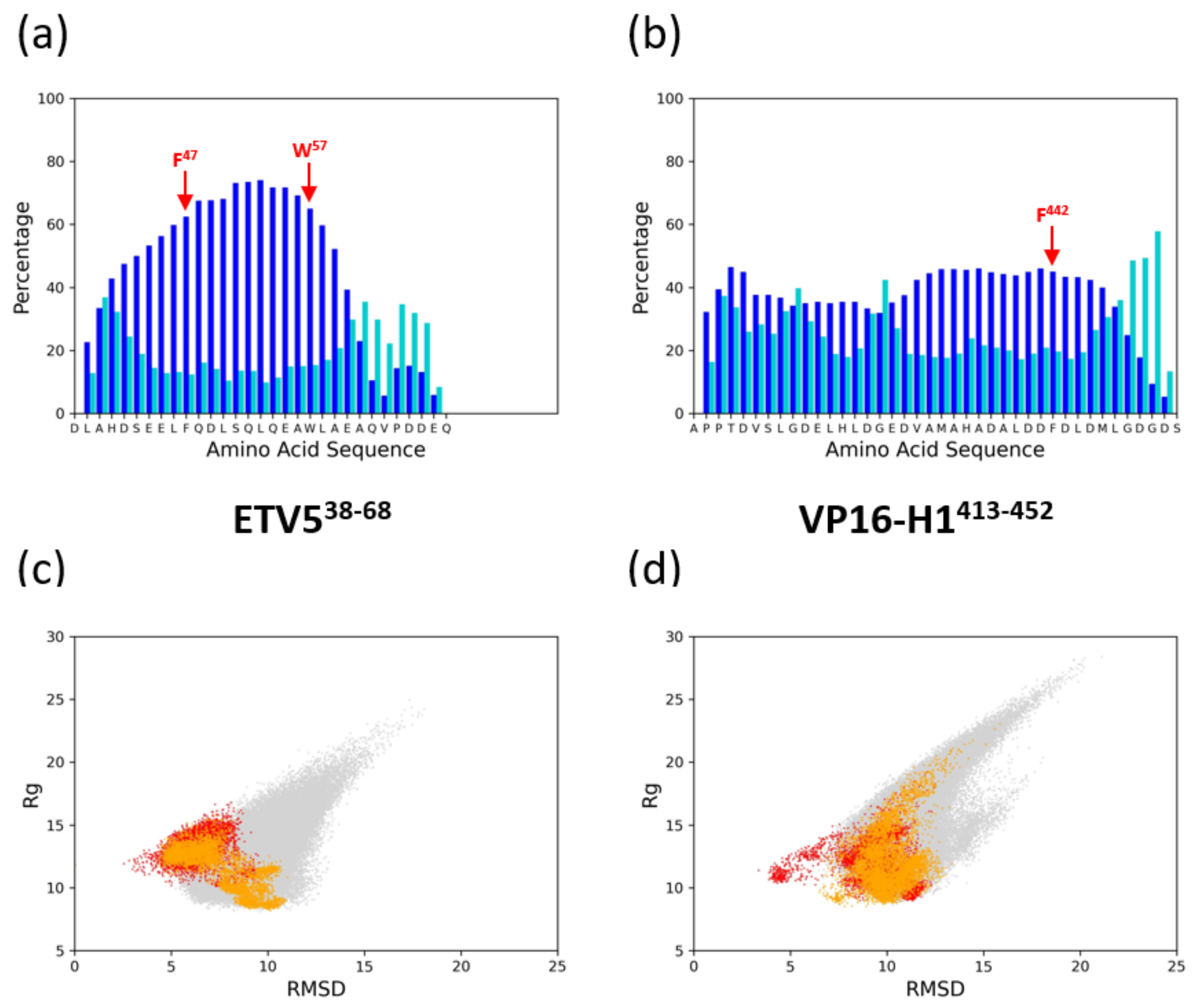
3.1. Structural Aspects of ETV538–68 and VP16-H1413–452 TADs Prior to Binding to Coactivators
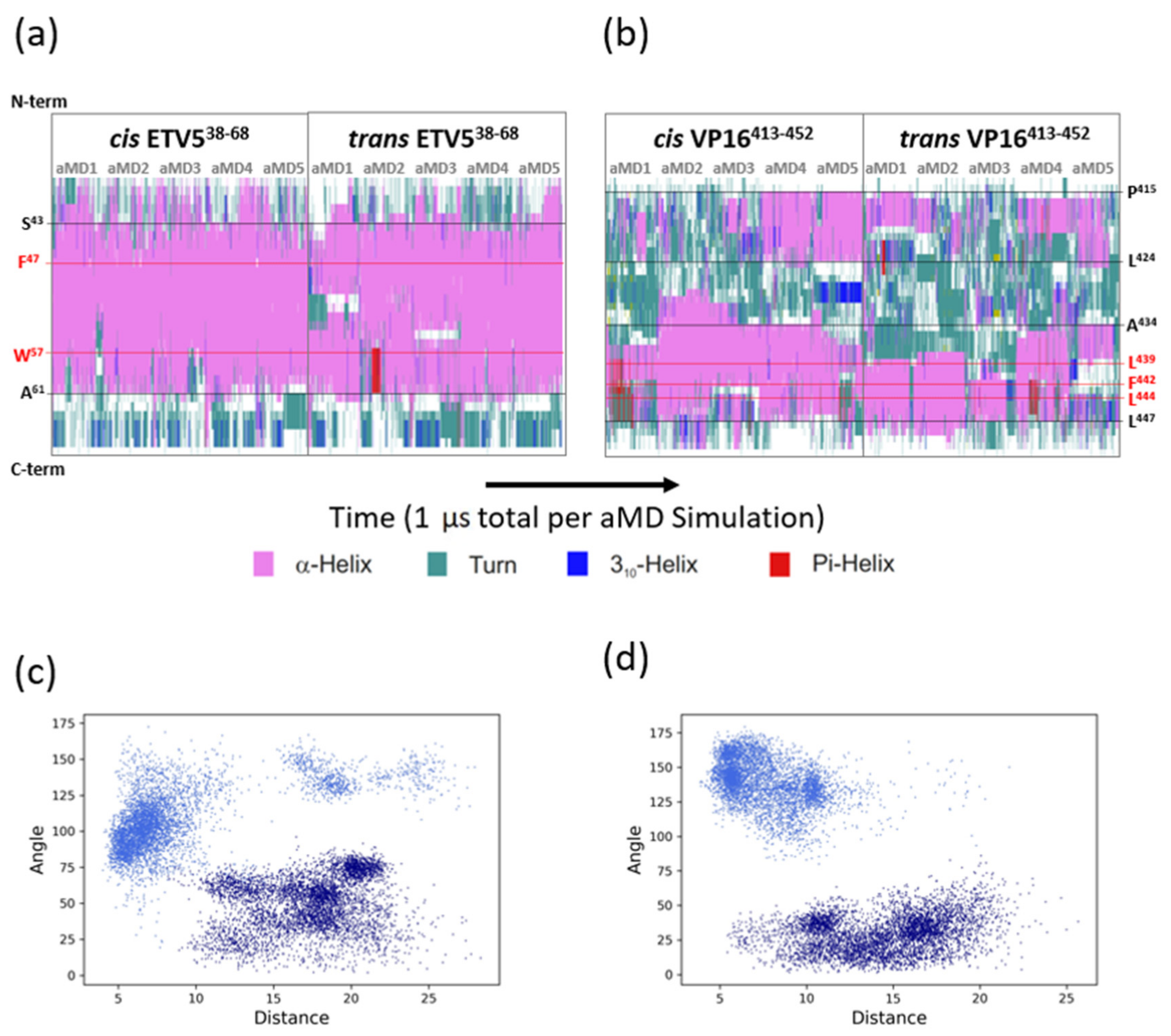
3.2. The ETV538–68 and VP16-H1413–452 TADs Interact with MED25 in an Orientation-Specific Manner
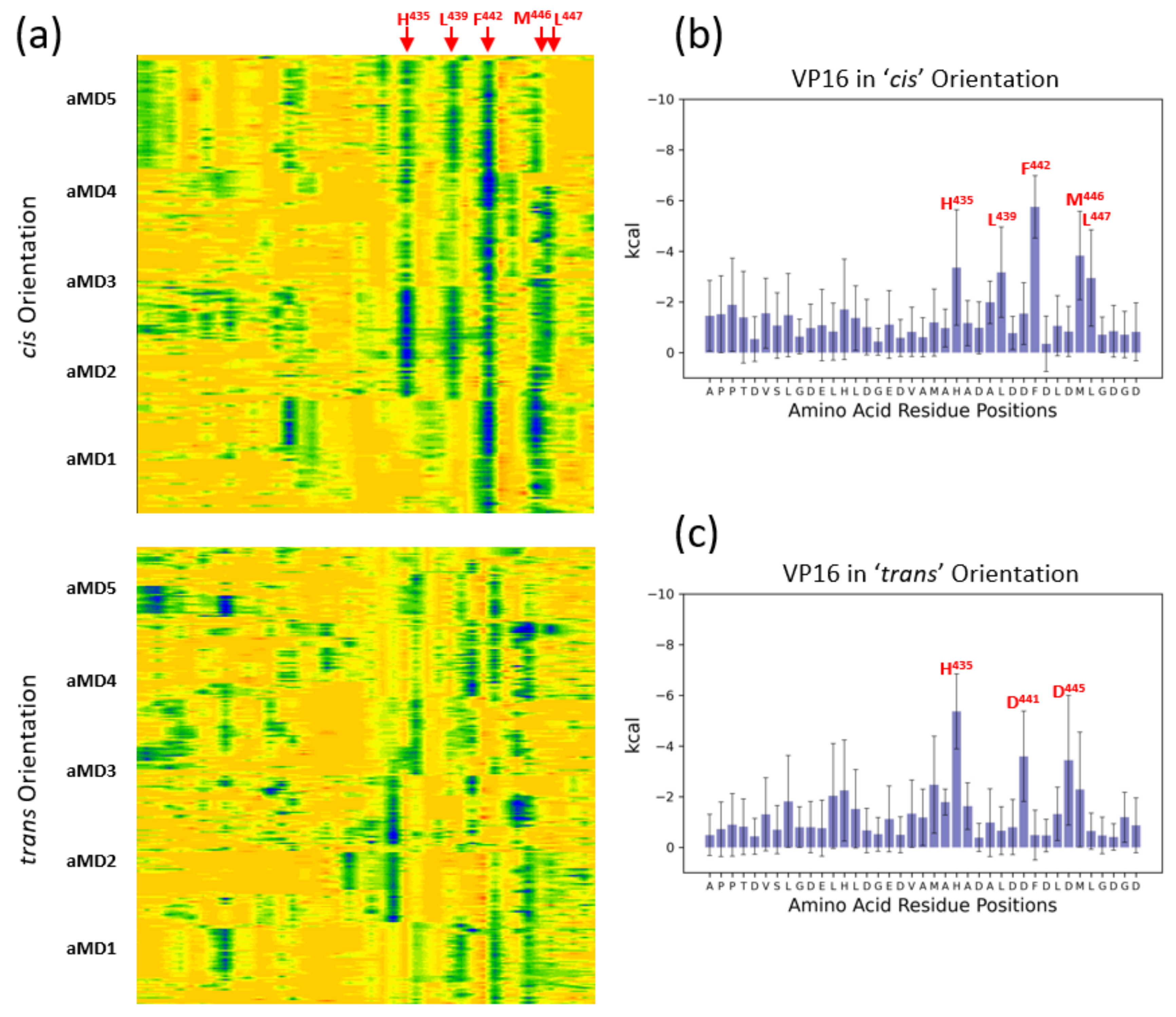
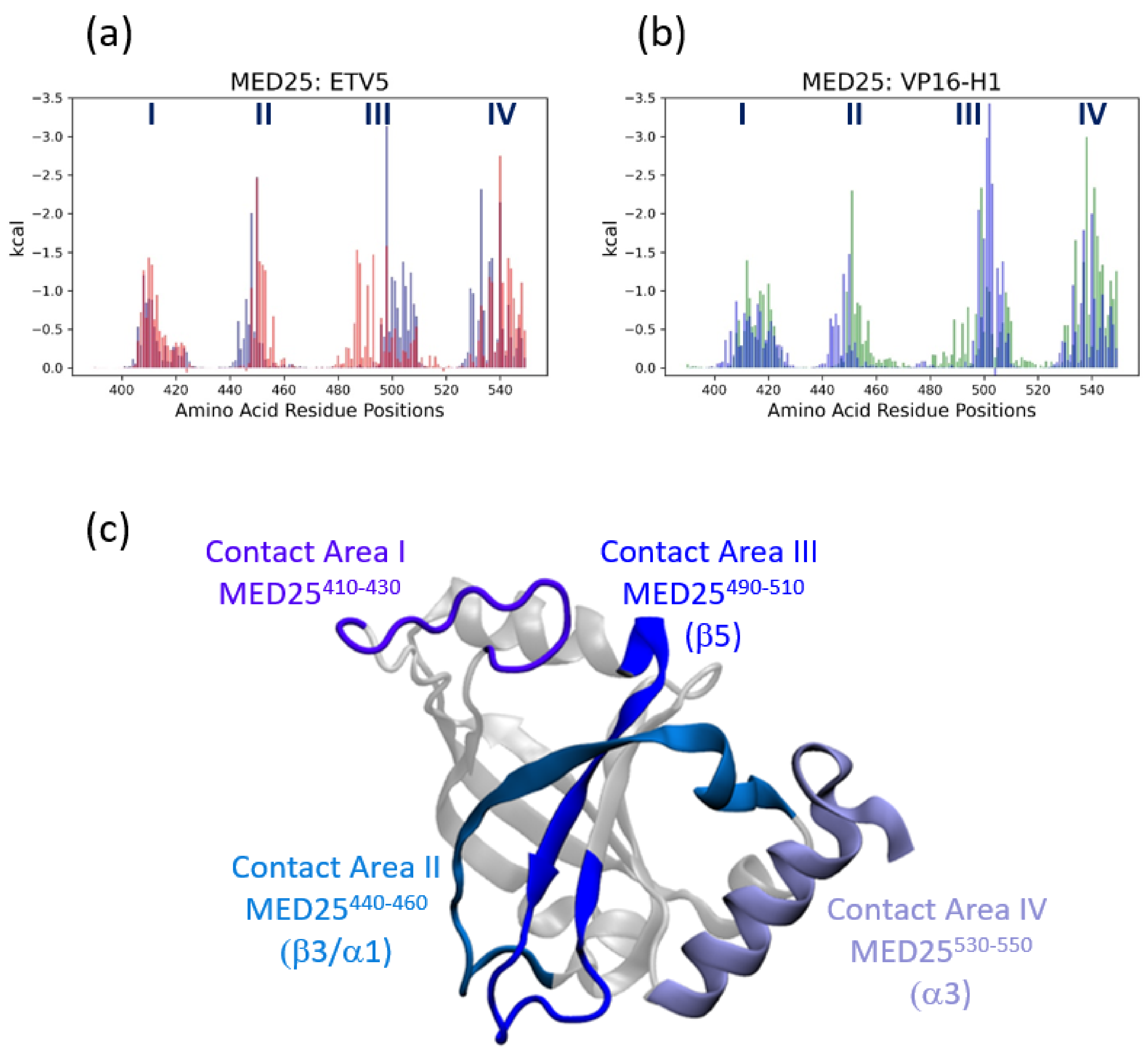
3.3. Energetic Aspects of Bidirectional Interactions of ETV538–68 and VP16-H1413–452—TADs with MED25
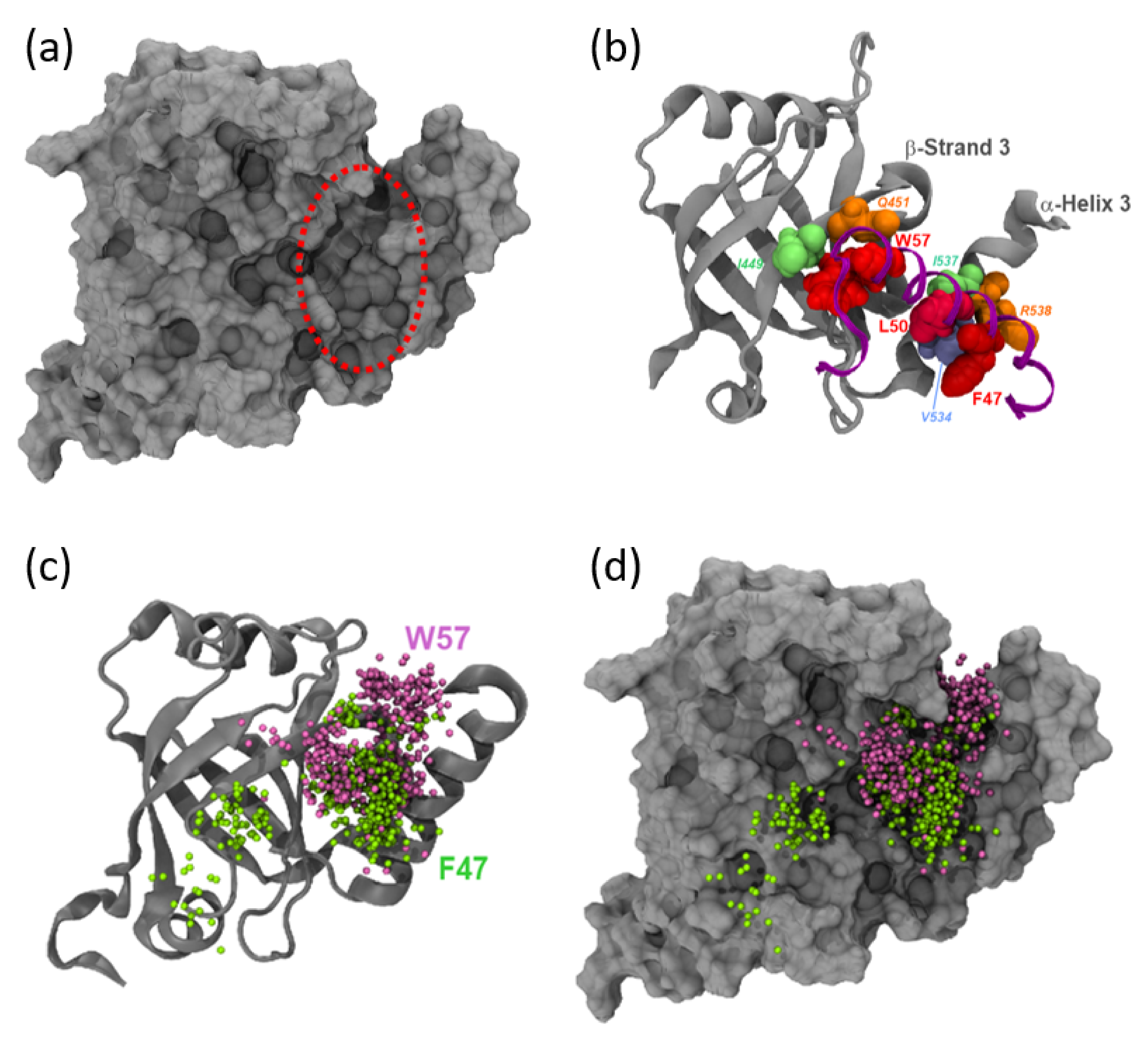
3.4. MED25 Coactivator Surfaces Interacting with ETV5- and VP16-H1 TADs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Et Biophys. Acta. 2015, 1849, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Piskacek, M.; Havelka, M.; Rezacova, M.; Knight, A. The 9aaTAD Transactivation Domains: From Gal4 to p53. PLoS ONE 2016, 11, e0162842. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J. Mammalian Mediator as a Functional Link between Enhancers and Promoters. Cell 2019, 178, 1036–1038. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.L.; Taatjes, D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef]
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, eaar3555. [Google Scholar] [CrossRef]
- Currie, S.L.; Doane, J.J.; Evans, K.S.; Bhachech, N.; Madison, B.J.; Lau, D.K.W. ETV4 and AP1 Transcription Factors Form Multivalent Interactions with three Sites on the MED25 Activator-Interacting Domain. J. Mol. Biol. 2017, 429, 2975–2995. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Heikaus, C.C.; Kisselev, L.; Vernon, R.; Herbig, E.; Pacheco, D. The Acidic Transcription Activator Gcn4 Binds the Mediator Subunit Gal11/Med15 Using a Simple Protein Interface Forming a Fuzzy Complex. Mol. Cell. 2011, 44, 942–953. [Google Scholar] [CrossRef]
- Henderson, A.R.; Henley, M.J.; Foster, N.J.; Peiffer, A.L.; Beyersdorf, M.S.; Stanford, K.D.; Sturlis, S.M.; Linhares, B.M.; Hill, Z.B.; Wells, J.A.; et al. Conservation of coactivator engagement mechanism enables small-molecule allosteric modulators. Proc. Natl. Acad. Sci. USA 2018, 115, 8960–8965. [Google Scholar] [CrossRef]
- Scholes, N.S.; Weinzierl, R.O. Molecular Dynamics of “Fuzzy” Transcriptional Activator-Coactivator Interactions. PloS Comput. Biol. 2016, 12, e1004935. [Google Scholar] [CrossRef]
- Borggrefe, T.; Yue, X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011, 22, 759–768. [Google Scholar] [CrossRef]
- Taatjes, D.J. Transcription Factor-Mediator Interfaces: Multiple and Multi-Valent. Journal of molecular biology. 2017, 429, 2996–2998. [Google Scholar] [CrossRef] [PubMed]
- Vojnic, E.; Mourao, A.; Seizl, M.; Simon, B.; Wenzeck, L.; Lariviere, L. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 2011, 18, 404–U29. [Google Scholar] [CrossRef] [PubMed]
- Milbradt, A.G.; Kulkarni, M.; Yi, T.; Takeuchi, K.; Sun, Z.Y.; Luna, R.E. Structure of the VP16 transactivator target in the Mediator. Nat. Struct. Mol. Biol. 2011, 18, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Lim, K.; Lee, M.K.; Chi, S.W. Structural Basis for the Interaction between p53 Transactivation Domain and the Mediator Subunit MED25. Molecules 2018, 23, 2726. [Google Scholar] [CrossRef]
- Kazan, K. The Multitalented MEDIATOR25. Front. Plant Sci. 2017, 8, 999. [Google Scholar] [CrossRef]
- Landrieu, I.; Verger, A.; Baert, J.L.; Rucktooa, P.; Cantrelle, F.X.; Dewitte, F. Characterization of ERM transactivation domain binding to the ACID/PTOV domain of the Mediator subunit MED25. Nucleic Acids Res. 2015, 43, 7110–7121. [Google Scholar] [CrossRef]
- Bontems, F.; Verger, A.; Dewitte, F.; Lens, Z.; Baert, J.L.; Ferreira, E. NMR structure of the human Mediator MED25 ACID domain. J. Struct. Biol. 2011, 174, 245–251. [Google Scholar] [CrossRef]
- Nicholas, T.R.; Strittmatter, B.G.; Hollenhorst, P.C. Oncogenic ETS Factors in Prostate Cancer. Adv. Exp. Med. Biol. 2019, 1210, 409–436. [Google Scholar]
- Defossez, P.A.; Baert, J.L.; Monnot, M.; de Launoit, Y. The ETS family member ERM contains an alpha-helical acidic activation domain that contacts TAFII60. Nucleic Acids Res. 1997, 25, 4455–4463. [Google Scholar] [CrossRef]
- Verger, A.; Baert, J.L.; Verreman, K.; Dewitte, F.; Ferreira, E.; Lens, Z. The Mediator complex subunit MED25 is targeted by the N-terminal transactivation domain of the PEA3 group members. Nucleic Acids Res. 2013, 41, 4847–4859. [Google Scholar] [CrossRef]
- Regier, J.L.; Shen, F.; Triezenberg, S.J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 1993, 90, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Shin, S.; Janknecht, R. ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim. Et Biophys. Acta. 2012, 1826, 1–12. [Google Scholar] [CrossRef]
- Oh, S.; Shin, S.; Song, H.; Grande, J.P.; Janknecht, R. Relationship between ETS Transcription Factor ETV1 and TGF-beta-regulated SMAD Proteins in Prostate Cancer. Sci. Rep. 2019, 9, 8186. [Google Scholar] [CrossRef]
- Roizman, B.; Zhou, G. The 3 facets of regulation of herpes simplex virus gene expression: A critical inquiry. Virology 2015, 479, 562–567. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.A.; Case, K.B.; Ben-Shalom, S.R.; Brozell, D.S.; Cerutti, T.E.; Cheatham, V.W.D. Amber 2020; University of California: San Francisco, CA, USA, 2020; Available online: https://ambermd.org/ (accessed on 17 August 2020).
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wires Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Pierce, L.C.; Salomon-Ferrer, R.; Augusto, F.O.C.; McCammon, J.A.; Walker, R.C. Routine Access to Millisecond Time Scale Events with Accelerated Molecular Dynamics. J. Chem. Theory Comput. 2012, 8, 2997–3002. [Google Scholar] [CrossRef]
- Boomsma, W.; Frellsen, J.; Harder, T.; Bottaro, S.; Johansson, K.E.; Tian, P.F. PHAISTOS: A framework for Markov chain Monte Carlo simulation and inference of protein structure. J. Comput. Chem. 2013, 34, 1697–1705. [Google Scholar] [CrossRef]
- Sullivan, S.S.; Weinzierl, R.O.J. Optimization of Molecular Dynamics Simulations of c-MYC (1-88)-An Intrinsically Disordered System. Life 2020, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Dahl, A.C.; Chavent, M.; Sansom, M.S. Bendix: Intuitive helix geometry analysis and abstraction. Bioinformatics 2012, 28, 2193–2194. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Kim, D.H.; Han, K.H. Transient Secondary Structures as General Target-Binding Motifs in Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2018, 19, 3614. [Google Scholar] [CrossRef]
- Piskacek, S.; Gregor, M.; Nemethova, M.; Grabner, M.; Kovarik, P.; Piskacek, M. Nine-amino-acid transactivation domain: Establishment and prediction utilities. Genom. 2007, 89, 756–768. [Google Scholar] [CrossRef]
- Erijman, A.; Kozlowski, L.; Sohrabi-Jahromi, S.; Fishburn, J.; Warfield, L.; Schreiber, J. A High-Throughput Screen for Transcription Activation Domains Reveals Their Sequence Features and Permits Prediction by Deep Learning. Mol. Cell. 2020, 78, 890–902. [Google Scholar] [CrossRef]
- Cress, W.D.; Triezenberg, S.J. Critical structural elements of the VP16 transcriptional activation domain. Science 1991, 251, 87–90. [Google Scholar] [CrossRef]
- Warfield, L.; Tuttle, L.M.; Pacheco, D.; Klevit, R.E.; Hahn, S. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc. Natl. Acad. Sci. USA 2014, 111, E3506–E3513. [Google Scholar] [CrossRef]
- Meyer, K.D.; Lin, S.C.; Bernecky, C.; Gao, Y.; Taatjes, D.J. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 2010, 17, 753–760. [Google Scholar] [CrossRef]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Pan, S.J.; Fan, X.; Breger, J. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.L.; Boeszoermenyi, A.; Vale-Silva, L.A.; Torelli, R.; Posteraro, B.; Sohn, Y.J. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 2016, 530, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Eletsky, A.; Szyperski, T.; Hay, J.; Ruyechan, W.T. Analysis of the varicella-zoster virus IE62 N-terminal acidic transactivating domain and its interaction with the human mediator complex. J. Virol. 2009, 83, 6300–6305. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Park, U.H.; Kim, E.J.; Um, S.J. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007, 26, 3545–3557. [Google Scholar] [CrossRef] [PubMed]
- Mittler, G.; Stuhler, T.; Santolin, L.; Uhlmann, T.; Kremmer, E.; Lottspeich, F. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003, 22, 6494–6504. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; de Beaumont, R.; Zhou, S.; Naar, A.M. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 2004, 101, 2339–2344. [Google Scholar] [CrossRef]
- Roupelieva, M.; Griffiths, S.J.; Kremmer, E.; Meisterernst, M.; Viejo-Borbolla, A.; Schulz, T. Kaposi’s sarcoma-associated herpesvirus Lana-1 is a major activator of the serum response element and mitogen-activated protein kinase pathways via interactions with the Mediator complex. J. Gen. Virol. 2010, 91, 1138–1149. [Google Scholar] [CrossRef]
- Cumbo, F.; Vergni, D.; Santoni, D. Investigating transcription factor synergism in humans. DNA Res. 2018, 25, 103–112. [Google Scholar] [CrossRef]
- Lens, Z.; Cantrelle, F.X.; Peruzzini, R.; Hanoulle, X.; Dewitte, F.; Ferreira, E. Solution Structure of the N-Terminal Domain of Mediator Subunit MED26 and Molecular Characterization of Its Interaction with EAF1 and TAF7. J. Mol. Biol. 2017, 429, 3043–3055. [Google Scholar] [CrossRef]
- Radhakrishnan, I.; PerezAlvarado, G.C.; Parker, D.; Dyson, H.J.; Montminy, M.R.; Wright, P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator: Coactivator interactions. Cell 1997, 91, 741–752. [Google Scholar] [CrossRef]
- Hua, Q.X.; Jia, W.H.; Bullock, B.P.; Habener, J.F.; Weiss, M.A. Transcriptional activator-coactivator recognition: Nascent folding of a kinase-inducible transactivation domain predicts its structure on coactivator binding. Biochemistry 1998, 37, 5858–5866. [Google Scholar] [CrossRef] [PubMed]
- Triezenberg, S.J. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995, 5, 190–196. [Google Scholar] [CrossRef]
- Odoux, A.; Jindal, D.; Tamas, T.C.; Lim, B.W.; Pollard, D.; Xu, W. Experimental and molecular dynamics studies showed that CBP KIX mutation affects the stability of CBP:C-Myb complex. Comput. Biol. Chem. 2016, 62, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Yazar, M.; Ozbek, P. Revisiting allostery in CREB-binding protein (CBP) using residue-based interaction energy. J. Comput. Aided Mol. Des. 2020, 34, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhou, W.; Guo, Y.; Wang, J.; Fu, W.; Sun, H. Molecular Dynamics Simulations Revealed the Regulation of Ligands to the Interactions between Androgen Receptor and Its Coactivator. J. Chem. Inf. Modeling 2018, 58, 1652–1661. [Google Scholar] [CrossRef] [PubMed]
- Piskacek, M.; Havelka, M.; Rezacova, M.; Knight, A. The 9aaTAD Is Exclusive Activation Domain in Gal4. PLoS ONE 2017, 12, e0169261. [Google Scholar] [CrossRef]
- Arnold, C.D.; Nemcko, F.; Woodfin, A.R.; Wienerroither, S.; Vlasova, A.; Schleiffer, A. A high-throughput method to identify trans-activation domains within transcription factor sequences. EMBO J. 2018, 37, e98896. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. The case for intrinsically disordered proteins playing contributory roles in molecular recognition without a stable 3D structure. F1000 Biol. Rep. 2013, 5, 1. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeffery, H.M.; Weinzierl, R.O.J. Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator. Biomolecules 2020, 10, 1205. https://doi.org/10.3390/biom10091205
Jeffery HM, Weinzierl ROJ. Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator. Biomolecules. 2020; 10(9):1205. https://doi.org/10.3390/biom10091205
Chicago/Turabian StyleJeffery, Heather M., and Robert O. J. Weinzierl. 2020. "Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator" Biomolecules 10, no. 9: 1205. https://doi.org/10.3390/biom10091205
APA StyleJeffery, H. M., & Weinzierl, R. O. J. (2020). Multivalent and Bidirectional Binding of Transcriptional Transactivation Domains to the MED25 Coactivator. Biomolecules, 10(9), 1205. https://doi.org/10.3390/biom10091205





